Abstract
Mycolic acids (MAs) are characteristic components of bacteria in the suborder Corynebacterineae, such as Mycobacterium. MAs are categorized into subclasses based on their functional bases (cyclopropane ring, methoxy, keto, and epoxy group). Since MAs have heterogeneity among bacterial species, analyzing of MAs are required in the chemotaxonomic field. However, their structural analysis is not easy because of their long carbon-chain lengths and several functional groups. In this study, total fatty acid (FA) methyl ester (ME) fraction of M. tuberculosis H37Rv was analyzed by matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry (TOFMS) with a spiral ion trajectory (MALDI spiral-TOFMS). The distributions of carbon-chain length and their relative peak intensities were confirmed with those obtained by analysis of each subclass fraction which was separated from total FA ME fraction using thin-layer chromatography (TLC). The observed major peaks were reliably assigned as MAs owing to the high mass accuracy (error<3 ppm). The types of MA subclasses, their distributions of carbon-chain lengths, their relative peak intensities, and the ratio of even- and odd-numbered carbon-chain MAs for the total FA ME fraction were consistent with those of MA subclass fractions. To visualize whole MAs, contour maps of relative peak intensities for whole MAs were created. The contour maps indicated the MA subclasses and their distributions of carbon-chains with relative peak intensities at a glance. Our proposed method allows simple characterization in a short time and thus enables the analysis of large numbers of samples, and it would contribute to the chemotaxonomy.
Keywords: mycolic acid, MALDI spiral-TOFMS, chemotaxonomy, contour map, Mycobacterium
INTRODUCTION
Mycolic acids (MAs) are the hallmark of bacteria in the suborder Corynebacterineae, which comprises genera such as Mycobacterium, Nocardia, Gordonia, Rhodococcus, Dietzia, and Corynebacterium. MAs are constructed of 2-alkyl 3-hydroxy fatty acids (FAs), the carbon-chains of which are among the longest in nature (C20–C90). MAs of the genus Mycobacterium have C60–C90 carbon-chains1,2) and are categorized into several subclasses on the basis of their functional groups, such as cyclopropane ring, methoxy, keto, and epoxy groups.3–6) MAs have been analyzed as important chemotaxonomic markers to characterize the suborder Corynebacterineae.7,8) In addition, inhibition of MA synthesis is attracting research interest as a target of antitubercular drugs,9–13) MA subclasses have been analyzed to verify the effects of medications.9,12–15)
Traditionally, thin-layer chromatography (TLC) has been widely used to separate MA subclasses on a functional basis.5,16) However, reference specimens are required for the identification of MA subclasses; the experiments are cumbersome, and large amounts of organic solvent are needed. The TLC method is therefore not optimal for analyzing large numbers of samples. High performance liquid chromatography (HPLC)17–19) and gas chromatography coupled with mass spectrometry (GC-MS) have also been used to analyze MA methyl esters (MEs).20) However, the resolution of HPLC is not sufficient to distinguish each molecular species.18) MA MEs need to be vaporized for GC-MS analysis; moreover, even if the MA MEs are trimethylsilylated to inhibit heat degradation it is difficult to volatilize them because of their high molecular weight and polar character. In addition, assignment of the many peaks corresponding to fragment ions requires special knowledge of mass spectrometry. Along with the advance of mass spectrometry, the structure elucidations of MAs of mycobacterium species have progressed especially in the field of clinical diagnosis. Electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) was applied for the identification of common clinical isolate of mycobacterial species.21) By using tandem mass spectrometry, not only molecular weight profiles but also molecular structural information was provided. This method would be reliable and potentially practical as an identification method for mycobacterial species. HPLC-ESI-MS/MS was applied to analyze of MAs, and it thought to be used as a diagnostic tool for rapid and sensitive identification of mycobacterial pathogens.22) However, these newly techniques require long time for HPLC; elution period of free MAs is about 25 to 50 min and the one of MAs derivatives is about 45 to 85 min.23) And, these newly developed techniques have not been applied in the field of chemotaxonomy.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS) has been used to analyze MAs. In the many cases, MALDI-TOFMS has been used to confirm the molecular species and the distribution of MAs in TLC fractions.24,25) These information were reported in the papers where proposal of the new species.26) However the sample preparation of MALDI-TOFMS is easy and mass-resolution is relatively high, extraction of purified subclass MA MEs from TLC plate was cumbersome. The diversity of functional groups and the number of double bonds of putative MAs makes it difficult to discriminate some subclass MAs with mass differences of only Δ0.036 Da by using conventional MALDI-TOFMS in the reflectron mode.27) Previously, we used high-resolution MALDI-TOFMS with a spiral ion trajectory28) to analyze the total FA ME fractions in three species of the genus Dietzia that have α-MAs with short carbon-chains.27) Because MALDI spiral-TOFMS has a spiral ion trajectory that gives high mass-resolution and high mass-accuracy, it enables the separation of adjacent peaks with a 0.036 Da mass difference. In our previous study, the high mass-resolution also enabled us to detect peaks corresponding to previously undetected oxygenated MAs. In addition, MAs were characterized by using relative peak intensities. Whole MA analysis by using the total FA ME fraction and MALDI spiral-TOFMS should contribute to progress in fields relevant to MAs, such as chemotaxonomy. However, since detailed analysis of the MA-subclasses had not been performed for the samples in our previous study using other methods, we consider reliability verification of the methods which characterize MAs by analysis of total FA ME fractions without prior separation is not enough: The reliability depends on the accuracy of the peak assignments in mass spectra as well as the results for the distribution of the carbon-chain length and the relative peak intensities. In this study, we used Mycobacterium tuberculosis H37Rv as a model strain because it is evident that this strain has three types of major MA subclasses and they have been extensively analyzed by using several approaches in other studies.24,29) The accuracy of the peak assignments were investigated by comparing observed mass and calculated mass for these three types of major MA subclasses. In addition, we also compared the distribution of the carbon-chain length and the relative peak intensities for those without TLC and those with TLC. Furthermore, we constructed contour maps of normalized relative peak intensities for whole MAs from M. tuberculosis, Mycobacterium bovis, Mycobacterium kyorinense, and Mycobacterium smegmatis to visualize the features of the whole MAs.
EXPERIMENTAL
Bacterial strains
M. tuberculosis H37Rv (ATCC 27294T) and M. smegmatis mc2155 (ATCC 700084) were purchased from the American Type Culture Collection (ATC C, Manassas, VA, USA). M. bovis BCG Tokyo 172 was supplied by Dr. Saburo Yamamoto of the Japan BCG Laboratory (Tokyo, Japan). M. kyorinense JCM 15038T was purchased from the Japan Collection of Microorganisms at Riken (JCM, Wako, Japan). M. tuberculosis was cultured at 37°C on 1% Ogawa media (Kyokuto Pharmaceutical Industrial Co., Ltd., Tokyo, Japan); the other strains were cultured at 37°C on 7H11 agar (Difco Laboratories, Detroit, MI) with 0.5% glycerol and 10% Middlebrook enrichment (Difco) containing oleic acid–albumin–dextrose–catalase (OADC). M. tuberculosis and M. bovis were grown for 2 to 3 weeks, and M. kyorinense and M. smegmatis were grown for 3 days.
Total FA ME fraction and TLC fractions
The heat-killed bacteria were treated with 10% KOH in methanol for 2 h at 90°C. Total FA fractions containing MAs were extracted with n-hexane. Total FA ME fractions were obtained by treatment with diazomethane. Whole MA MEs were analyzed by TLC on Silicagel G (Uniplate; 20×20 cm, 250 μm; Analtech, Inc., Newark, DE), which was developed with n-hexane–diethyl ether (90 : 15, v/v; three runs) and visualized by spraying with 20% sulfuric acid in ethanol and heating at 180°C for 3 min. TLC fractions were obtained until a single spot was obtained.11)
MALDI spiral-TOFMS
Mass analysis of the samples was performed with MALDI spiral-TOFMS (JMS-S3000, JEOL, Tokyo, Japan) with a flight length of ca. 17 m and eight cycles of the spiral trajectory to achieve high mass-resolution.28) Ions generated by irradiation with a 349-nm Nd:YLF laser were accelerated at 20 kV. The settings for delay time and grid voltage were optimized to stay constant at ∆M<ca. 0.03 Da at full width at half maximum (FWHM) over the range of m/z 800 to 1500. Sample preparation for mass spectrometry has been described previously.27) The matrix was 10 mg/mL 2,5-dihydroxybenzoic acid in tetrahydrofuran (THF). A sodium iodide solution of 1 mg/mL in THF was used as a cationization agent. MALDI mass spectra were observed in positive spiral mode by averaging 1500 individual laser shots. Three or five mass spectra for each sample were collected.
Mass calibration was performed as follows. Provisional peak assignment was performed by internal calibration using a poly (methyl methacrylate) (PMMA) standard (peak-top molecular weight, Mp=1310) purchased from Polymer Laboratories (Church Stretton, UK). Self calibration was further performed by using major peaks assigned to MAs as internal reference peaks. MAs were assigned on the basis of the errors within 5 ppm between the calculated masses as [M+Na]+ ions and the observed masses in the MALDI mass spectra. Data were processed by using monoisotopic peaks. Mass spectral data were processed with Polymerix software (Sierra Analytics, Modesto, CA, USA).
Contour maps
Contour maps of relative peak intensities corresponding to MAs were constructed as functions of carbon-chain length (x-axis) and MA subclass (y-axis). The relative peak intensities were normalized so that the strongest peak intensity of each sample was 100%. Contour maps were constructed by using relative peak intensities of more than 1%.
RESULTS AND DISCUSSION
Analysis of MA subclasses in TLC fractions
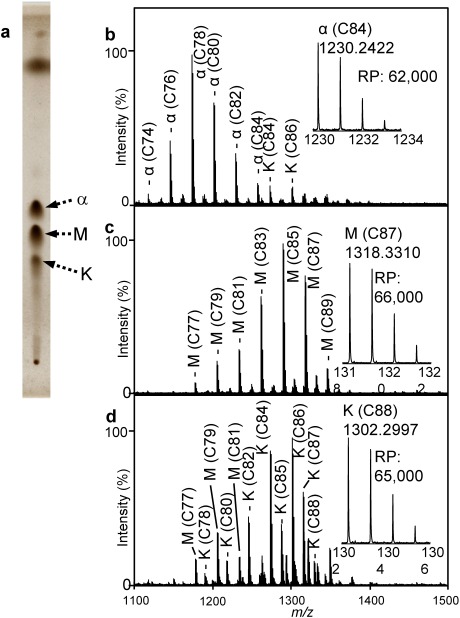
MA subclasses in the TLC fractions were analyzed by using MALDI spiral-TOFMS. Three MA subclasses (α-, methoxy-, and keto-MA) were detected in the total FA ME fraction of M. tuberculosis on TLC (Fig. 1a). MA subclass fractions were analyzed by using MALDI spiral-TOFMS. Major peaks were assigned as α-MA (C76–C84, Fig. 1b), methoxy-MA (C77–C89, Fig. 1c), and keto-MA (C78–C88, Fig. 1d). The most abundant molecular species were C78, α-MA; C85, methoxy-MA; and C86, keto-MA. Here, methoxy-MA components in Fig. 1c are named M1. In addition, as shown in Fig. 1d, minor peaks corresponding to methoxy-MAs (C77–C81) were observed in the keto-MA fraction. Here, these methoxy-MA components are named M2. This was presumably due to the presence of structural isomers that had the same molecular formula and different retention factor (Rf) values on TLC. These results suggest that complete separation of MA into subclasses by TLC is not easy.
Fig. 1. MA subclasses in M. tuberculosis.
TLC of MA ME (a). MALDI mass spectra of α-MA ME fraction (b), methoxy-MA ME fraction (c), and keto-MA ME fraction (d). RP indicates mass resolving power (FWHM). α, M, and K indicate α-, methoxy-, and keto-MAs, respectively.
Assignment of MALDI mass spectral peaks observed in the total FA ME fraction of M. tuberculosis
To assess the peak assignment of MAs observed in the mass spectra of the total FA ME fraction, we compared the observed and calculated masses of MAs. Table 1 summarizes the assigned major MAs for the total FA ME fraction, together with the calculated and observed masses before the self calibration with standard deviations (S.D.) and the errors in Da and in ppm. The mass accuracy of the major MAs was greater than 3 ppm before self calibration.
Table 1. Major MAs assigned to the total FA ME fraction from M. tuberculosis.
| MA subclass | CCL | m/z Values | Errora | |||
|---|---|---|---|---|---|---|
| Calculated [M+Na]+ | Observeda [M+Na]+ | ±S.D. | in Da | in ppm | ||
| α-MA | ||||||
| 76 | 1146.148 | 1146.149 | 0.002 | −0.001 | −0.5 | |
| 78 | 1174.180 | 1174.180 | 0.001 | 0.000 | 0 | |
| 80 | 1202.211 | 1202.211 | 0.001 | 0.000 | 0 | |
| 82 | 1230.242 | 1230.243 | 0.001 | −0.001 | −0.7 | |
| 84 | 1258.273 | 1258.273 | 0.003 | 0.000 | 0 | |
| Methoxy-MA | ||||||
| 79 | 1206.206 | 1206.206 | 0.001 | 0.000 | −0.2 | |
| 81 | 1234.237 | 1234.238 | 0.001 | −0.001 | −0.5 | |
| 83 | 1262.268 | 1262.269 | 0.001 | −0.001 | −0.5 | |
| 85 | 1290.300 | 1290.300 | 0.001 | 0.000 | 0.2 | |
| 87 | 1318.331 | 1318.330 | 0.001 | 0.001 | 0.5 | |
| 89 | 1346.362 | 1346.362 | 0.002 | 0.000 | 0.1 | |
| Keto-MA | ||||||
| 80 | 1218.206 | 1218.208 | 0.001 | −0.002 | −2.0 | |
| 82 | 1246.237 | 1246.238 | 0.001 | −0.001 | −1.0 | |
| 84 | 1274.268 | 1274.269 | 0.000 | −0.001 | −0.9 | |
| 85 | 1288.284 | 1288.287 | 0.000 | −0.003 | −2.5 | |
| 86 | 1302.300 | 1302.301 | 0.001 | −0.001 | −0.6 | |
| 87 | 1316.315 | 1316.318 | 0.001 | −0.003 | −2.0 | |
| 88 | 1330.331 | 1330.333 | 0.002 | −0.002 | −1.5 | |
a: Average values from five mass spectra. The observed masses were obtained by internal calibration using PMMA as a standard reagent (Fig. S1). (CCL=carbon-chain length; S.D.=standard deviation.)
High mass-resolution and mass-accuracy are not required when MAs are purified into each subclass by TLC. However, as discussed previously,27) they are required when the MAs are analyzed by using the total FA ME fraction, because the calculated masses for α′-/keto-MA or methoxy-/dicarboxy-MA differ by only 0.036 Da. The mass accuracy in this study (error<±0.003) seemed to be enough to achieve correct peak assignments.
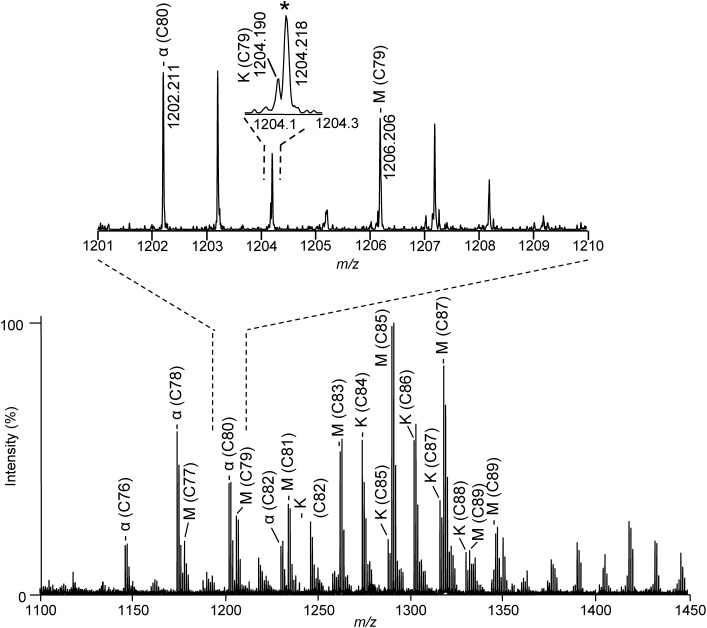
The mass spectrum of the total FA ME fraction from M. tuberculosis is shown in Fig. 2. In the total FA ME fraction, α-MAs were observed in the range of m/z 1100–1250; methoxy- and keto-MAs were observed in the range of m/z 1150–1350. Importantly, we detected a minor peak corresponding to keto-MA with C79 at m/z 1204.190 adjacent to the major peak for an isotope of α-MAs with C80 at m/z 1204.218 (see the enlarged spectrum in Fig. 2).
Fig. 2. MALDI mass spectrum of the total FA ME fraction from M. tuberculosis.
α, M, and K indicate α-, methoxy-, and keto-MAs, respectively. Asterisk indicates the isotope peak corresponding to α-MA with C80.
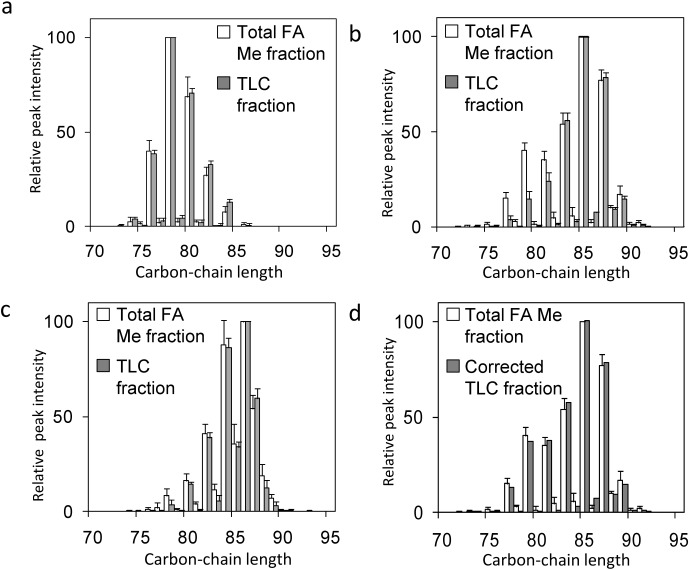
The distribution of carbon-chain lengths and the distribution of relative peak intensities for each MA subclass observed by analysis of the total FA ME fraction were compared with those of the TLC fractions (Figs. 3a–c). The distribution of carbon-chain lengths in each MA subclass was identical between the TLC fractions and the total FA ME fraction, as follows: α-MA, C74–C84; methoxy-MA, C77–C89; and keto-MA, C79–C89. The distributions of relative peak intensities in α-MA (Fig. 3a) and keto-MA (Fig. 3c) were identical between those for the TLC fractions and those for the total FA ME fraction. On the other hand, the distribution of relative peak intensities in methoxy-MA was not identical between those for the TLC fraction and those for the total FA ME fraction (Fig. 3b).
Fig. 3. Comparisons of relative peak intensities of MA subclasses from M. tuberculosis.
α-MA (a), methoxy-MA (b), keto-MA (c), and corrected methoxy MA (d). Peak intensities were normalized such that the maximum peak intensity was 100%.
The discrepancies of the peak intensities in Fig. 3b were explained as follows. As shown in Fig. 1d, some methoxy-MAs were detected in the keto-MA fraction (M2 component). This means that only M1 component existed in methoxy-MA fraction in Fig. 3b, while sum of M1 and M2 components were shown in total FA ME fraction. Actually, there were two plateaus of intensity, at about C79 and C85 (Fig. 3b, white) for methoxy-MA in the total FA ME fraction.
The original peak intensity distribution for TLC fraction was also speculated as follows. The ratio of maximum peak intensity for M1 and that for keto-MA is evaluated from total FA Me fraction data in Table S1, which is 100 : 66.4. The ratio of maximum peak intensity for M2 and that of keto-MA is evaluated from TLC fraction data in Table S2, which is 33.6 : 100. From these ratios, peak intensity ratio of M1 and M2 is speculated to be 100 : 22.3. The M2 component detected in keto-MA fraction is added to M1 component so as the ratio to be 100 : 22.3 (Fig. 3d corrected TLC fraction), and compared with the peak intensity distribution for total FA Me fraction. Figure 3d shows good agreement of these peak intensity distributions.
The distributions of the relative peak intensities showed the same tendency in each MA subclass. In addition, the ratios of even- and odd-numbered carbon-chain MAs obtained by analysis of the total FA ME fraction were in good agreement with those obtained for the TLC fractions. Thus MALDI spiral-TOFMS characterization of the total FA ME fraction is accurate and reliable.
Characterization of MAs for three species of Mycobacterium by using total FA ME fractions
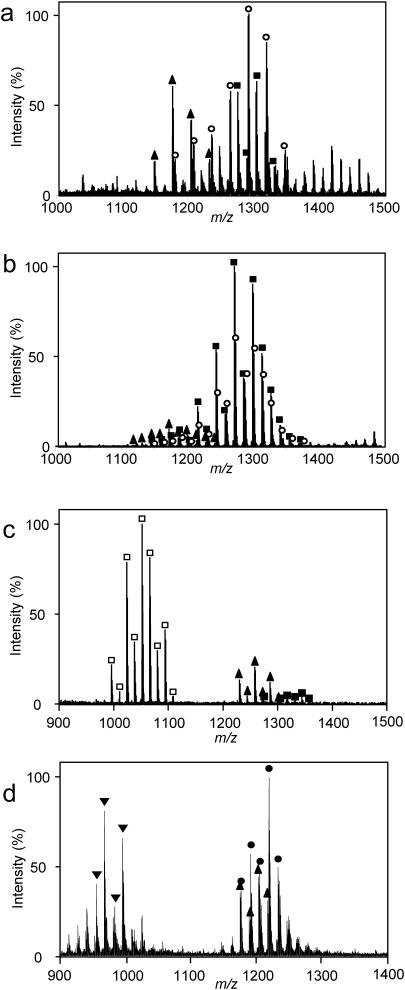
We characterized whole MAs of slow-growing M. tuberculosis, M. bovis, and M. kyorinense and of fast-growing M. smegmatis by analyzing the total FA ME fractions. Figure 4 showed MALDI mass spectra of four species of Mycobacterium, whose characteristic diversities in MA subclasses were determined in other studies. Comparing of the MALDI mass spectrum of M. tuberculosis and M. bovis, the detected MA subclasses were the same, but the relative peak intensities for α- and methoxy-MAs for M. tuberculosis were stronger than those of M. bovis (Figs. 3a and b). In the MALDI mass spectrum of M. bovis, weak peaks corresponding to α-MAs were observed in the range of m/z 1100–1250. Peaks corresponding to methoxy- and keto-MAs were clearly observed in the range of m/z 1150–1400 (Fig. 4b). The distributions of carbon-chain lengths were α-MAs, C76–C80; methoxy-MAs, C81–C89; and keto-MAs, C77–C90. In the total FA ME fraction of M. kyorinense, peaks with high intensities corresponding to dicarboxy-MA were observed in the range of m/z 1000–1100, and weak peaks corresponding to α- and keto-MAs were observed in the range of m/z 1200–1400 (Fig. 4c). The distributions of carbon-chain lengths were α-MAs, C82–C86; keto-MAs, C82–C90 with weak peaks; and dicarboxy-MAs, C62–C70. Notably, peaks that we assigned as dicarboxy-MAs corresponding to wax-MAs lost their long carbon-chain secondary alcohols (C18–C22) in the process of methyl esterification.30,31) The MALDI mass spectrum of the total FA ME fraction from M. smegmatis displayed a typical pattern of rapid growth that produced both even- and odd-numbered homologs; peaks corresponding to α′-MAs were observed in the range of m/z 900–1000, and peaks corresponding to α- and epoxy-MAs were observed in the range of m/z 1100–1300 (Fig. 4d). The distributions of carbon-chain lengths were α′-MAs, C59–C65; α-MAs, C75–C81; and epoxy-MAs, C74–C83. These results for the major MA subclasses agreed with the results obtained by using TLC and MALDI-TOFMS.25,26,32)
Fig. 4. MALDI mass spectra of total FA fractions of M. tuberculosis (a), M. bovis (b), M. kyorinense (c), and M. smegmatis (d).
Closed triangles (▲), inverted closed triangles (▼), open circles (○), closed squares (■), open squares (□), and closed circles (●) indicate α-, α′-, methoxy-, keto-, dicarboxy-, and epoxy-MAs, respectively.
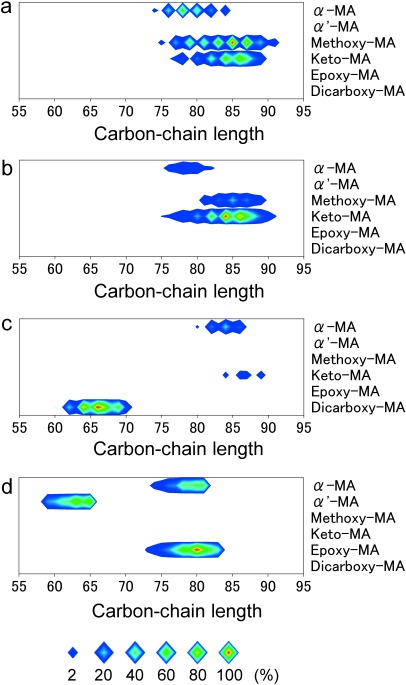
A wealth of information on MALDI mass spectra exists; however, valuable information corresponding to all the MAs in each sample was, until now, unable to be viewed at a glance. We made contour maps for the normalized relative peak intensities of all MAs so as to visualize the MA subclasses and their distribution of carbon-chain lengths (Figs. 5a–d, Tables S1–4). In the figures, the size and color of each rhomboid dot indicate the peak intensity.
Fig. 5. Contour maps of relative peak intensities for the whole MA contents of M. tuberculosis (a), M. bovis (b), M. kyorinense (c), and M. smegmatis (d).
Data are average peak intensities (a: n=5; b, c, and d: n=3). Size and color of each dot indicate peak intensity.
The existence of MA subclasses in the total FA ME fraction, the distribution of carbon-chain lengths for each MA subclass, and their peak intensities were visualized by the contour maps at a glance. The ratios of even- or odd-numbered MAs were also visualized. For example, the strongest observed peaks corresponding to whole MAs for each sample strain were methoxy-MA with C85 for M. tuberculosis; keto-MA with C84 for M. bovis; dicarboxy-MA with C66 for M. kyorinense; and epoxy-MA with C80 for M. smegmatis. Our contour maps of relative peak intensity for whole MAs enabled us to successfully visualize important information such as the variation of MA subclasses, the distribution of carbon-chain lengths for each MA subclass, and the distribution of relative peak intensities, including the ratio of odd-numbered and even-numbered MAs for each subclass. Although the relative peak intensities of each MA subclass are not necessarily indicative of composition, the contour maps are useful for practical visualization of whole MAs.
In this study, four strains of Mycobacterium which have typical types of MAs were analyzed. This number is too small to show the ability of identifying mycobacterium species in the clinical laboratory. In addition, only cultured colonies of Mycobacterium species were analyzed, suggesting this study is insufficient to use it at clinical laboratory, since some of mycobacterium species are known as a slow grower. However, several methods which uses sputum or liquid culture media have already been attempt to identify mycobacterium species in clinical laboratory. Future studies using MALDI spiral-TOFMS for total FA ME fraction for these samples are needed.
CONCLUSION
Analysis of the MAs in the total FA ME fraction by using MALDI spiral-TOFMS yielded details of the distributions of carbon-chain lengths, the ratio of even- and odd-numbered MAs, and the relative peak intensities for each MA subclass. The contour maps of relative peak intensities enabled clear visualization of the complicated whole MA. The high resolution and high accuracy of MALDI spiral-TOF enabled us to easily and accurately characterize whole MAs in short time. We consider that this high-throughput method is valuable for revealing the diversity of MAs at the genus or intraspecific level and for analyzing the culture condition–dependent composition of MAs. This method should be a valuable tool for chemotaxonomy of the suborder Corynebacterineae.
SUPPORTING INFORMATION
Original data on internal calibration and the MALDI mass spectra of the three species of the genus Mycobacterium.
Acknowledgments
This study was supported in part by a research grant from the Institute for Fermentation, Osaka (IFO), Japan.
Mass Spectrom (Tokyo) 2015; 4(1): A0035
- CCL
carbon-chain length
- MA
mycolic acid
- ME
methyl ester
- PMMA
poly (methyl methacrylate)
- THF
tetrahydrofuran
SUPPORTING INFORMATION
References
- 1) K. Kaneda, S. Imaizumi, I. Yano. Distribution of C22-, C24- and C26-alpha-unit-containing mycolic acid homologues in mycobacteria. Microbiol. Immunol. 39: 563–570, 1995. [DOI] [PubMed] [Google Scholar]
- 2) M. Watanabe, Y. Aoyagi, M. Ridell, D. E. Minnikin. Separation and characterization of individual mycolic acids in representative mycobacteria. Microbiology 147: 1825–1837, 2001. [DOI] [PubMed] [Google Scholar]
- 3) D. E. Minnikin, S. M. Minnikin, G. Dobson, M. Goodfellow, F. Portaels, L. Vandenbreen, D. Sesardic. Mycolic acid patterns of four vaccine strains of Mycobacterium bovis BCG. J. Gen. Microbiol. 129: 889–891, 1983. [DOI] [PubMed] [Google Scholar]
- 4) M. Daffe, M. A. Laneelle, C. Asselineau, V. Levyfrebault, H. David. Taxonomic interest of mycobacterial fatty-acids: Proposal of a method for analysis. Ann. Microbiol. 134B: 241–256, 1983. [PubMed] [Google Scholar]
- 5) C. E. Barry 3rd, R. E. Lee, K. Mdluli, A. E. Sampson, B. G. Schroeder, R. A. Slayden, Y. Yuan. Mycolic acids: Structure, biosynthesis and physiological functions. Prog. Lipid Res. 37: 143–179, 1998. [DOI] [PubMed] [Google Scholar]
- 6) M. Watanabe, Y. Aoyagi, H. Mitome, T. Fujita, H. Naoki, M. Ridell, D. E. Minnikin. Location of functional groups in mycobacterial meromycolate chains: The recognition of new structural principles in mycolic acids. Microbiology 148: 1881–1902, 2002. [DOI] [PubMed] [Google Scholar]
- 7) M. Goodfellow, D. E. Minnikin, C. Todd, G. Alderson, S. M. Minnikin, M. D. Collins. Numerical and chemical classification of Nocardia amarae. J. Gen. Microbiol. 128: 1283–1297, 1982. [DOI] [PubMed] [Google Scholar]
- 8) M. D. Collins, M. Goodfellow, D. E. Minnikin. A survey of the structures of mycolic acids in Corynebacterium and related taxa. J. Gen. Microbiol. 128: 129–149, 1982. [DOI] [PubMed] [Google Scholar]
- 9) R. C. Hartkoorn, C. Sala, J. Neres, F. Pojer, S. Magnet, R. Mukherjee, S. Uplekar, S. Boy-Roettger, K.-H. Altmann, S. T. Cole. Towards a new tuberculosis drug: Pyridomycin—nature’s isoniazid. EMBO Mol. Med. 4: 1032–1042, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10) M. T. Gler, V. Skripconoka, E. Sanchez-Garavito, H. Xiao, J. L. Cabrera-Rivero, D. E. Vargas-Vasquez, M. Gao, M. Awad, S.-K. Park, T. S. Shim, G. Y. Suh, M. Danilovits, H. Ogata, A. Kurve, J. Chang, K. Suzuki, T. Tupasi, W.-J. Koh, B. Seaworth, L. J. Geiter, C. D. Wells. Delamanid for multidrug-resistant pulmonary tuberculosis. N. Engl. J. Med. 366: 2151–2160, 2012. [DOI] [PubMed] [Google Scholar]
- 11) A. Bhatt, V. Molle, G. S. Besra, W. R. Jacobs Jr., L. Kremer. The Mycobacterium tuberculosis FAS-II condensing enzymes: Their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol. Microbiol. 64: 1442–1454, 2007. [DOI] [PubMed] [Google Scholar]
- 12) L. Y. Gao, F. Laval, E. H. Lawson, R. K. Groger, A. Woodruff, J. H. Morisaki, J. S. Cox, M. Daffe, E. J. Brown. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: Implications for therapy. Mol. Microbiol. 49: 1547–1563, 2003. [DOI] [PubMed] [Google Scholar]
- 13) D. Barkan, Z. Liu, J. C. Sacchettini, M. S. Glickman. Mycolic acid cyclopropanation is essential for viability, drug resistance, and cell wall integrity of Mycobacterium tuberculosis. Chem. Biol. 16: 499–509, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14) M. S. Glickman. The mmaA2 gene of Mycobacterium tuberculosis encodes the distal cycloproplane synthase of the alpha-mycolic acid. J. Biol. Chem. 278: 7844–7849, 2003. [DOI] [PubMed] [Google Scholar]
- 15) D. Sambandan, D. N. Dao, B. C. Weinrick, C. Vilcheze, S. S. Gurcha, A. Ojha, L. Kremer, G. S. Besra, G. F. Hatfull, W. R. Jacobs Jr. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. mBio 4: e00222-13, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16) A. Bhatt, N. Fujiwara, K. Bhatt, S. S. Gurcha, L. Kremer, B. Chen, J. Chan, S. A. Porcelli, K. Kobayashi, G. S. Besra, W. R. Jacobs Jr. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 104: 5157–5162, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17) W. R. Butler, D. G. Ahearn, J. O. Kilburn. High-performance liquid chromatography of mycolic acids as a tool in the identification of Corynebacterium, Nocardia, Rhodococcus, and Mycobacterium species. J. Clin. Microbiol. 23: 182–185, 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18) W. R. Butler, L. S. Guthertz. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin. Microbiol. Rev. 14: 704–726, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19) J. A. Kellogg, D. A. Bankert, G. S. Withers, W. Sweimler, T. E. Kiehn, G. E. Pfyffer. Application of the Sherlock Mycobacteria Identification System using high-performance liquid chromatography in a clinical laboratory. J. Clin. Microbiol. 39: 964–970, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20) G. O. Guerrant, M. A. Lambert, C. W. Moss. Gas-chromatographic analysis of mycolic acid cleavage products in mycobacteria. J. Clin. Microbiol. 13: 899–907, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21) S. H. Song, K. U. Park, J. H. Lee, E. C. Kim, J. Q. Kim, J. Song. Electrospray ionization-tandem mass spectrometry analysis of the mycolic acid profiles for the identification of common clinical isolates of mycobacterial species. J. Microbiol. Methods 77: 165–177, 2009. [DOI] [PubMed] [Google Scholar]
- 22) G. Shui, A. K. Bendt, I. A. Jappar, H. M. Lim, M. Laneelle, M. Herve, L. E. Via, G. H. Chua, M. W. Bratschi, S. Z. Z. Rahim, A. L. T. Michelle, S.-H. Hwang, J.-S. Lee, S.-Y. Eum, H.-K. Kwak, M. Daffe, V. Dartois, G. Michel, C. E. Barry III, M. R. Wenk. Mycolic acids as diagnostic markers for tuberculosis case detection in humans and drug efficacy in mice. EMBO Mol. Med. 4: 27–37, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23) G. Shui, A. K. Bendt, K. Pethe, T. Dick, M. R. Wenk. Sensitive profiling of chemically diverse bioactive lipids. J. Lipid Res. 48: 1976–1984, 2007. [DOI] [PubMed] [Google Scholar]
- 24) F. Laval, M. A. Laneelle, C. Deon, B. Monsarrat, M. Daffe. Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal. Chem. 73: 4537–4544, 2001. [DOI] [PubMed] [Google Scholar]
- 25) T. Naka, S. Maeda, M. Niki, N. Ohara, S. Yamamoto, I. Yano, J. Maeyama, H. Ogura, K. Kobayashi, N. Fujiwara. Lipid phenotype of two distinct subpopulations of Mycobacterium bovis bacillus calmette-guerin Tokyo 172 substrain. J. Biol. Chem. 286: 44153–44161, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26) M. Okazaki, K. Ohkusu, H. Hata, H. Ohnishi, K. Sugahara, C. Kawamura, N. Fujiwara, S. Matsumoto, Y. Nishiuchi, K. Toyoda, H. Saito, S. Yonetani, Y. Fukugawa, M. Yamamoto, H. Wada, A. Sejimo, A. Ebina, H. Goto, T. Ezaki, T. Watanabe. Mycobacterium kyorinense sp. nov., a novel, slow-growing species, related to Mycobacterium celatum, isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 59: 1336–1341, 2009. [DOI] [PubMed] [Google Scholar]
- 27) K. Teramoto, T. Tamura, S. Hanada, T. Sato, H. Kawasaki, K. Suzuki, H. Sato. Simple and rapid characterization of mycolic acids from Dietzia strains by using MALDI spiral-TOFMS with ultra high mass-resolving power. J. Antibiot. 66: 713–717, 2013. [DOI] [PubMed] [Google Scholar]
- 28) T. Satoh, T. Sato, J. Tamura. Development of a high-performance MALDI-TOF mass spectrometer utilizing a spiral ion trajectory. J. Am. Soc. Mass Spectrom. 18: 1318–1323, 2007. [DOI] [PubMed] [Google Scholar]
- 29) Y. Fujita, T. Naka, T. Doi, I. Yano. Direct molecular mass determination of trehalose monomycolate from 11 species of mycobacteria by MALDI-TOF mass spectrometry. Microbiology 151: 1443–1452, 2005. [DOI] [PubMed] [Google Scholar]
- 30) M. Watanabe, A. Ohta, S. Sasaki, D. E. Minnikin. Structure of a new glycolipid from the Mycobacterium avium–Mycobacterium intracellulare complex. J. Bacteriol. 181: 2293–2297, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31) J. Astola, M. Munoz, M. Sempere, P. Coll, M. Luquin, P. L. Valero-Guillen. The HPLC-double-cluster pattern of some Mycobacterium gordonae strains is due to their dicarboxy-mycolate content. Microbiology-Sgm 148: 3119–3127, 2002. [DOI] [PubMed] [Google Scholar]
- 32) N. Fujiwara, T. Naka, M. Ogawa, R. Yamamoto, H. Ogura, H. Taniguchi. Characteristics of Mycobacterium smegmatis J15cs strain lipids. Tuberculosis (Edinb.) 92: 187–192, 2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.