Abstract
Understanding the genetic and molecular bases of the ability to distinguish self from nonself (allorecognition) and mechanisms underlying evolution of allorecognition systems is an important endeavor for understanding cases where it becomes dysfunctional, such as in autoimmune disorders. In filamentous fungi, allorecognition can result in vegetative or heterokaryon incompatibility, which is a type of programmed cell death that occurs following fusion of genetically different cells. Allorecognition is genetically controlled by het loci, with coexpression of any combination of incompatible alleles triggering vegetative incompatibility. Herein, we identified, characterized, and inferred the evolutionary history of candidate het loci in the filamentous fungus Neurospora crassa. As characterized het loci encode proteins carrying an HET domain, we annotated HET domain genes in 25 isolates from a natural population along with the N. crassa reference genome using resequencing data. Because allorecognition systems can be affected by frequency-dependent selection favoring rare alleles (i.e., balancing selection), we mined resequencing data for HET domain loci whose alleles displayed elevated levels of variability, excess of intermediate frequency alleles, and deep gene genealogies. From these analyses, 34 HET domain loci were identified as likely to be under balancing selection. Using transformation, incompatibility assays and genetic analyses, we determined that one of these candidates functioned as a het locus (het-e). The het-e locus has three divergent allelic groups that showed signatures of positive selection, intra- and intergroup recombination, and trans-species polymorphism. Our findings represent a compelling case of balancing selection functioning on multiple alleles across multiple loci potentially involved in allorecognition.
Keywords: Neurospora crassa, heterokaryon incompatibility, balancing selection, self/nonself recognition, programmed cell death, vegetative incompatibility, allorecognition, trans-species polymorphism
Introduction
Allorecognition is the ability to distinguish self from nonself between cells and tissues of members of the same species and has been documented across the tree of life. Allorecognition occurs in a variety of biological processes, ranging from tissue-transplant fusion to immune defense (Boehm 2006). In jawed vertebrates, cell surface molecules encoded by the major histocompatibility complex (MHC) determine compatibility of donors for organ transplants (Benacerraf and Germain 1978). Allorecognition systems are also present in early branching metazoans, including the cnidarian Hydractinia symbiolongicarpus (Cadavid et al. 2004) and the tunicate Botryllus schlosseri (Voskoboynik et al. 2013). In these two species, asexually expanding colonies can engage in a natural transplantation reaction, whereby self-recognition leads to fusion of colonies (i.e., formation of parabionts sharing vascular systems), whereas interactions between genetically incompatible colonies result in programmed cell death and rejection. An analogous allorecognition system is present in the other main group of opisthokonts, the Fungi. Recent advances in the genetic dissection of fungal allorecognition illustrate that insights from fungal models may illuminate mechanisms and evolutionary events underlying allorecognition systems and more generally, immunity-related recognition mechanisms and signaling pathways (Glass and Dementhon 2006; Paoletti and Saupe 2009; Daskalov et al. 2015). The dissection of the genetic and molecular basis as well as the evolutionary history of allorecognition systems may lead to a better understanding of dysfunctional allorecognition phenomena, including autoimmune disorders.
In many species of filamentous fungi, the somatic body is composed of an interconnected, multinucleate syncytial structure. Filaments (hyphae) within growing colonies are capable of fusing, which creates an interconnected hyphal network that makes up the fungal individual. Hyphal fusion is not restricted to an individual colony but can also occur between colonies of the same species, irrespective of their genetic backgrounds, leading to the presence of genetically different nuclei in a common cytoplasm (heterokaryon) (Read et al. 2010). Within a heterokaryon, allorecognition processes determine the fate of the fused cell: Interactions between compatible genotypes lead to a heterokaryon indistinguishable from a wild type (WT) colony, whereas heterokaryotic cells resulting from the fusion of incompatible genotypes are rapidly compartmentalized and undergo a type of programmed cell death, termed vegetative (or heterokaryon) incompatibility (Glass and Dementhon 2006; Paoletti and Saupe 2009). The induction of vegetative incompatibility in filamentous fungi is extremely rapid; incompatible heterokaryotic cells are compartmentalized and die approximately 30 min postfusion (Biella et al. 2002; Glass and Kaneko 2003).
Genetic analyses of vegetative incompatibility in filamentous fungi, such as in Neurospora crassa (Glass and Dementhon 2006), Podospora anserina (Saupe 2000), Aspergillus nidulans (Pal et al. 2007), and the plant pathogen Cryphonectria parasitica (Cortesi and Milgroom 1998) showed that incompatibility is genetically controlled by multiple, unlinked het (for heterokaryon) or vcg (for vegetative compatibility group) loci. Vegetative incompatibility is suppressed during the sexual cycle allowing for production of viable progeny irrespective of the somatic compatibility of the parental strains. Molecular characterization of het loci in N. crassa, P. anserina, and C. parasitica showed that these loci often encode proteins containing a domain of unknown biochemical function, labeled the HET domain (Pfam06985) (Glass and Dementhon 2006; Zhang et al. 2014). The HET domain contains approximately 150 amino acids (Smith et al. 2000; Espagne et al. 2002) and has no identified cellular or biochemical function other than its association with vegetative incompatibility. In N. crassa, at least 11 loci are functional in allorecognition (Perkins 1988), with two to three allelic specificities occurring at each locus. Thus, in natural recombining populations of N. crassa, at least 211 different het genotypes are potentially possible. It is therefore believed that viable somatic fusion between genetically different colonies is virtually excluded in nature (Muirhead et al. 2002). Three het loci have been molecularly characterized in N. crassa (mat/tol, un-24/het-6 and het-c/pin-c). The tol, het-6, and pin-c loci all encode HET domain proteins (Glass and Dementhon 2006).
Genes involved in allorecognition often display elevated allelic diversity, along with trans-species polymorphism, supporting the idea that these loci are under balancing selection (Charlesworth 2006). In fungi, the role of balancing selection in shaping allorecognition systems is supported by the finding that phenotypic classes with alternative het specificities are equally frequent in Neurospora spp. and P. anserina populations, and by the fact that some loci exhibit trans-species polymorphism (Bastiaans et al. 2014; Wu et al. 1998; Powell et al. 2001; Powell et al. 2007; Hall et al. 2010). Vegetative incompatibility in filamentous fungi has been shown to prevent various forms of somatic parasitism (e.g., cases in which one genotype drains reproductive resources from the other), and to reduce the risk of transmission of infectious cytoplasmic elements and mycoviruses (Debets et al. 1994; Biella et al. 2002; Zhang et al. 2014). Under this scenario, balancing selection acts through frequency-dependent selection favoring rare alleles, as individuals carrying rare alleles are incompatible with most of the population, and are thus more efficiently protected against infectious cytoplasmic elements or exploitation by aggressive genotypes (Muirhead et al. 2002). However, it is also hypothesized that fungal allorecognition systems represent cases of exaptation (Gould and Erba 1982), whereby components of a yet-unidentified system of defense against pathogens are being reused by natural selection for the recognition of conspecifics (Paoletti and Saupe 2009). Under this hypothesis, the diversification of het loci predates their function in allorecognition, and results from the selection of new allelic variants insensitive to pathogen effector proteins. Consistent with both models, signatures of diversifying selection have been found in a number of het genes in Neurospora spp. and P. anserina (Wu et al. 1998; Powell et al. 2001; Paoletti et al. 2007; Powell et al. 2007; Chevanne et al. 2010; Hall et al. 2010; Bastiaans et al. 2014). What remains unclear is the nature of the evolutionary forces underlying the appearance and maintenance of het alleles in fungi, the reconciliation of signatures of diversifying selection with the observation of a limited number of incompatibility alleles (typically two or three), and the persistence of incompatibility alleles through speciation events.
Identification and characterization of additional fungal het loci and analyses of evolutionary mechanisms underlying their diversification will enable an understanding of the origin and evolution of allorecognition systems in eukaryotic species, as well as insights into the molecular mechanisms underlying recognition. However, cloning het loci by conventional genetic methods is laborious due to the high degree of allelic variability at het loci among different strains in populations. Herein, we used a combination of bioinformatics, population genomics, and functional analyses to identify, characterize, and infer the evolutionary history of additional candidate het loci in N. crassa. As all het loci in N. crassa encode proteins containing an HET domain, we analyzed genetic variation at these loci based on genomic data from a wild population. From these analyses, we identified 34 HET domain loci that displayed elevated levels of allelic variability, an excess of intermediate frequency alleles, and deep gene genealogies consistent with a history of balancing selection, in sharp contrast to the vast majority of genes encoded in the N. crassa genome. To gain insight into the diversification of HET domain loci over evolutionary time, we compared the organization and content of HET domain genes among species related at different evolutionary distances. Further, using transformation, compatibility assays, and genetic analyses, we showed that one of these candidate HET domain loci, NCU09037, was functional as a het locus. Alleles at NCU09037 (het-e) fell into three very divergent groups (termed haplogroups) that showed signatures of positive selection, intra- and interhaplogroup recombination, and evidence of trans-species polymorphism. Our work illustrates how bioinformatics and evolutionary approaches to identify candidate allorecognition genes using a species with population genomic resources can lead to hypotheses regarding function that can be subsequently tested by molecular and genetic methods. Our analyses provide insights into the role of natural selection and recombination in the diversification of allorecognition systems and represent a compelling case of balancing selection functioning on multiple alleles across multiple loci that are potentially involved in self/nonself recognition.
Results
Identification of HET Domain Loci in N. crassa Isolates
The majority of het genes previously characterized in N. crassa and other Sordariomycetes encode proteins containing an HET domain (Glass and Dementhon 2006; Zhang et al. 2014). We therefore mined the N. crassa FGSC 2489 reference genome (Galagan et al. 2003) for this specific domain to identify all candidate het loci. BLAST searches using a consensus HET domain protein sequence identified 69 HET domain-encoding loci (fig. 1), two of which were not identified in the latest annotated version of the reference genome. One of these two HET domain genes produces an antisense transcript to NCU10839 (Xue et al. 2014), and the second locus was named HET069.
Fig. 1.
HET domain genes in Neurospora crassa. The N. crassa sequenced strain (FGSC 2489; Galagan et al. 2003) has 69 HET domain loci. Within the N. crassa Louisiana population, 73 HET domain loci were mapped to the N. crassa FGSC 2489 genome. The numbers at the top and bottom of each LG indicate the position (in bp). The newly identified HET domain loci in the Louisiana strains (HET070, HET071, HET072, and HET073) identified from de novo assemblies were mapped based on their conserved neighbor sequences. The 34 loci with the most variable alleles among the Louisiana population determined by PCA are highlighted in red and blue colors (fig. 2). Black color indicates the 33 HET domain loci present, but not variable, in all strains. Green color indicates HET domain loci only present in some strains and that were not polymorphic. HET073 (blue color) was not present in the genome of FGSC 2489, but was polymorphic in the Louisiana strains that contained HET073.
To determine whether the distribution and number of HET domain loci were conserved among wild isolates of N. crassa, we generated whole-genome sequence from a subset of 25 strains from a collection of 112 strains (supplementary table S1, Supplementary Material online; Sequence read archive study accession number: SRP052921) from a single Louisiana population (Palma-Guerrero et al. 2013). The reference genome strain, FGSC 2489, is a member of this population. The predicted genome size of the 25 isolates was approximately the same as for the reference strain (∼42 Mb), with the exception of one strain, P4476, whose genome was predicted to be only approximately 36 Mb. The number of genes predicted in the genome of the 25 isolates ranged from 9,615 to 9,658 (supplementary table S1, Supplementary Material online); the predicted number of genes in P4476 was only marginally lower (9,433 genes).
In order to ascertain orthologous relationships between the sets of genes identified in the reference genome sequence and in de novo assemblies of resequenced genomes, we performed synteny analysis based on whole-genome alignments combined with comparisons of gene content using reciprocal tBLASTn and BLASTp searches. We used the PROMER program in the MUMMER3 package (Kurtz et al. 2004), which performs alignment on the six-frame amino acid translation of the DNA input sequence. The tBLASTn and BLASTp searches were used to identify homologous sequences between de novo assemblies and genes identified in the FGSC 2489 reference genome. Genes in syntenic regions and for which identity between query and subject sequences was maximal and above 70% amino acid identity were considered as orthologous. Using this approach, orthologs of 95–97% of the 9,730 genes predicted in FGSC 2489 were identified in the genomes of the wild isolates.
The repertoire and location of HET domain loci were almost identical between the 25 de novo genome assemblies and the reference FGSC 2489 genome. Only three HET domain loci identified in FGSC 2489 (NCU09954, NCU17026, and NCU17077) were not present in the genomes of all resequenced Louisiana isolates (NCU09954, NCU17026, and NCU17077 were detected in only 17, 23, and 14 strains, respectively). Four HET domain loci, termed HET070, HET071, HET072, and HET073 (supplementary fig. S1, Supplementary Material online) were absent in the FGSC 2489 genome, but were present in the re-sequenced genomes, with frequencies 2/25, 14/25, 3/25, and 25/25, respectively. The presence of these HET domain loci in the wild isolates as compared with the reference genome was associated with rearrangements/deletions/insertions, although the surrounding gene order was highly conserved (supplementary fig. S1, Supplementary Material online). The remaining HET domain loci were conserved and syntenic in the genomes of all 25 wild isolates and the genome of FGSC 2489.
Genomic Organization of HET Domain Genes in Neurospora and Related Ascomycete Species
To assess whether loci encoding HET domain proteins were clustered or evenly distributed across all seven chromosomes, we determined the genomic coordinates of the 73 identified HET domain loci on the linkage groups (LG) of reference strain FGSC 2489. Visual inspection of the genomic distribution of HET domain loci suggested nonrandom distribution and clustering of these genes at the ends of LG, although deviations from the null hypothesis of uniform distribution were only significant for LG 1, 6, and 7 (fig. 1; supplementary table S2, Supplementary Material online).
To gain insight into the diversification of HET domain genes over evolutionary time, we determined the organization of HET domain loci in other species and compared gene order and content between species. We first mapped HET domain loci in the genome of the related species N. tetrasperma (FGSC 2508 mat A). Of the 73 HET domain loci identified in N. tetrasperma, 69 were present and syntenic between N. tetrasperma and N. crassa and tended to be clustered toward the ends of LGs (supplementary fig. S2A, Supplementary Material online). Four HET domain loci were specific to N. tetrasperma, whereas four of the 73 N. crassa HET domain loci were not identified in the N. tetrasperma reference genome (supplementary table S3, Supplementary Material online). The null hypothesis of random uniform distribution could be rejected in most cases with tests conducted at the scale of chromosome arms (supplementary table S2, Supplementary Material online).
We also evaluated the organization, order, and content of HET domain loci in the more distantly related fungi P. anserina (Sordariomycete) and A. oryzae (Eurotiomycete). In P. anserina, 125 HET domain loci were identified, of which 38 were present on chromosome 5 (supplementary fig. S2B, Supplementary Material online). Deviations from the null hypothesis of random uniform distribution for this species were significant for most chromosomes, including chromosome 5 (supplementary table S2, Supplementary Material online). In A. oryzae, 43 HET domain loci were identified and some displayed a trend toward clustering at the ends of chromosomes, with deviations from the null hypothesis of random uniform distribution being significant for chromosomes 2, 3, 5, and 7 (supplementary fig. S2C and table S2, Supplementary Material online). Despite the general trend toward a nonrandom distribution of HET domain loci in the Neurospora spp, P. anserina, and A. oryzae genomes, we did not observe synteny among predicted HET domain loci between the four species, except for one HET domain locus, NCU09629, which was conserved and syntenic between Neurospora species and P. anserina.
Genetic Variability of HET Domain Genes
In many species, allorecognition genes are under balancing selection, which maintains different allelic lines at selected loci (Charlesworth 2006). Previously characterized fungal vegetative incompatibility genes, such as the HET domain loci pin-c and het-6 in N. crassa (Smith et al. 2000; Kaneko et al. 2006), het-D and het-E in P. anserina (Espagne et al. 2002), and vic6 and vic7 in C. parasitica (Choi et al. 2012), display high levels of allelic variability. To determine whether any of the 73 HET domain loci in N. crassa were plausible allorecognition genes, we estimated sequence variability at these loci among Louisiana isolates, and assessed signatures of balancing selection by comparing several summaries of variability at HET domain loci versus a set of genes representing the genomic background. Five summary statistics were computed for each of the 73 HET domain loci and for the full reference gene set in the 25 Louisiana isolates (8,621 genes containing at least one SNP, and with sample size >6) (supplementary table S4, Supplementary Material online). Results were visualized by PCA, where the first principal component (PC1) mostly represented variation in terms of S (segregating sites standardized by sequence length), π (the average number of nucleotide differences between sequence pairs), and KSmax (the maximum number of synonymous substitutions between sequences pairs). Loci with high PC1 values displayed high levels of sequence diversity. The second principal component (PC2) corresponded to variation in Tajima’s D (a measure of skewness of the allele frequency spectrum) and the number of amino acid sequences based on nonsynonymous substitutions. A high positive Tajima’s D value at a given locus indicates the presence of alleles at an intermediate frequency in a population, which is consistent with balancing selection and limited recombination among alleles causing the evolution of highly divergent haplogroups. Visual inspection of PCA results revealed 34 of the 73 HET loci falling outside the main cloud representing 8,621 reference genes (fig. 2; genes with PC1 < −8.0 or PC2 > 2.5); all these genes were also in the top 5% for at least three of the five summary statistics (supplementary table S5, Supplementary Material online). Two HET domain loci with high levels of variability (NCU03494, pin-c and NCU03533, het-6) out of the 34 identified confer vegetative incompatibility in N. crassa and show evidence of balancing selection (Powell et al. 2007; Hall et al. 2010). Although a few highly variable genes in the N. crassa genome that are not HET domain loci may have been missed by our approach based on short-read sequencing, these results nonetheless show that over half of the HET domain loci in N. crassa displayed high allelic diversity and/or divergent alleles, unlike the vast majority of the remaining genes in the genome, which is consistent with balancing selection acting at these loci. The remaining 32 HET domain loci are considered to be the best candidates for functioning as allorecognition loci in N. crassa. Visual inspection of neighbor-joining gene genealogies built for these 32 HET domain loci revealed that 15 of them displayed topologies showing at least two long-diverged haplogroups (supplementary fig. S3, Supplementary Material online).
Fig. 2.
PCA of variability at HET domain loci relative to a set of reference genes. PCA was used to visualize the variability at HET domain loci (black dots) relative to a set of 8,621 reference genes representing the background level of genomic diversity (blue dots; reference loci with at least one polymorphism and a sample size >6 were included in the analysis). The first two principal components (PC1 and PC2) summarize variance at five summary statistics of genetic variability (supplementary tables S4 and S5, Supplementary Material online). PC1 is impacted by variation at S, π, and KSmax. PC2 is impacted by variation in the number of protein variants and in Tajima’s D values. Alleles at loci in the top left corner of the plot exhibited high levels of sequence variability and allelic variants tended to cluster within a few haplogroups of intermediate frequency. Thirty-four HET domain loci with PC1 < −8.0 or PC2 > 2.5 are considered the best candidates for being under long-term balancing selection.
Presence and Similarity of HET Domain Genes in Fungi, Plants, and Protists
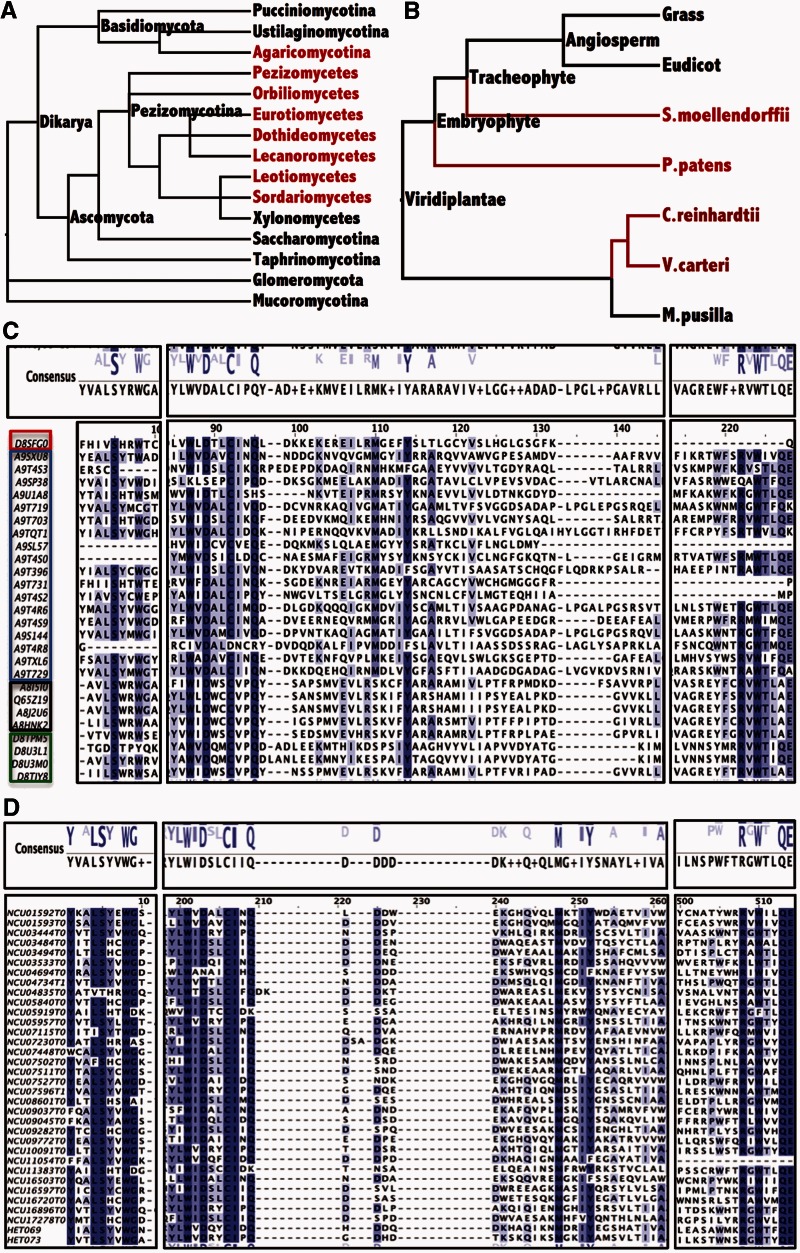
HET domains genes are present (but variable in number) in a large number of filamentous ascomycete species (Fedorova et al. 2005), but are notably absent from the Saccharomycotina (Saccharomyces yeasts and relatives), the Taphrinomycotina (Schizosaccharomyces yeasts and its relatives), and the Xylonomycetes, a recently discovered class of ascomycetes that are associated as endophytes in rubber trees (Gazis et al. 2012) (fig. 3A). However, HET domain genes are present in the genomes of the Basidiomycete group Agaricomycotina, which includes most of the mushroom-forming and ectomycorrizal species, although conservation of HET domain genes among members of this group is also variable (Van der Nest et al. 2014). Although absent from the genomes of most plant species, HET domain genes are present in the green algae Chlamydomonas reinhardtii (4 HET domain loci) and Volvox carteri (4 HET domain loci), the basal vascular plant species, Selaginella moellendorffii (1 HET domain locus) and the bryophyte (moss) species (Physcomitrella patens) (18 HET domain loci) (fig. 3B). The approximately 150-amino acid HET domain consensus sequence in nonfungal species was very similar to that the consensus generated by analysis of HET domain genes from N. crassa (fig. 3C and D) and included conservation of key amino acid residues that define this domain.
Fig. 3.
HET domain presence and similarity in fungal, protist, and plant species. (A) HET domain genes can be identified in most members of the Pezizomycotina (red), with the exception of the Xylonomycetes. HET domain genes were not identified in the Saccharomycotina or the Taphrinomycotina (black). HET domain genes were identified in members of the Agaricomycotina (Basidiomycota) but were not identified in other members of Basidiomycota or in basal fungal groups, Glomeromycota and Mucoromycotina (black). Tree based on modifications of MycoCosm (Grigoriev et al. 2014). (B) HET domain genes were identified in some photosynthetic protists (in red), such as green algae Chlamydomonas reinhardtii and Volvox carteri. HET domain genes were also identified in the genome of the lycophyte Selaginella moellendorffii and the moss Physcomitrella patens. The tree in (B) is based on Phytozome v10 (Goodstein et al. 2012). (C) HET domain amino acid sequence alignments in plant and algal species. The species from top to bottom are Selaginella moellendorffii (red box), Physcomitrella patens subsp. patens (blue box), Chlamydomonas smithii (black box), and Volvox carteri (green box). Boxes on top show consensus sequence for the three conserved blocks in the HET domain. The blue letters above the line show conserved amino acids, with larger letters indicating more conservation, whereas black letters below the lines summarize amino acids based on alignment. Conserved residues in the amino acid alignment are colored blue (darker blue, more conserved, lighter blue, less conserved). (D) Amino acid alignment of the 34 polymorphic HET domain loci in Neurospora crassa. Box on top shows the consensus sequence for the three conserved blocks in HET domain. Blue letters above the lines show conserved amino acids, with larger letters indicating more conservation, whereas the letters below the lines summarize amino acids based on alignment. The boxes at the bottom show the alignment with more highly conserved residues colored dark blue, whereas less conserved residues are colored lighter blue.
Candidate HET Domain Gene NCU09037 Corresponds to a Previously Mapped het Locus, het-e, with Three Haplogroups
One of the identified HET domain genes, NCU09037, showed substantial polymorphism and evidence for long-term balancing selection (S = 0.43/bp, π = 0.19/bp, D = 2.8, KSmax = 0.88/bp, 20 protein variants; in top 0.1% for all statistics but KSmax) and mapped close to a previously genetically characterized het locus named het-e (Wilson and Garnjobst 1966). The protein sequence alignment of NCU09037 from 26 Louisiana isolates showed that NCU09037 has several indels, including a large indel of the HET domain in some strains (fig. 4A and supplementary fig. S4A, Supplementary Material online). To assess whether NCU09037 was het-e, we designed allele-specific primers and amplified the NCU09037 gene in het-e tester strains (FGSC 2489 and FGSC 2658) and het-E tester strains (FGSC 2603 and FGSC 1424) (Wilson and Garnjobst 1966; Xiang and Glass 2004). The het-e tester strains contained the longer allele of NCU09037 (het-e1; see below), whereas the het-E tester strains contained the shorter NCU09037 allele (het-e2; see below) (supplementary fig. S4B, Supplementary Material online). Thus, based on map position, PCR results, and functional analyses (see below), we refer to NCU09037 as het-e.
Fig. 4.
Protein alignment and phylogenetic tree for the three haplogroups of het-e in populations of Neurospora crassa and Neurospora discreta. (A) Alignment of protein sequences for three het-e (NCU09037) haplogroups in N. crassa and two het-e haplogroups in N. discreta. The HET domain region is highlighted by black boxes. The red highlighted regions show het-e1 haplogroup-specific regions, the black highlighted regions show het-e2 haplogroup-specific regions, and the blue-highlighted regions show het-e3 haplogroup-specific regions. The black colored amino acid residues are conserved among alleles in all three haplogroups. The purple highlighted residues indicate sites under positive selection in the het-e2 alleles. The scale on top of the alignment shows amino acid position, whereas the numbers left and right to the alignment show the absolute position of amino acids in the corresponding HET-E sequence. In N. crassa, the HET-E1 sequence shown is from strain FGSC 2489, HET-E2 is from strain P4468, and HET-E3 is from strain JW75. In N. discreta, the HET-E1 sequence shown is from CAWE978 and HET-E2 is from CAMH1105. (B) Bayesian tree of het-e nucleotide sequences built by MrBayes 3.2 (Ronquist et al. 2012). CDSs of all het-e haplogroups (red for het-e1, black for het-e2, and blue for het-e3) from 26 N. crassa Louisiana strains (supplementary table S1, Supplementary Material online), and from 11 strains from a N. discreta population (in green boxes) were used to build the phylogenetic tree. Black bar indicates substitution rate. The HET domain gene with the highest E-value homologous sequence (Pa_2_9350; evalue = 1.85e-056; amino acid identity 46%) to HET-E1 in Podospora anserina was used as outgroup. (C) Cartoon of the structure of the het-e locus in N. crassa (left) versus that of N. discreta (right). In N. discreta, the het-e2 CDS is followed by part of het-e1 sequence, which occurs in all strains of N. discreta included in this study. The red boxes are for het-e1, the black for het-e2, and the blue for het-e3. (D) Venn diagram showing the distribution of different het-e haplogroups in the N. crassa Louisiana population (26 strains; left) and from the N. discreta CA/WA population (11 strains; right). Numbers of het-e alleles in the population sample are indicated below the allele name.
Based on indel composition and genealogical relationships among sequences, HET-E in different isolates of the Louisiana population clustered into three distinct groups (fig. 4A and supplementary fig. S4A, Supplementary Material online). The haplogroup showing highest identity to NCU09037 in the reference genome (FGSC 2489; more than 90% amino acid (aa) identity, 676 amino acids, and 2,031 bp CDS) is termed het-e1. A second het-e haplogroup, het-e2 (478 amino acids and 1,437 bp CDS) showed only 30% amino acid identity to HET-E1 and contained a large deletion within the HET domain region (fig. 4A). The third haplogroup, het-e3, showed only 26% amino acid identity to HET-E1 (432 amino acids and 1,269 bp CDS) and had an even larger deletion of the HET domain region (fig. 4A and supplementary fig. S4A, Supplementary Material online). HET-E2 and HET-E3 were more similar to each other (75% amino acid identity) than either was to HET-E1. Among the 26 Louisiana isolates with genome sequence, strains bearing het-e1 and het-e2 were equally frequent (n = 11 and 12 individuals, respectively), whereas the het-e3 haplogroup was less abundant (n = 3) (fig. 4D).
Recombination, Trans-species Polymorphism, and Positive Selection at het-e
A characteristic of loci under long-term balancing selection is the presence of trans-species polymorphism (Klein et al. 1993; Muirhead et al. 2002). Trans-specific polymorphism has been reported in het-c/pin-c (Kaneko et al. 2006; Hall et al. 2010) and het-6 in N. crassa (Mir-Rashed et al. 2000). We therefore evaluated whether het-e1, het-e2, and/or het-e3 or other new het-e haplogroups occurred in a population of the related species, N. discreta, which diverged 7–10 Ma (Corcoran et al. 2014). Sequences at the het-e locus were obtained for a population of N. discreta from California/Washington, which belongs to the N. discreta phylogenetic species 4 subgroup B (PS4B) (Gladieux P, Taylor JW, unpublished data) (Dettman et al. 2006). Only two het-e haplogroups were identified in N. discreta PS4B (het-e1 and het-e2) (fig. 4). The het-e2 allele CDS in the N. discreta strains was followed by a region highly similar to the 3′-end of het-e1.
Alignments and phylogenetic analyses of the full set of het-e sequences from the two species revealed trans-species polymorphism: The het-e1 sequences from N. discreta were more similar to het-e1 of N. crassa, than to het-e2 from other N. discreta isolates from the same population (fig. 4B). Average nucleotide divergence between homologous portions of heterospecific het-e1 and het-e2 alleles was 0.251/bp, which is almost three times greater than the genomic average for CDSs (0.086/bp; 30 randomly chosen orthologous nuclear genes), suggesting that het-e haplogroups have been carried as trans-specific polymorphisms for at least 21 My before present (estimated divergence between N. crassa and N. discreta, 7 My before present; Corcoran et al. 2014). Phylogenetic analysis also revealed population subdivision within each of the haplogroups, with sequences from either species clustering in different groups within the het-e haplogroups. Strong subdivision was also supported by the numbers of fixed differences: 105 and 164 variants were fixed between the two species for haplogroups het-e1 and het-e2. No shared polymorphism was found between species for each haplogroup, which is consistent with expectations under a model of strict isolation between species (Clark 1997) and rules out introgression as a cause for trans-specific polymorphism (expected numbers of shared polymorphisms between species based on numbers of segregating sites within species: 0.053 and 0.0 for het-e1 and het-e2, respectively).
The role of intra-allelic recombination in the evolution of het-e was also assessed by applying the four-gamete test (Hudson and Kaplan 1985) at the scale of haplogroups for each species. In the N. crassa het-e1 haplogroup, three pairs of sites (378-901, 434-901, and 479-901) showed all four gametic combinations, providing evidence that at least one recombination event between sites 479 and 901. The four-gamete test also revealed all four gametic combinations for 37 pairs of sites in N. crassa het-e2, implying at least three recombination events between sites 16-127, 190-426, and 426-831. No evidence of recombination was found in N. discreta haplogroups.
We tested for positive selection by comparing models assuming, or not, the presence of a proportion of sites evolving under positive selection along the branch carrying sequences from a given haplogroup (branch-site model of Test 2, in PAML; Yang 2007). Analyses suggested positive selection on the het-e2 haplogroup only, although the test was only marginally significant (2Δln L = 3.61, df = 2, P = 0.0573; supplementary table S6, Supplementary Material online). Within het-e2, 15 sites were identified as potentially being under positive selection, all with probabilities above 0.9 (supplementary fig. S4A, Supplementary Material online).
To better understand the evolutionary history of het-e, we searched for homologs of het-e in the genomes of other filamentous ascomycete species. Comparisons of gene order and content showed a clear shared synteny between genomic regions that included the het-e locus in N. crassa and other Sordariaceae fungi (N. tetrasperma, N. discreta and S. macrospora), but no shared synteny was observed at the scale of the order Sordariales (three Sordariaceae species and P. anserina) or at higher level taxonomic ranks. Reciprocal BLASTp and tBLASTn searches further confirmed that het-e orthologs were only present in members of the Sordariaceae (fig. 5). The protein sequences of N. discreta, S. macrospora, and N. tetrasperma showed at least 80% amino acid identity to the orthologous HET-E haplogroup in N. crassa.
Fig. 5.
Genomic analysis of synteny in the het-e region in filamentous ascomycete species. The phylogenetic tree (left) was derived from Hall et al. (2010). Syntenic genes are marked by colors, and boxes of the same color are orthologs and syntenic. het-e is indicated by a black box. White ovals mark genes predicted in genomes of species shown, but which are not within the genetic interval shown surrounding het-e. Boxes show gene organization, not the actual gene size. Three genes were conserved and syntenic in all species (NCU09035, alcohol dehydrogenase; NCU09034, mandelate racemase; NCU09033, C6 transcription factor). Checkered brown boxes show conservation of genes between species within the het-e genetic interval in species outside of the Sordariomycetes. Gene 1–11 stands for genes within the het-e syntenic region: NCU09043, NCU09042, NCU09041, NCU09040, NCU09039, NCU09038, NCU09036, NCU09035, NCU09034, NCU09033, and NCU09032, respectively. The Neurospora tetrasperma reference genome contains het-e2 allele at het-e, whereas N. discreta and Sordaria macrospora reference genomes have the het-e1 allele.
het-e Is Functional in Allorecognition and Vegetative Incompatibility
The signature of balancing selection suggested that NCU09037 (het-e) functions to confer vegetative incompatibility in N. crassa. To test this hypothesis, we used auxotrophic markers and a strain carrying a deletion of NCU09037 (constructed in FGSC 2489; Colot et al. 2006) to construct isogenic strains that differed only in allelic specificity at het-e. We first crossed the het-e deletion into a his-3; pan-2 or his-3; pyr-4 background. We then placed an epitope-tagged het-e1, het-e2 or het-e3 allele at the his-3 locus in the het-e deletion strain, resulting in strains carrying het-e1, het-e2 or het-e3 alleles with pan-2 or pyr-4 auxotrophic markers. Heterokaryons using auxotrophic markers that carry alleles of different HET specificity are aconidial and show greatly inhibited growth and cell death (Hutchison et al. 2009). Forced heterokaryons between the Δhet-e strain and strains carrying het-e1, het-e2 or het-e3 alleles exhibited normal WT morphology and growth rates (fig. 6A). However, forced heterokaryons between strains bearing alternate het-e alleles (het-e1 + het-e2; het-e1 + het-e3; het-e2 + het-e3) were aconidial and showed greatly inhibited growth (fig. 6A).
Fig. 6.
het-e functions as a het locus in Neurospora crassa, and which is dependent upon vib-1. (A) Strains with compatible and incompatible het-e alleles were used to forced heterokaryons using auxotrophic markers. The recipient strain for constructing strains carrying het-e2 or het-e3 alleles was a Δhet-e1 mutant derived from the sequenced strain FGSC 2489. (A1) A forced heterokaryon (his-3::Pccg1-het-e1-gfp; pan-2 + Δhet-e1; his-3; pyr-4) showed a compatible growth phenotype indistinguishable from a WT homokaryon. (A2) A forced heterokaryon carrying different het- e alleles (het-e1 and het-e3) (het-e1; his-3 + his3::Pccg1-het-e3-gfp; pan-2) showed a typical incompatible phenotype of inhibited growth. (A3) A forced heterokaryon (his3::Pccg1-het-e2-gfp; pan-2 + Δhet-e1; his-3; pyr-4) showed a compatible phenotype. (A4) A forced heterokaryon between strains carrying different het-e alleles (het-1 and het-e2) (het-e1; his-3 + his3::Pccg1-het-e2-gfp; pan-2) showed an incompatible phenotype. (A5) A forced heterokaryon (his3::Pccg1-het-e3-gfp; pan-2 + Δhet-e1; his-3; pyr-4) showed a compatible phenotype. (A6) A forced heterokaryon between strains carrying different het-e alleles (het-e2 and het-e3) (his-3::Pqa2-het-e2; pyr-4 + his3::Pccg1-het-e3-gfp; pan-2) showed an incompatible phenotype. (B) het-e mediated heterokaryon incompatibility is suppressed by deletions in vib-1. (B1) A forced heterokaryon with identical het-e alleles (his-3::Pccg1-het-e1-gfp; thr-2 + het-e1; pan-2) showed a compatible phenotype. (B2) A forced heterokaryon between strains that have different het-e alleles (het-e1 and het-e2) (het-e1; pan-2 + his3::Pccg1-het-e2-gfp; thr-2) showed a typical incompatible phenotype. (B3) A forced heterokaryon between strains with identical het-e (het-e1) alleles in a homozygous vib-1 deletion background (his-3::Pccg1-het-e1-gfp; Δvib-1; thr-2 + het-e1; Δvib-1; pan-2) showed a compatible phenotype. (B4) A forced heterokaryon between strains carrying different het-e alleles (het-e1 and het-e2) in vib-1 deletion background (his3::Pccg1-het-e2-gfp; Δvib-1; thr-2 + het-e1; Δvib-1; pan-2) showed a compatible phenotype. (C) RT-PCR showing het-e expression levels in a WT strain (FGSC 2489) versus a vib-1 deletion strain. The expression level of het-e in FGSC 2489 was set arbitrarily to one. Error bars indicate standard deviation.
The vib-1 locus encodes an NDT80-like transcription factor that is required for mediating incompatibility by genetic differences at het-c/pin-c, het-6/un-24, and mat-A/mat-a/tol (Xiang and Glass 2004; Dementhon et al. 2006; Hutchison and Glass 2010). We therefore asked whether deletion of vib-1 also suppressed het-e-mediated incompatibility by the introduction of the vib-1 deletion into our het-e1 and het-e2 strains. As shown in figure 6B, forced heterokaryons were indistinguishable between Δvib-1 strains of identical het-e specificity (het-e1 + het-e1) or of different het-e specificity (het-e1 + het-e2), indicating that deletions of vib-1 suppressed vegetative incompatibility in strains bearing allelic differences het-e. Previously, it was observed that strains bearing a deletion of vib-1 showed a reduction in expression of HET domain genes, such as pin-c and het-6 (Dementhon et al. 2006). To test whether a deletion of vib-1 also affected the expression of het-e, we performed quantitative RT-PCR experiments from Δvib-1 cultures grown for 16 h at 30 °C in minimal medium as compared with WT (FGSC 2489) cultures. As shown in figure 6C, expression of het-e showed a significant reduction in expression levels in the Δvib-1 mutant relative to the WT parental strain. These compatibility assays and genetic tests indicate that NCU09037 functions in allorecognition and vegetative incompatibility in N. crassa.
Determining the localization of HET proteins is important as it informs studies investigating the type of ligand or cellular processes responsible for triggering programmed cell death. HET-E has no other identifiable domains, other than the HET domain. To assess cellular localization of HET-E, we constructed Green Fluorescent Protein (GFP)- or mCherry-tagged het-e alleles under the regulation of the ccg-1 promoter. GFP or mCherry fluorescence was not sufficient to assess localization of HET-E in live cells, although HET-E1-, HET-E2- and HET-E3-GFP-tagged proteins could be detected through Western blots using anti-GFP antibodies (fig. 7A), and tagged proteins were functional in vegetative incompatibility tests (fig. 6). To assess subcellular localization of epitope-tagged HET-E, we performed sucrose gradient fractionation of disrupted cells. As controls, we used a strain bearing cytoplasmic GFP and anti-GFP antibodies for cytoplasmic fractions, antibodies to ERV25, an endoplasmic reticulum (ER)-localized protein (Palma-Guerrero et al. 2014), and antibodies to a vacuolar and plasma membrane localized protein, PMA-1 (Fajardo-Somera et al. 2013). Cytoplasmic GFP was detected in fractions 1–2 (cytoplasmic fraction), ERV25 was enriched in fractions 3–4 (endomembranes), and PMA-1 was enriched in fractions 7–10 (endomembranes and plasma membrane). In gfp-het-e1 strains, GFP-HET-E was enriched in fractions 2–6 (fig. 7B). These data showed that HET-E is associated with the endomembranes of the cell as well as in the cytoplasm, suggesting that signaling or disruption of endomembrane function in the cell may be associated with HET-E-mediated vegetative incompatibility.
Fig. 7.
HET-E is localized to cytoplasmic and membrane fractions. (A) Western blot showing GFP tagged het-e alleles are expressed and translated into protein and have different molecular weight relative to cytoplasmic GFP. Molecular weight markers are shown in the left lane. Lane 1 indicates cytoplasmic GFP. Lanes marked 2, 3, or 4 indicate HET-E1, HET-E2, and HET-E3 fused with GFP, respectively. (B) Subcellular fractionation of a Neurospora crassa vegetative culture grown for 16 h in VMM (Vogel 1956). T is for total protein used as a positive control for Western blot analyses (rightmost lane). PMA-1 is a plasma membrane and vacuolar membrane-localized marker (Bowman and Bowman 1988; Fajardo-Somera et al. 2013) and ERV25 is an ER-localized marker (Palma-Guerrero et al. 2014). Antibodies used are noted to the left and the genotypes of strains (top panel: his3::Pccg1-het-e1-gfp and bottom panel: his3::Pccg1-gfp). HET-E1-GFP and GFP was detected using anti-GFP antibodies. PMA-1 was detected using anti-PMA-1 antibodies (kindly provided by Dr K. Allen, Yale School of Medicine) and ERV25 was detected using anti-ERV25 peptide antibodies (Palma-Guerrero et al. 2014).
Discussion
Identification and Characterization of a New HET Domain Locus Involved in Allorecognition
Here we tested the hypothesis that new allorecognition loci that are functional in vegetative incompatibility (het loci) could be identified by population genomic analysis of genes containing an HET domain, which is a predicted cell-death effector. We tested this hypothesis on a highly polymorphic HET domain locus (NCU09037) identified in our search and which showed evidence of balancing selection. In support of our hypothesis, we showed that strains that were isogenic, except for different NCU09037 alleles, showed a typical incompatibility phenotype. Based on map position, we named NCU09037 as het-e, which was one of the first genetically characterized het loci in N. crassa (Wilson and Garnjobst 1966). het-e has three haplogroups, but two of them (het-e2 and het-e3) have a large deletion in the HET domain. Based on the strong incompatibility reaction in (het-e2 + het-e3) heterokaryons, it is apparent that an entire HET domain is not required for het-e-mediated vegetative incompatibility. We also showed that incompatible interactions mediated by het-e, like all of the vegetative incompatibility interactions identified in N. crassa, are suppressed by a deletion of the transcription factor vib-1 (Xiang and Glass 2004; Lafontaine and Smith 2012; this study). It has been shown recently that VIB-1 plays a role in nutrient signaling and is essential for the utilization of plant biomass by N. crassa as well as the production of extracellular proteases (Hutchison and Glass 2010; Xiong et al. 2014). In the plant-symbiotic fungus Epichloe festucae, a vib-1 homolog is essential for the production of antifungal compounds (Niones and Takemoto 2015). It is unclear how nutrient signaling, the secretion of extracellular enzymes, and production of antifungal compounds might be linked to vegetative incompatibility, but a role for these processes in defense against parasites is feasible (see below).
Diversification of HET Domain Genes in Fungi
We determined the organization, order, and content of HET domain genes in Neurospora spp. and other Ascomycete species to gain insight into the diversification of HET domain genes over evolutionary time. The HET domain loci in N. crassa, N. tetrasperma, and the more distantly related species, P. anserina and A. oryzae were not randomly distributed, but tended to be clustered at the ends of chromosomes, and thus might be a general feature of highly variable genomic features in fungi (Gladieux et al. 2014). The recombination rate at the end of chromosomes is much higher than other regions of the chromosome (Jensen-Seaman et al. 2004), and telomeric regions in fungi (and other eukaryotes) are highly dynamic and undergo rearrangements or insertions/deletions (Cohn et al. 2006; McEachern 2007). It is likely that the clustering of HET domain genes to ends of chromosomes facilitates generation of allelic variability at HET domain loci as well as new HET domain genes, some of which may undergo selection to function in allorecognition and vegetative incompatibility. The identification of HET domain genes showing variable conservation in the N. crassa Louisiana population and which were associated with deletion, insertion or inversion events (supplementary fig. S1, Supplementary Material online) supports the hypothesis that HET domain genes emerge due to genome instability events.
HET domain loci, although common in the genomes of filamentous ascomycete species, are highly variable and synteny is not conserved (Fedorova et al. 2005; this study). These observations support the hypothesis that HET domain loci encode a unique gene set that is subject to high variability in fungal genomes, perhaps as a consequence of evolutionary selection for functions in allorecognition. HET domain genes are also present in some basidiomycete species (Van der Nest et al. 2014), as well as lower plant and some protist species. It will be of great interest to determine whether alleles at HET domain genes in species outside of filamentous ascomycete fungi are highly variable within populations and show evidence of balancing selection. If so, the allorecognition function of HET domain genes could be widely conserved.
Biological Processes and Molecular Mechanisms Underlying the Genetic Variability of Alleles at HET Domain Loci
Genes involved in allorecognition display patterns of elevated genetic variability (Boehm 2006), for example, at the vertebrate MHC (Garrigan and Hedrick 2003), immunoglobulin genes (Su and Nei 1999), plant disease resistance genes (Bakker et al. 2006), and plant self-incompatibility genes (Vekemans and Slatkin 1994). The extreme allelic diversity observed at N. crassa HET domain loci relative to the rest of the genome is consistent with their proposed role in allorecognition. Studies of individual het loci in other fungi provide circumstantial evidence for high sequence variability, although sequence variation was not explicitly measured nor were levels of variation at het genes compared with the rest of the genome (e.g., Wu and Glass 2001; Hall et al. 2010; Bastiaans et al. 2014). Our study thus represents the first systematic survey of sequence variability at HET domain loci and candidate het genes in fungi. Using population genomic analyses, we identified a set of 34 HET domain genes that were clearly more variable than the genomic background, representing a highly dynamic fraction of the genome. This is the first report of the existence of such a fraction of hidden “hyperdiversity” within fungal genomes and provides a striking example of intragenomic heterogeneity in rates of molecular evolution.
There is accumulating evidence that fungal het genes contain multiple codons under positive diversifying selection (Wu and Glass 2001; Paoletti and Clave 2007; Hall et al. 2010; Bastiaans et al. 2014; this study), allowing the maintenance of selectively distinct alleles by a form of rare allele advantage (Takahata and Nei 1990). In N. crassa, trans-species polymorphism is a feature of all molecularly characterized het loci (mating type idiomorphs (Glass et al. 1988); un-24/het-6 (Powell et al. 2007); het-c/pin-c (Wu et al. 1998; Hall et al. 2010); het-e (this study)). In the case of het-e, signatures of repeated adaptive evolution (in terms of having a proportion of sites with dN/dS > 1) were only in evidence for het-e2. These data suggest that new het-e alleles, and potentially new functionally different alleles, preferentially enter the population and cause turnover of existing alleles following the appearance and selection of new mutations. Our analyses provide evidence for a contribution of recombination within but also between haplogroups in intrahaplogroup polymorphism, as previously observed for pin-c (Hall et al. 2010).
Although diversifying and balancing selection appear to be driving the appearance and maintenance of alleles at het loci in fungi, the nature of the underlying selective force remains to be determined (Paoletti and Saupe 2009). Classically, it is proposed that new and rare compatibility types emerge and rise in frequency because their carriers are less susceptible to exploitation or infection by parasitic agents. An alternative, although not mutually exclusive hypothesis, is that allorecognition genes evolved as a byproduct of pathogen-driven divergence of host defense genes (Paoletti and Saupe 2009; Bastiaans et al. 2014). Within our set of 34 hyperdiverse HET domain genes, 15 were subdivided into two or three long-divergent haplogroups, whereas the remaining displayed multiple alleles without evidence for clustering of sequences in a limited number of highly divergent groups. These data are consistent with a role of the HET domain genes with two or three haplogroups as het loci, given that modeling work predicts that most incompatibility loci will have two or three phenotypic classes of compatibility, under conditions of strong selection, weak mutation, and limited number of loci (Muirhead et al. 2002). Eleven het loci have been identified in N. crassa based on crosses with translocation strains, which often carry duplications for large parts of chromosomes and from limited crosses between laboratory strains (Perkins 1988), making it likely that not all het loci have been identified. It is possible that the remaining 19 hyperdiverse HET domain genes may be involved in defense against fungal parasites (bacteria, fungi, nematodes, or viruses). Such genes may be part of the pool of genes involved in recognition and response to heterospecific nonself that can be occasionally recruited to specifically function in the recognition of conspecifics in N. crassa, providing an example of exaptation, which describes a shift in the function of a structure or gene during evolution (Gould and Erba 1982). As shown in this study, the genetic, molecular, and genomic resources available for N. crassa provide unparalleled resources by which to investigate the molecular function of allorecognition genes and their evolutionary relationships with parasite defense mechanisms in filamentous fungi, and more broadly in other multicellular eukaryote species.
Materials and Methods
Strains and Growth Conditions
We sequenced genomes of 25 N. crassa isolates randomly selected from Louisiana population whose structure is described in Palma-Guerrero et al. (2013) (supplementary table S1, Supplementary Material online) (Sequence read archive study accession number: SRP052921). Reference strains FGSC 2489, FGSC 9716 (his-3 a), FGSC 6103 (his-3 A), FGSC 11308 (Δvib-1::hph a), and FGSC 12034 (Δhet-e::hph A) were obtained from the Fungal Genetic Stock Center. FGSC 12034 was changed to mat a background BH22 through crossing. KD02-14 (his-3; pan-2 A) and KD02-15 (his-3; pyr-4 A) were used to cross with BH22 to obtain BH10 (his-3; pan-2; Δhet-e::hph a) and BH11 (his-3; pyr-4; Δhet-e::hph a.). All strains were grown on Vogel’s minimal medium (VMM) (Vogel 1956) and crossed on Westergaard’s medium (Westergaard and Mitchell 1947) unless otherwise noted. Transformations were performed as previously described (Margolin et al. 1997).
For incompatibility assays, strains with different auxotrophic markers were grown on VMM with corresponding required nutrients to obtain fresh conidia (approximately 1 week). Equal amounts of conidia from the two auxotrophic strains were mixed and inoculated into the center of a minimal medium plate. The growth phenotype (compatible or incompatible) was recorded the third day after inoculation. Photos were captured by Bio-Rad ChemiDoc XRS using white light and edited by Image Lab 4.
Neurospora discreta strains (CAWE967, CAWE973, CAWE978, CAWE990, CAWE992, CAMH1105, CAMH1108, CAMH1110, WACL1050, WACL1070, and WACL1073) were collected in Weaverville (CA), Morgan Hill (CA), and Lake Chelan (WA) by D.J. Jacobson. These isolates cluster within phylogenetic species 4, subgroup B (Gladieux P, Taylor JW, unpublished data) (Dettman et al. 2006).
Neurospora crassa Whole-Genome Resequencing Analyses
The genomes of 25 N. crassa strains from Louisiana were searched for additional HET domain genes and the levels of genetic variation at candidate loci were characterized. Genomic DNA was extracted following a standard protocol with proteinase K and RNase digestion to avoid protein and RNA contamination. Purified genomic DNA (10 ng/µl in 300 µl volume) was sheared by Bioruptor@D300 (21 cycles, 30 s off and 15 s on); sheared DNA had a peak at 500–600 bp sized fragments. TruSeq DNA LT Sample Prep kit (Illumina) was used to build paired-end DNA-seq libraries.
Preliminary analyses indicated extensive size variation (insertion/deletions) at HET domain genes, so we used an approach based on whole-genome alignment, instead of mapping reads onto a reference genome, to identify single nucleotide polymorphisms (SNPs) in genes. Sequencing reads were processed using PRINSEQ (Schmieder and Edwards 2011) and custom scripts to remove polymerase chain reaction (PCR) duplicates, poor quality reads, and orphan reads. Filtered sequencing reads were cleaned using cutadapt (https://github.com/marcelm/cutadapt, last accessed June 22, 2015) to remove adapters and sorted by FASTQCOMBINEPAIREDEND.PY to make paired-end files (https://github.com/enormandeau/Scripts/blob/master/fastqCombinePairedEnd.py, last accessed June 22, 2015). Sequencing reads were assembled using the short read assembler SOAPDENOVO2 (Luo et al. 2012). Gene prediction was carried out using AUGUSTUS (Hoff and Stanke 2013) and protein sequences from FGSC 2489 as query sequences (supplementary table S1, Supplementary Material online).
Identification of HET Domain Genes and Synteny Analyses
HET domain sequences predicted in the N. crassa reference genome (Broad Institute N. crassa database) were aligned to de novo assemblies using BLAST+ (Basic Local Alignment Search Tool) (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/bla st+/). HET domain sequences were aligned using MACSE (Ranwez et al. 2011). Multiple alignments were visualized using JALVIEW (http://www.jalview.org). Genome assemblies of N. tetrasperma, N. discreta, and P. anserina were obtained from the U.S. Department of Energy Joint Genome Institute database (http://genome.jgi-psf.org/programs/fungi/index.jsf). Sordaria macrospora genome assembly was downloaded from http://fungidb.org/fungidb/ and the A. oryzae genome assembly was obtained from Comparative Database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html).
Synteny of identified HET domain loci in genomic sequences of the 25 wild N. crassa isolates, N. tetrasperma and N. discreta versus reference genome sequence FGSC 2489 was tested by whole chromosome comparisons using MUMmer3 (Kurtz et al. 2004). Synteny analyses for het-e used FungiDB (http://fungidb.org/fungidb/). It was further evaluated by reciprocal BLAST searches in the genomes from the different species. Genes encoded by HET domain loci were considered orthologous if they showed significant amino acid identity and were also syntenic. However, orthology could not be established for some genes, either because they were not predicted or not correctly predicted, or because shared synteny could not be established based on whole-genome alignments due to elevated levels of interallelic divergence or sequencing gaps. For these genes, we used tBLASTn to confirm gene prediction and to identify potential homologous sequences, and subsequently used neighboring genes to establish shared synteny. If a gene fell into a region that was syntenic with other genomes, and the identity of the sequence in this region was the highest in reciprocal homology searching, the homologous sequences were considered as orthologs.
To identify HET domain genes in other species, we searched Pfam database (http://pfam.xfam.org). The obtained HET domain sequences in protist and plant species were aligned to JGI Phytozome 10.1 database (http://phytozome.jgi.doe.gov/pz/portal.html#) by BLAST to confirm the corresponding gene loci sequences. The extracted HET domain sequences were aligned and viewed by JALVIEW (http://www.jalview.org) to N. crassa HET domain sequences for confirmation. Non-HET portion of the genes was excluded from the multiple sequence alignments.
To determine whether HET domain loci were randomly distributed across chromosomes or chromosome arms according to a uniform distribution, Kendall–Sherman tests (Mielke and Berry 2001) were conducted. Statistical significance was determined using 1,000 data sets simulated under the null hypothesis of a random uniform distribution. For N. crassa, chromosomal arms were defined using the positions of centromeres determined in Smith et al. (2011). For N. tetrasperma, we determined the coordinates of centromeres assuming synteny with N. crassa (Smith et al. 2011). Approximate coordinates of centromeres were determined using the AspGD genome browser for A. oryzae (http://www.aspergillusgenome.org/) and a genetic map for P. anserina (Marcou et al. 1993).
Signatures of Balancing Selection at HET-Domain Genes
HET domain genes were scanned for signatures of balancing selection by comparing them to a reference set of 8,621 genes (out of the 9,730 genes identified in the N. crassa genome), after removal of genes without SNPs (because of no coverage, or no polymorphism) and of genes with too small a sample size (N < 6). Summary statistics were calculated using the BIO++ (Dutheil et al. 2006) and EGGLIB python/C++ libraries (De Mita and Siol 2012): The number of segregating sites standardized by sequence length S (a measure of polymorphism), the average number of nucleotide differences between sequence pairs π (a measure of nucleotide diversity), Tajima’s D (which measures skews in allele frequency distributions), and the maximum number of synonymous substitutions between sequence pairs KSmax (a measure of gene tree depth). The position of HET domain genes in the reference gene space was based on combinations of their summary statistics and was visualized by PCA (principal component analysis) in the R environment. Neighbor-joining gene genealogies were constructed for all candidate het loci using the F84 model of nucleotide substitution, as implemented in the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html).
Phylogenetic Analysis and Positive Selection Analyses of het-e Haplogroups
Multiple alignments for het-e sequences that excluded the highly diverse 3′-end of the coding sequence (CDS) as well as alignments that included these highly diverse sequences were used for phylogenetic inference. As the two methods produced the same tree topology, only alignments including the 3′-end of the CDS of the HET-E protein were used for analyses. Codon alignments were carried out using MACSE (Ranwez et al. 2011). Phylogenetic trees were inferred using MrBayes 3.2 (Ronquist et al. 2012) with four separate chains run for 1,000,000 generations, a burn-in value of 10,000, and sampling every 100 generations.
Codon-based-likelihood models implemented in the PAML package (Yang 2007) were used to test for positive selection by recurrent changes in amino acids of the het-e gene and the different het-e haplogroups. We used the “branch-site” model included in Test 2 (Zhang et al. 2005). Test 2 compares a model in which the branch under consideration (i.e., the branch corresponding to a given haplogroup) is evolving without constraint (dN/dS = 1) to a model in which this branch has sites evolving under positive selection (dN/dS > 1).
Allele Constructions
To test the function of the het candidate locus NCU09037 (het-e), we introduced different NCU09037 alleles into the his-3 locus of an N. crassa ΔNCU09037 deletion strain. The promoter of qa-2 (NCU06023) was amplified using primers pQA-2 For NotI (5′-GCG GCC GCA GAG TCC TTG ACA AGC AGT-3′) and pQA-2 Rev XbaI (5′-TCT AGA TGT GTT TGG TAC CTC TG-3′) and cloned into pMF272 (Freitag et al. 2004) using NotI and XbaI (New England Biolabs). The het-e1, het-e2 and het-e3 alleles were amplified from FGSC 2489, P4468 and JW75, respectively, using primers het-e1For (5′-CGCTCTAGAATGGTTCCTAGA CCTCC-3′), het-E1Rev (5′-CGCTTAATTAATACCAGCATAATTTTCTGGC-3′), het-e2For (5′-CGCTCTAGAATGGTCTTAAGATCCTCACACAAACC-3′), het-e2Rev (5′-CGCGGATCCGAGTGGAGCAAGCTGTTGGAT-3′), het-e3For (5′-CGCTCTAGAATGGTCCTAGGATCCCCAAACA-3′), and het-e3Rev (5′-CGCCCCGGGGCGACGAATGGATAAGGTTACAGG-3′). The het-e2 allele was cloned into the pMF272 pqa-2 by amplification using the primers het-e2 XbaI (5′-CGC TCT AGA ATG GTC CTA AGA TCC CC-3′), and het-e2 PacI (5′-CGC TTA ATT AAT ATT TTC AAA GAC CTT TC-3′). To construct mCherry tagged het-e alleles, we used a plasmid TSL84C with mCherry (Jonkers et al. 2014) and the primers/procedure described above.
RNA Extraction, Quantitative Reverse-Transcription PCR, and Cell Fractionation
Reverse-transcription (RT)-PCR was used to determine whether strains bearing a vib-1 deletion (encoding an NDT80-like transcription factor) showed a reduction in expression of HET domain genes. RNA extraction and quantitative RT-PCR (Q RT-PCR) were performed as previously described (Hutchison et al. 2012). Briefly, RNA was extracted using the TRIzol reagent (Invitrogen), purified using a Qiagen RNeasy kit, and treated with Ambion Turbo DNase. RNA quality was assessed using gel electrophoresis and a Nanodrop (Thermo Scientific). Q RT-PCR was performed using a Qiagen QuantiTect SYBR Green RT-PCR kit and an ABI 7300 machine, according to the manufacturer’s instructions. Reactions were performed in triplicate, and actin was used as the endogenous control. Data were analyzed using the ABI 7300 software. Q RT-PCR primers used to amplify actin were actin-F and actin-R from Dementhon et al. (2006), and the primers for het-e were NCU09037-FOR (5′-TACCCCCAATCTTCACTCT-3′) and NCU09037-REV (5′-GTTGTCGGCTTGGTTGATG-3′).
The subcellular localization of HET-E protein was determined by cell fractionation using sucrose gradients, as GFP or mCherry fluorescent signals were not detected in live cells. Sucrose gradient fractionation was conducted following protocols described in Palma-Guerrero et al. (2014). Western blot analysis was conducted using anti-GFP antibody (1:1,000 dilution; Roche LOT11814460001). Anti-PMA1 antibody (1:3,000 dilution) was kindly provided by Dr K. Allen (Yale School of Medicine) and anti-ERV25 antibody (1:1,000 dilution) kindly provided by Dr T. Starr (Energy Biosciences Institute, University of California, Berkeley).
Supplementary Material
Supplementary figures S1–S4 and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a National Institutes of Health Grant (R01GM060468) to N.L.G. and a Marie Curie postdoctoral fellowship awarded to P.G. (FP7-PEOPLE-2010-IOF-No.273086). The authors thank JW Taylor (University of California, Berkeley) for sharing unpublished data, David Kowbel (UCB) for assistance with computational analyses, and Juliet Welch and Dr Jens Heller (UCB) for experimental assistance.
References
- Bakker EG, et al. 2006. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18:1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaans E, et al. 2014. Natural variation of heterokaryon incompatibility gene het-c in Podospora anserina reveals diversifying selection. Mol Biol Evol. 31:962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benacerraf B, Germain RN. 1978. The immune response genes of the major histocompatibility complex. Immunol Rev. 38:70–119. [DOI] [PubMed] [Google Scholar]
- Biella S, et al. 2002. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc Biol Sci. 269:2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T. 2006. Quality control in self/nonself discrimination. Cell 125:845–858. [DOI] [PubMed] [Google Scholar]
- Bowman EJ, Bowman BJ. 1988. Purification of vacuolar membranes, mitochondria, and plasma membranes from Neurospora crassa and modes of discriminating among the different H+-ATPases. Methods Enzymol. 157:562–573. [DOI] [PubMed] [Google Scholar]
- Cadavid LF, et al. 2004. An invertebrate histocompatibility complex. Genetics 167:357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevanne D, et al. 2010. WD-repeat instability and diversification of the Podospora anserina hnwd non-self recognition gene family. BMC Evol Biol. 10:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GH, et al. 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG. 1997. Neutral behavior of shared polymorphism. Proc Natl Acad Sci U S A. 94:7730–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, Liti G, Barton DBH. 2006. Telomeres in fungi. In: Sunnerhagen PaPJ, editor. Comparative genomics using fungi as models. Springer Berlin Heidelberg. p. 101–130. [Google Scholar]
- Colot HV, et al. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 103:10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran P, et al. 2014. A global multilocus analysis of the model fungus Neurospora reveals a single recent origin of a novel genetic system. Mol Phylogenet Evol. 78:136–147. [DOI] [PubMed] [Google Scholar]
- Cortesi P, Milgroom MG. 1998. Genetics of vegetative incompatibility in Cryphonectria parasitica. Appl Environ Microbiol. 64:2988–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, et al. 2015. Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol. 13:e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, Siol M. 2012. EggLib: processing, analysis and simulation tools for population genetics and genomics. BMC Genet. 13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debets F, et al. 1994. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 26:113–119. [DOI] [PubMed] [Google Scholar]
- Dementhon K, et al. 2006. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot Cell. 5:2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, et al. 2006. Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia 98:436–446. [DOI] [PubMed] [Google Scholar]
- Dutheil J, et al. 2006. Bio++: a set of C++ libraries for sequence analysis, phylogenetics, molecular evolution and population genetics. BMC Bioinformat 7:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E, et al. 2002. HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina . Genetics 161:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo-Somera RA, et al. 2013. The plasma membrane proton pump PMA-1 is incorporated into distal parts of the hyphae independently of the Spitzenkorper in Neurospora crassa. Eukaryot Cell. 12:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova ND, et al. 2005. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics 6:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, et al. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 41:897–910. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Hedrick PW. 2003. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57:1707–1722. [DOI] [PubMed] [Google Scholar]
- Gazis R, et al. 2012. Culture-based study of endophytes associated with rubber trees in Peru reveals a new class of Pezizomycotina: Xylonomycetes. Mol Phylogenet Evol. 65:294–304 [DOI] [PubMed] [Google Scholar]
- Gladieux P, et al. 2014. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol. 23:753–773. [DOI] [PubMed] [Google Scholar]
- Glass NL, Dementhon K. 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol. 9:553–558. [DOI] [PubMed] [Google Scholar]
- Glass NL, et al. 1988. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 241:570–573. [DOI] [PubMed] [Google Scholar]
- Glass NL, Kaneko I. 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucl Acids Res. 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Erba ES. 1982. Exaptation—a missing term in the science of form. Paleobiology 8:4–15. [Google Scholar]
- Grigoriev IV, et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42:D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, et al. 2010. Evolution and diversity of a fungal self/nonself recognition locus. PLoS One 5:e14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Stanke M. 2013. WebAUGUSTUS—a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 41:W123–W128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison E, et al. 2009. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiology 155:3957–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison EA, et al. 2012. Diversification of a protein kinase cascade: IME-2 is involved in nonself recognition and programmed cell death in Neurospora crassa . Genetics 192:467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison EA, Glass NL. 2010. Meiotic regulators Ndt80 and ime2 have different roles in Saccharomyces and Neurospora. Genetics 185:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, et al. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W, et al. 2014. HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa. PLoS Genet. 10:e1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko I, et al. 2006. Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172:1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, et al. 1993. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 11:269–295. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Smith ML. 2012. Diverse interactions mediate asymmetric incompatibility by the het-6 supergene complex in Neurospora crassa. Fungal Genet Biol. 49:65–73. [DOI] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcou D, et al. 1993. Genetic Map of Podospora anserina. In: O’Brien S, editor. Genetic maps. 6th ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. p. 92-93. [Google Scholar]
- Margolin BS, et al. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newsl. 34:34–36 [Google Scholar]
- McEachern MJ. 2007. Telomeres: guardians of genomic integrity or double agents of evolution? In: Noseck J, Tomaska L, editors. Origins and evolution of telomeres. Landes Austin (TX): Bioscience. [Google Scholar]
- Mielke P, Berry K. 2001. Permutation methods: a distance function approach. New York: Springer Verlag. [Google Scholar]
- Mir-Rashed N, et al. 2000. Molecular and functional analyses of incompatibility genes at het-6 in a population of Neurospora crassa. Fungal Genet Biol. 30:197–205. [DOI] [PubMed] [Google Scholar]
- Muirhead CA, et al. 2002. Multilocus self-recognition systems in fungi as a cause of trans-species polymorphism. Genetics 161:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niones JT, Takemoto D. 2015. VibA, a homologue of a transcription factor for fungal heterokaryon incompatibility, is involved in antifungal compound production in the plant-symbiotic fungus Epichloe festucae. Eukaryot Cell. 14:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal K, et al. 2007. Sexual and vegetative compatibility genes in the aspergilli. Stud Mycol. 59:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J, et al. 2013. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 9:e1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J, et al. 2014. Identification and characterization of LFD1, a novel protein involved in membrane merger during cell fusion in Neurospora crassa. Mol Microbiol. 92:164–182. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Clave C. 2007. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot Cell. 6:2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, et al. 2007. Genesis of a fungal non-self recognition repertoire. PLoS One 2:e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Saupe SJ. 2009. Fungal incompatibility: evolutionary origin in pathogen defense? BioEssays 31:1201–1210. [DOI] [PubMed] [Google Scholar]
- Perkins DD. 1988. Main features of vegetative incompatibility in Neurospora crassa. Fungal Genet Newsl. 35:44–46. [Google Scholar]
- Powell AJ, Jacobson DJ, Natvig DO. 2001. Allelic diversity at the het-c locus in Neurospora tetrasperma confirms outcrossing in nature and reveals an evolutionary dilemma for pseudohomothallic ascomycetes. J Mol Evol. 52:94–102. [DOI] [PubMed] [Google Scholar]
- Powell AJ, Jacobson DJ, Natvig DO. 2007. Ancestral polymorphism and linkage disequilibrium at the het-6 region in pseudohomothallic Neurospora tetrasperma. Fungal Genet Biol. 44:896–904. [DOI] [PubMed] [Google Scholar]
- Ranwez V, et al. 2011. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One 6:e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read ND, et al. 2010. Hyphal fusion. In: Borkovich KA, Ebbole D, editors. Cellular and molecular biology of filamentous fungi. Washington (DC): ASM Press; p. 260–273. [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe SJ. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 64:489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, et al. 2011. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 31:2528–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, et al. 2000. Vegetative incompatibility in the het-6 region of Neurospora crassa is mediated by two linked genes. Genetics 155:1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Nei M. 1999. Fifty-million-year-old polymorphism at an immunoglobulin variable region gene locus in the rabbit evolutionary lineage. Proc Natl Acad Sci U S A. 96:9710–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Nei M. 1990. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124:967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Nest MA, et al. 2014. Distribution and evolution of het gene homologs in the basidiomycota. Fungal Genet Biol. 64:45–57. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Slatkin M. 1994. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics 137:1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ. 1956. A convenient growth medium for Neurospora. Microbial Genet Bull. 13:42–43. [Google Scholar]
- Voskoboynik A, et al. 2013. Identification of a colonial chordate histocompatibility gene. Science 341:384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M, Mitchell HK. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am J Bot. 34:573–577. [Google Scholar]
- Wilson JF, Garnjobst L. 1966. A new incompatibility locus in Neurospora crassa. Genetics 53:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. 1998. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc Natl Acad Sci U S A. 95:12398–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Glass NL. 2001. Identification of specificity determinants and generation of alleles with novel specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa. Mol Cell Biol. 21:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Glass NL. 2004. The control of mating type heterokaryon incompatibility by vib-1, a locus involved in het-c heterokaryon incompatibility in Neurospora crassa. Fungal Genet Biol. 41:1063–1076. [DOI] [PubMed] [Google Scholar]
- Xiong Y, et al. 2014. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet. 10:e1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, et al. 2014. Transcriptional interference by antisense RNA is required for circadian clock function. Nature 514:650–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang DX, et al. 2014. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 197:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.