Abstract
Panorama Plus (Natera), a single-nucleotide polymorphism- (SNP-) based approach that relies on the identification of maternal and fetal allele distributions, allows the detection of common aneuploidies and also incorporates a panel of 5 microdeletions including Di George syndrome. We report here the first case of Di George syndrome detected by NIPT in Italy; blood was drawn at 12 weeks' gestation. The patient had an amniocentesis to confirm the diagnosis by MLPA (multiplex ligation-dependent probe amplification) and an ultrasound aimed to detect the features associated with the syndrome. A right aortic arch and suspect of thymus atrophy were detected, but not other severe malformations typical of the disease. The patient terminated the pregnancy at 17 weeks. NIPT allowed an early screening of Di George syndrome. As the patient was at low risk, it is likely that an ultrasound would have missed the condition.
1. Introduction
Numerous studies have validated the accuracy of noninvasive prenatal testing (NIPT) using fetal cell-free DNA (cffDNA) to assess the risk of fetal aneuploidies early in pregnancy [1].
cffDNA is the most effective method of screening for trisomy 21, with DR of more than 99% and FPR of about 0.1%. [2–4]. There are two different approaches in analyzing the cffDNA: quantitative and single-nucleotide polymorphism- (SNP-) based methods. The quantitative approach works better for twin pregnancies and donor egg pregnancies, but the SNP-based methods offer a wider spectrum, including the highly sensitive and specific detection of triploidy, vanishing twins, and, so far, 5 microdeletions.
Microdeletion of chromosome 22q11.2 or 22q11.2 deletion syndrome (22q11.2 DS) occurs in 1 : 2000 live births [5]. Up to 93% of cases occur de novo, whereas the remaining 7% of deletion cases are inherited from a parent [6]. Its phenotypic expression is variable and includes Di George syndrome (OMIM 188400) and velocardiofacial syndrome (OMIM 192430). Patients with 22q11.2 deletion syndrome can suffer from congenital heart disease (CHD), cleft palate, velopharyngeal insufficiency with hypernasal speech, hypocalcemia, dysmorphic facial features, and mental retardation [6]. CHD is found in approximately 75% of patients with 22q11.2 DS and typically constitutes conotruncal malformations such as interrupted aortic arch type B, tetralogy of Fallot (TOF), truncus arteriosus communis, and pulmonary atresia with ventricular septal defect (VSD) [7–9].
Even if the samples size of Di George cases collected so far in the various studies is relatively small, the performance of the SNP method is quite impressive. For example, in a study by Wapner et al. [5] the DR for 22q11.2 deletion was 97.8% (45/46) at a false positive rate of 0.76% (3/397). The positive predictive value (PPV) was estimated to be 5.3% based on the incidence of the disease being about 1 : 2000. It is also worth noting that this value is quite similar to that quoted for the combined test as reported in the SURUSS study (1 : 22 or 4.5%) [10]; therefore the potential impact on a routine screening program is very effective.
2. Case Presentation
A 38-year-old primipara woman was referred for counseling at 15 weeks of gestation because of a high risk finding for Di George syndrome with the estimated risk of 1 : 19 by a SNP-based NIPT performed at 12 weeks' gestation (Panorama Plus, Natera Inc.; San Carlos, California). CffDNA from maternal plasma was amplified at 4,128 SNPs, including 672 SNPs from a 2.91 Mb in the 22q11.2 region, and sequenced. The risk calculation was performed by using the Next-generation Aneuploidy Test Using SNPs (NATUS) algorithm, which involves applying a Bayesian-based maximum likelihood statistical method to the SNP data to determine the number of copies of the 22q11.2 region present in the genome of the fetus, and the likelihood of the determination, expressed as a risk score. Her husband was 45 years old. She and her husband were healthy and nonconsanguineous. There was no family history of congenital malformations. She denied any recent infections or exposure to teratogens during this pregnancy. She was referred to our centre, and amniocentesis was performed for cytogenetic and molecular analysis. DNA extracted from the amniotic fluid was analyzed by MLPA (multiplex ligation-dependent probe amplification) using the SALSA MLPA kit for Di George syndrome, velocardiofacial syndrome, and cat eye syndrome (MRC-Holland, Amsterdam, Netherlands) according to the manufacturer's protocol. The MLPA showed a deletion in the Di George syndrome critical region of chromosome 22 low copy number repeat (LCR). The deletion was determined to have an extension from 2,058 Mb to 3,117 Mb. Ultrasonographic examination was performed at 17 weeks' gestation using a Voluson E8 and RAB 4–8P (4D, 4–8 MHz Convex) probe (GE Healthcare, Milwaukee, WI) which revealed a singleton fetus with right aortic arch (Figure 1). The amount of amniotic fluid and fetal growth were normal. Other internal organs were unremarkable. The fetus did not show any cleft lip or palate. The woman requested a termination of pregnancy (TOP) only after the US scan revealed some of the clinical features of the syndrome.
Figure 1.
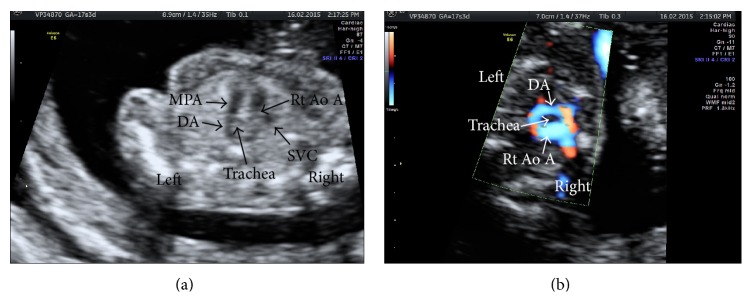
Fetal echocardiography at 17 weeks' gestation. Two-dimensional gray-scale (a) and high definition color Doppler (b) images in three-vessel and trachea view. The right aortic arch (Rt Ao A) and ductus arteriosus (DA), forming classic “U” shape. MPA, main pulmonary artery; SVC, superior vena cava.
3. Discussion
NIPT for aneuploidy was introduced in Italy a couple of years ago, giving women an alternative to serum screening and ultrasound, with considerably higher sensitivity and specificity to detect trisomies 21, 18, and 13, as well as monosomy X. In 2014, an extended panel for microdeletions NIPT screening was also introduced [5]. Unlike NIPT for aneuploidy, which was an improvement in care, NIPT for microdeletions represents an entirely new offering, as there were previously no methods which could detect microdeletions noninvasively in the first trimester. In fact, at the present, only the invasive search for microdeletions is available and, in Italy, the test is reserved only for those fetuses with an abnormal scan or abnormal nuchal translucency (NT) >3 mm, or in the presence of familiar history, and it is generally performed around 20 weeks' gestation.
This case is the first detection of fetal Di George syndrome in Italy using NIPT in the first trimester. Di George syndrome was detected using a SNP-based NIPT test (Panorama Plus). Panorama Plus uses an algorithm termed NATUS that analyzes up to 28,000 SNPs and determines mutually exclusive probability of a fetal aneuploidy (trisomies 21, 18, and 13 and monosomy X), fetal microdeletion (Di George syndrome, Cri-du-chat syndrome, Prader-Willi syndrome, Angelman syndrome, and 1p36 deletion syndrome) or euploidy. The DR and FPR of aneuploidies [11, 12] and microdeletions [5] have been reported in several studies elsewhere. It is important to note that NIPT is only a screening test and all high-risk results should be confirmed by an invasive, diagnostic testing method.
Multiple options exist for diagnosis of microdeletions from invasive testing samples, such as FISH (fluorescence in situ hybridization), MLPA (multiplex ligation-dependent probe amplification), aCGH (microarray-based comparative genomic hybridization), and next generation sequencing [13, 14]. FISH analyses for microdeletion syndromes have successfully been replaced by MLPA [15–17]. MLPA is a novel, commercially available PCR-based technique analysis. It has several advantages in comparison with FISH analysis: the ability to analyze several loci simultaneously, reliable detection of duplications, and the facility to obtain the result within a similar turnaround time of 1 to 2 days. MLPA is a new technique for multiple quantification of copy numbers at specific target sequences. The ability of Panorama Plus to detect the 22q11.2 microdeletion noninvasively in the first trimester allowed for earlier decision making. In this case, the termination of pregnancy was delayed because the patients chose to pursue additional confirmation by way of an ultrasound scan that revealed a right aortic arch (without an aberrant left subclavian) and a hypoaplasia of the thymus.
Abbreviations
- NIPT:
Noninvasive prenatal testing
- SNP:
Single-nucleotide polymorphism
- cffDNA:
Cell-free DNA
- TOF:
Truncus arteriosus communis
- VSD:
Ventricular septal defect
- PPV:
Positive predictive value
- NATUS:
Next-generation Aneuploidy Test Using SNPs
- MLPA:
Multiplex ligation-dependent probe amplification
- TOP:
Termination of pregnancy
- Rt Ao A:
Right aortic arch
- DA:
Ductus arteriosus
- MPA:
Main pulmonary artery
- SVC:
Superior vena cava.
Consent
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review.
Conflict of Interests
The authors declare that they have no competing interests.
References
- 1.Quezada M. S., Gil M. M., Nicolaides K. H., Francisco C., Oròsz G. Screening for trisomies 21, 18 and 13 by cell-free DNA analysis of maternal blood at 10-11 weeks' gestation and the combined test at 11–13 weeks. Ultrasound in Obstetrics and Gynecology. 2015;45(1):36–41. doi: 10.1002/uog.14664. [DOI] [PubMed] [Google Scholar]
- 2.Palomaki G. E., Kloza E. M., Lambert-Messerlian G. M., et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genetics in Medicine. 2011;13(11):913–920. doi: 10.1097/gim.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 3.Norton M. E., Brar H., Weiss J., et al. Non-Invasive Chromosomal Evaluation (NICE) study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. American Journal of Obstetrics & Gynecology. 2012;207(2):137.e1–137.e8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi D. W., Lamar Parker R., Wentworth J., et al. DNA sequencing versus standard prenatal aneuploidy screening. The New England Journal of Medicine. 2014;370(9):799–808. doi: 10.1056/nejmoa1311037. [DOI] [PubMed] [Google Scholar]
- 5.Wapner R. J., Babiarz J. E., Levy B., et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. American Journal of Obstetrics and Gynecology. 2015;212(3):332.e1–332.e9. doi: 10.1016/j.ajog.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 6.Kobrynski L. J., Sullivan K. E. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. The Lancet. 2007;370(9596):1443–1452. doi: 10.1016/s0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- 7.Cohen E., Chow E. W. C., Weksberg R., Bassett A. S. Phenotype of adults with the 22q11 deletion syndrome: a review. American Journal of Medical Genetics. 1999;86(4):359–365. doi: 10.1002/(sici)1096-8628(19991008)86:4lt;359::aid-ajmg10>3.0.co;2-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldmuntz E., Clark B. J., Mitchell L. E., et al. Frequency of 22q11 deletions in patients with conotruncal defects. Journal of the American College of Cardiology. 1998;32(2):492–498. doi: 10.1016/s0735-1097(98)00259-9. [DOI] [PubMed] [Google Scholar]
- 9.Bassett A. S., Chow E. W. C., Husted J., et al. Clinical features of 78 adults with 22q11 deletion syndrome. American Journal of Medical Genetics. 2005;138(4):307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wald N. J., Rodeck C., Hackshaw A. K., Rudnicka A. SURUSS in perspective. BJOG. 2004;111(6):521–531. doi: 10.1111/j.1471-0528.2004.00193.x. [DOI] [PubMed] [Google Scholar]
- 11.Nicolaides K. H., Syngelaki A., Gil M., Atanasova V., Markova D. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenatal Diagnosis. 2013;33(6):575–579. doi: 10.1002/pd.4103. [DOI] [PubMed] [Google Scholar]
- 12.Pergament E., Cuckle H., Zimmermann B., et al. Single-nucleotide polymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstetrics and Gynecology. 2014;124(2):210–218. doi: 10.1097/aog.0000000000000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C.-P., Su Y.-N., Lin S.-Y., et al. Rapid aneuploidy diagnosis by multiplex ligation-dependent probe amplification and array comparative genomic hybridization in pregnancy with major congenital malformations. Taiwanese Journal of Obstetrics and Gynecology. 2011;50(1):85–94. doi: 10.1016/j.tjog.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Mademont-Soler I., Morales C., Soler A., et al. MLPA: a prenatal diagnostic tool for the study of congenital heart defects? Gene. 2012;500(1):151–154. doi: 10.1016/j.gene.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes T., Kirchhoff M., Lind A.-M., Vestergaard Larsen G., Schwartz M., Lundsteen C. Computer-assisted prenatal aneuploidy screening for chromosome 13, 18, 21, X and Y based on multiplex ligation-dependent probe amplification (MLPA) European Journal of Human Genetics. 2005;13(2):171–175. doi: 10.1038/sj.ejhg.5201307. [DOI] [PubMed] [Google Scholar]
- 16.Rooms L., Reyniers E., Wuyts W., et al. Multiplex ligation-dependent probe amplification to detect subtelomeric rearrangements in routine diagnostics. Clinical Genetics. 2006;69(1):58–64. doi: 10.1111/j.1399-0004.2005.00545.x. [DOI] [PubMed] [Google Scholar]
- 17.Fernández L., Lapunzina P., Arjona D., et al. Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clinical Genetics. 2005;68(4):373–378. doi: 10.1111/j.1399-0004.2005.00493.x. [DOI] [PubMed] [Google Scholar]


