Abstract
Mass spectrometric proteomics is an effective approach for identifying and quantifying histone post-translational modifications (PTMs) and their binding proteins, especially in the cases of methylation and acetylation. However, another vital PTM, phosphorylation, tends to be poorly quantified because it is easily lost and inefficiently ionized. In addition, PTM binding proteins for phosphorylation are sometimes resistant to identification because of their variable binding affinities. Here, we present our efforts to improve the sensitivity of detection of histone H4 tail peptide phosphorylated at serine 1 (H4S1ph) and our successful identification of an H4S1ph binder candidate by means of a chemical proteomics approach.
Our nanoLC-MS/MS system permitted semi-quantitative label-free analysis of histone H4 PTM dynamics of cell cycle-synchronized HeLa S3 cells, including phosphorylation, methylation, and acetylation. We show that H4S1ph abundance on nascent histone H4 unmethylated at lysine 20 (H4K20me0) peaks from late S-phase to M-phase. We also attempted to characterize effects of phosphorylation at H4S1 on protein–protein interactions. Specially synthesized photoaffinity bait peptides specifically captured 14-3-3 proteins as novel H4S1ph binding partners, whose interaction was otherwise undetectable by conventional peptide pull-down experiments.
This is the first report that analyzes dynamics of PTM pattern on the whole histone H4 tail during cell cycle and enables the identification of PTM binders with low affinities using high-resolution mass spectrometry and photo-affinity bait peptides.
Keywords: cell cycle, cross-linking, ETD, histone, epigenetics
INTRODUCTION
Histone post-translational modifications (PTMs) play vital roles in various cellular processes, and their distributions, combinations, and intermolecular interactions constitute the so-called ‘histone code.’ 1) To decipher the histone code, precise mapping and quantification of histone PTMs are essential, as is the determination of the consequences of the PTMs in the context of chromatin. A deeper understanding of such epigenetic principles will, in turn, promote the development of methods for beneficial intervention in the human epigenome.
Mass spectrometry is now the best option for studying PTMs. However, the robust identification and quantification of multiple PTMs on a protein remains challenging, and histones are among the most complicated target.2) The largest number of combinatorial PTM patterns on histones that has been detected was achieved by means of a combination of two-dimensional liquid chromatography (LC) and top-down proteomics in a thorough study.3) Sensitivity and throughput can be improved by shortening the target peptide sequences, but this sacrifices combinatorial information on PTMs. For histone H4, a middle-down analysis with digestion by AspN proteinase covers the whole N-terminal tail, which contains most of the rewritable histone code.4,5) As a further complication, the coincidence of multiple PTMs on a protein can also hamper quantification. So we improved quantitative accuracy by adopting effective means collectively; optimization of device materials in the LC-MS/MS system,6) careful LC maintenance and operation, thorough alignment of retention time between LC-MS datasets,7) and strict quality control of the whole experiments.
We have chosen the cell cycle as a model of dynamically changing biological process, because cellular PTM states are globally affected throughout the cycle. For example, in S-phase, histone H4K5/K12 acetylation is supposed to be elevated along with the synthesis of nascent histones.8) And it is well known that H4K20 monomethylation is accomplished in G2/M-phase.9)
PTMs cause switching of cellular signaling with the aid of interacting proteins.10,11) Our next challenge is to characterize the relationships between PTM states and any resulting cellular phenomena. To achieve a better understanding of the role of individual PTMs, we need to discover how a PTM affects the chromatin state, such as recruitment of PTM-binding proteins, change in electrical charge, or conformational change of histone tail. The pull-down proteomic approach has been quite successful in identifying high-affinity binders such as trimethyllysine readers12–14) owing to their low KD values (sometimes in the sub-micromolar range). In contrast, reader proteins for other PTMs, such as acetyllysine or phosphoserine/threonine, show quite variable binding affinities15) and their KD values sometimes exceed 102 μM. To overcome those limitations, which are inherent to conventional pull-down analysis as a result of variations in affinity, crosslink-assisted pull-down has recently emerged as a new technique.16–21) Unlike formaldehyde or amine crosslinkers, which are added extraneously to the lysate, crosslinkers incorporated directly into peptide baits can capture ‘true’ binding partner covalently.
Here, we present our workflow for analyzing the combinatorial PTM dynamics of histone H4, extracting significant PTM changes, and identifying the resulting protein–protein interactions.
First, we evaluated our label free semi-quantification method by examining the well-defined cell cycle-dependent PTM of the H4 tail. We showed that the technique was robust and that our equipment had advantages in detection of phosphorylation, which accounted for only 1% of all the H4 tail ion signals. Despite this low abundance, our label-free semi-quantification technique successfully characterized the PTM dynamics in the cell cycle. Next, we developed a highly sensitive and specific crosslinking-assisted pull-down method to capture weak PTM–protein interaction by using specially designed photoreactive peptide baits, and we identified a potential H4S1ph-interacting protein.
EXPERIMENTAL
Cell culture and cell cycle synchronization
Human epithelial cervix carcinoma cell line HeLa S3 cells obtained from the RIKEN BRC Cell Bank were grown in suspension in RPMI-1640 medium modified with HEPES and L-glutamine (Sigma-Aldrich, St. Louis, MO, USA) and supplemented with 10% fetal bovine serum (FBS), 2 mM GlutaMAX I (Gibco Life Technologies, Grand Island, NY, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cultures were maintained at a density of 6×105 to 8×105 cells/mL and synchronized by double thymidine blocking.22) Briefly, 2 mM thymidine (Sigma-Aldrich) was added to the media, which were kept for 20 h (1st block). This was followed by thymidine-free culture for 12 h, succeeded by a second treatment with 2 mM thymidine for 15 h. After the second portion of thymidine had been removed (G1/S release), the cells were collected sequentially at two-hour intervals. Each sample was checked for its synchrony and cell cycle profile (DNA content) by means of flow cytometry with propidium iodide staining. Samples for histone PTM analysis were collected by centrifugation and quickly washed with cold phosphate-buffered saline (PBS). The resulting cell pellets were immediately frozen in liquid N2 and stored at −80°C before histone extraction.
Extraction of histones
Histones were isolated by using the Histone Purification Mini Kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. Protein concentrations in the eluted samples were measured by means of the 660 nm Protein Assay (Pierce, Thermo Fisher Scientific, Rockford, IL, USA).
AspN endoproteinase digestion
Purified histone (2.5 μg) or bovine histone (Roche Applied Science, Mannheim, Germany) were denatured and reduced with 2 mM tris(2-carboxyethyl)phosphine (TCEP; Pierce) in 16% acetonitrile at 50°C for 15 min. The samples were then alkylated with 5 mM iodoacetamide (Sigma-Aldrich) at room temperature for one hour in darkness. The histones were then digested with 50 ng of AspN endoproteinase (Sequencing Grade; Roche) in 85 mM phosphate buffer (pH 8.0) at 30°C overnight. The resulting peptides were desalted with a C-tip (AMR Inc., Tokyo, Japan) and dissolved in 0.2% aqueous trifluoroacetic acid (TFA) containing 2% acetonitrile for subsequent liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Liquid chromatography-tandem mass spectrometry
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed by using an LTQ Orbitrap Elite ETD mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The micro-high-performance liquid chromatography/nanospray ionization MS/MS system consisted of a Paradigm dual-solvent-delivery system (Michrom BioResources, Inc., Auburn, CA, USA) for HPLC, a HTS PAL auto sampler (CTC Analytics, Zwingen, Switzerland), and a LTQ Orbitrap Elite ETD mass spectrometer (Thermo Fisher Scientific) equipped with a nanospray ionization (NSI) source (AMR). All devices were connected by nanoscale fused-silica capillaries (nanoViper; Thermo Fisher Scientific). Digested histones were automatically injected from a high-durability syringe (AMR) into a micro-precolumn C18 PepMap 100 Peptide Trap cartridge (5×0.3 mm i.d.; Thermo Fisher Scientific) on a Cheminert PAEK injector valve (CTC) for concentration and desalting. After desalting with 0.2% aqueous TFA containing 2% acetonitrile, the sample was loaded into a reversed-phase L-column2 micro C18 column (3 μm, 200 Å, 150×0.2 mm i.d.; CERI, Tokyo, Japan) for separation. Mobile phase A was 2% aqueous acetonitrile and mobile phase B was 90% aqueous acetonitrile, both containing 0.1% formic acid. The gradient conditions in the chromatographic run were set up as follows: B 4% (0 min)→10% (40 min)→28% (100 min)→95% (102 min)→95% (110 min)→4% (112–120 min). Eluent from the HPLC at a flow rate of 1.0–1.2 μL/min was introduced into the mass spectrometer by the NSI interface through an injector valve with the peptide trap cartridge and the column. An NSI needle (PicoTip FS360-50-30, New Objective Inc., Woburn, MA, USA) attached directly to the reversed-phase column was used as the NSI interface, and the voltage was 2.0 kV. The capillary was heated to 200°C. Assist nitrogen gas was used by the T3 Spray (AMR). The mass spectrometer was operated in a data-dependent acquisition mode in which MS acquisition in the mass range m/z 430–1000 was automatically switched to MS/MS acquisition under automated control of Xcalibur software (Thermo Fisher Scientific). MS scans were selected by the Orbitrap, with a resolution R=240,000, and those in the subsequent MS/MS scans were selected by an ion trap operated in automated gain control (AGC) mode, where the AGC values were 1×106 and 1×104 for full MS and MS/MS, respectively. For fragmentation, electron transfer dissociation (ETD) and collision induced dissociation were used. For ETD, the four most abundant ions of the full MS scan was selected as precursor ions and subjected to MS/MS scan with an isolation width of m/z 3.0; the reaction-time parameter was set at 100 ms. Dynamic exclusion was enabled with a repeat count of two over a duration of 10 s, an exclusion window of 25 s, and an exclusion mass width of 5 ppm. For CID, normalized collision energy parameter was set at 35. Other parameters were set same as ETD.
Label-free semi-quantification
Peptides were detected and quantified by using Progenesis QI software (version 2.0; Nonlinear Dynamics Ltd., Newcastle-upon-Tyne, UK). Retention time alignment was manually fine-tuned after the autoalignment. We then performed peak detection to select +2 to +6 ions as peptide peaks. A merged peak list generated by Progenesis QI was searched against the MASCOT database search (version 2.5.1; Matrix Science Ltd., London, UK). Data were analyzed using JMP Pro 10 software (version 10.0.2; SAS Institute Inc., North California, USA).
Database searching
Tandem mass (MS/MS) spectra were extracted by Proteome Discoverer version 1.3 (Thermo Fisher Scientific). All MS/MS datasets were analyzed by using MASCOT software (version 2.5.1; Matrix Science Ltd., London, UK). MASCOT was set up to search Swissprot_2014_08.fasta (selected for Homo sapiens; 20,194 entries or other mammalia; 13,077 for Bovine histones) with a fragment-ion mass tolerance of 0.80 Da and a parent ion tolerance of 5 ppm. The maximum number of missed cleavage sites was set to 1. Carbamidomethylation (57.021464) of cysteine was specified as a fixed modification. Citrullination (0.984016) of arginine, oxidation (15.994915) of methionine, methylation (14.01565) of lysine and arginine, dimethylation (28.0313) of lysine and arginine, trimethylation (42.04695) of lysine, acetylation (42.010565) of lysine and the peptide N-terminus, and phosphorylation (79.966331) of serine, threonine, and tyrosine were specified as variable modifications. Criteria for peptides identification were MASCOT significance threshold expected score<0.05.
Bait peptide synthesis
Standard Fmoc amino-acids were purchased from Novabiochem (Merck KGaA, Darmstadt Germany), including the amino acids of which the side chains were protected such as Arg(Pbf), Asp(OtBu), Lys(Boc), Ser(tBu), and Tyr(tBu). Fmoc-4-azide-L-phenylalanine {Fmoc-Phe(4-N3)-OH}, Fmoc-O-benzyl-L-phosphoserine [Fmoc-Ser{PO(OBzl)OH}-OH], and Fmoc-4-benzoyl-L-phenylalanine (Fmoc-Bpa-OH) were purchased from Watanabe Chemical Industries (Hiroshima, Japan). 3-(3-Methyl-3H-diaziren-3-yl)propanoic acid (4,4-azopentanoic acid) was chemically synthesized as previously reported.23) Peptides were synthesized by a standard Fmoc solid-phase peptide synthesis method in an automated peptide synthesizer (ResPep SL; intavis AG, Cologne, Germany) on a scale of 10 mmol of TentaGel rink-amide resin (0.23 mmol/g loading: intavis). Briefly, the coupling reaction was completed by activation of the amino acids with (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyBOP) and N-methylmorpholine (NMM). Fmoc removal was carried out by treatment with 20% piperidine/N,N-dimethylformamide. Phosphoserine and 4,4-azopentanoic acid were manually reacted with the resin by double coupling with (1H-benzotriazol-1-yloxy)(dimethylamino)-N,N-dimethylmethaniminium hexafluorophosphate (HBTU), 1-hydroxybenzotriazole (HOBt), and N,N-diisopropylethylamine (DIEA) for 30 min. Cleavage from the resin was performed by treatment with 95% TFA, 2.5% water, and 2.5% triisopropylsilane (TIPS) for six hours on ice. The crude peptides were obtained by precipitation with cold diethyl ether and centrifugation. The supernatant was decanted and the pellet was washed twice with cold diethyl ether. The crude peptides were dissolved in aqueous acetonitrile containing 0.1% TFA then purified by reverse-phase HPLC (RP-HPLC) on a C18 column (AR-300; Nacalai Tesque, Kyoto, Japan) with a 30-min linear gradient of acetonitrile/water containing 0.1% TFA. Peptide identity was confirmed by nanoLC-MS: m/z 655.82 [M+4H]4+, calculated m/z 655.82 [M+4H]4+ for dzH4S1; m/z 675.81 [M+4H]4+, calculated m/z 675.81 [M+4H]4+ for dzH4S1ph; m/z 460.25 [M+4H]4+, calculated m/z 460.25 [M+4H]4+ for H4S1-Bpa; m/z 480.25 [M+4H]4+, calculated m/z 480.25 [M+4H]4+ for H4S1ph-Bpa. All masses are monoisotopic.
Biotinylation of peptide probes by click chemistry cycloaddition
Photoaffinity peptides with C-terminal azide groups were treated with equimolar amounts of dibenzylcyclooctyne–PEG12–biotin conjugate (Jena Bioscience, Jena, Germany) to form C-terminally biotinylated baits for subsequent affinity purification. The cycloaddition reaction mixtures were incubated overnight at 37°C. The identity of the reaction products was confirmed by HPLC and MALDI-TOF MS: m/z 3695.76 [M−N2+H]+, calculated m/z 3696.08 [M−N2+H]+ for dzH4S1-PEG12-biotin; m/z 3775.52 [M−N2+H]+, calculated m/z 3776.06 [M−N2+H]+ for dzH4S1ph-PEG12-biotin; m/z 2941.45 [M+H]+, calculated m/z 2941.42 [M+H]+ for H4S1-Bpa-PEG12-biotin; m/z 3021.48 [M+H]+, calculated m/z 3021.40 [M+H]+ for H4S1ph-Bpa-PEG12-biotin. The calculated masses are average masses.
MALDI-TOF MS
MALDI-TOF MS was performed by using a MALDI-7090 mass spectrometer (Shimadzu, Kyoto, Japan). Synthetic Peptides were spotted onto the HCCA Pre-spotted μFocus MALDI plate 384 circles 900 μm (HST Inc., NJ) and air-dried. All analyses were carried out in the positive reflector mode. Mass spectra were externally calibrated using ProteoMass™ Peptide and Protein MALDI-MS Calibration Kit (Sigma-Aldrich).
Photocrosslinking and streptavidin affinity enrichment of biotinylated proteins
Probes were incubated for 15 min at 4°C with 2 mL of HeLa S3 whole-cell lysate (5 mg/mL) in ProNET LIVE! cell lysis buffer PBS (ESI Source Solutions, Woburn, MA, USA) supplied with Complete EDTA-free protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor cocktail (Roche). 14-3-3 binding inhibition experiments were conducted with the addition of R18 peptide (Sigma-Aldrich) at the concentration of 10 μM. The samples were then irradiated at 365 nm by using a Bio-Link BLX-E UV lamp (Vilber Lourmat, Marne La Vallée, France) for 10 min on ice. The irradiated lysate was incubated with Dynabeads streptavidin C1 (Invitrogen, Carlsbad, CA, USA) for 1.5 h at 4°C with gentle rotation. It was then washed sequentially with PBS containing 0.8% sodium dodecylsulfate (SDS), PBS containing 0.1% Tween 20 (PBST), 5 M NaCl containing 0.1% Tween 20, and finally with PBST again. The enriched proteins were eluted by boiling with 1× LDS sample buffer (Novex, San Diego, CA, USA) containing 50 mM dithiothreitol (DTT) at 96°C for 3 min.
Detection of crosslinked products, and sample preparation for protein identification
Each eluted protein sample was split into two; one portion was used for Western detection and the other was subjected to in-gel digestion followed by mass spectrometric analysis. The samples were separated on NuPAGE 10% Bis-Tris gel (Invitrogen) and transferred onto poly(vinylidene difluoride) (PVDF) membranes by using iBlot apparatus (Invitrogen). The membranes were blocked with Tris-buffered saline (TBS) containing 0.1% Tween 20 and 1% bovine serum albumin (BSA), then hybridized with streptavidin–poly(horseradish peroxidase) conjugate (SA-HRP; Pierce Protein Biology Products, Rockford, IL, USA) in Can Get Signal Immunoreaction Enhancer Solution 1 (Toyobo Corp., Osaka, Japan) for two hours. The proteins were then detected by using Luminata Forte Western HRP substrate (EMD Millipore, Bellerica, MA, USA) with a LAS-3000 imager (FUJIFILM, Tokyo, Japan). The same samples separated simultaneously in other wells on the gel were subsequently fixed in a 50% methanol–7% acetic acid solution. The gel was stained with SYPRO Ruby protein gel stain (Molecular Probes, Eugene, OR), and the gel regions with the corresponding molecular weight to the chemiluminescent bands observed on SA-HRP detection were excised and sliced into 1 mm3 pieces.
Mass spectrometric identification of in-gel digested proteins
The excised pieces were digested with trypsin according to a previous method.24) Digested sample were analyzed by LC-MS, and the MS/MS data were processed with the MASCOT database-search software as previously described.25,26)
RESULTS
Dynamics of H4 modification during the cell cycle
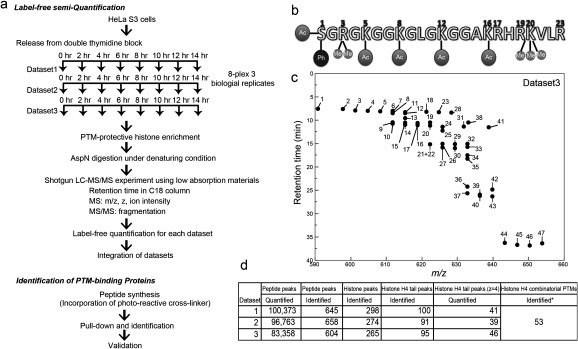
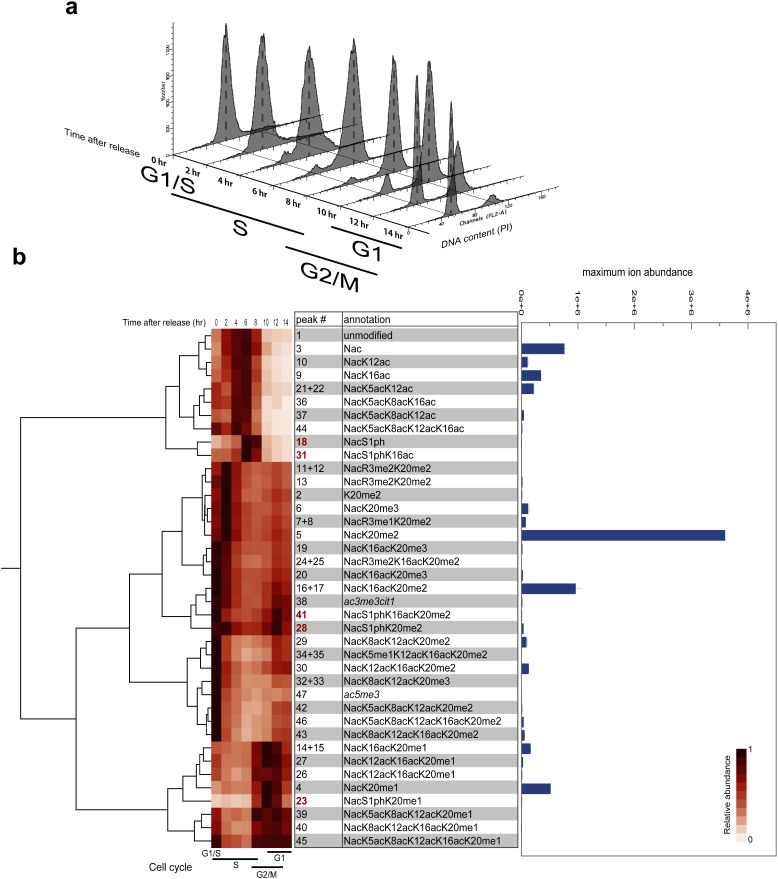
We synchronized the cell cycle in HeLa S3 cells and we examined the dynamics of modification changes (Fig. 1a). Cells were sampled at 8 time points of two-hour intervals from G1/S to G1. Cell synchronization was determined by flow cytometry, where cells at each time point showed roughly concentrated phase as follows: 0 h: G1/S; 2 h: early-S; 4 h: middle-S; 6 h: late-S; 8 h: S/G2; 10 h: G2/M; 12 h: M/G1; 14 h: G1 (Fig. 2a). Histones were then extracted using a PTM-protective extraction kit based on the previously reported protocol27) and digested by AspN to give 23-residue-long histone H4 N-terminal tail peptides (Fig. 1b) under the denaturing conditions, including reduction and carbamidomethylation of cysteine residues, because this step was found to be essential to enhance digestion. The digested proteins (∼500 ng) were injected into the LC-MS/MS system. Not to lose phosphorylated peptides during LC, all devices were connected by fused-silica capillaries, and metal devices were avoided.6,28) Histone H4 tail peptide ions (+3, +4, +5) with various masses as a result of acetylation, methylation, or phosphorylation (Fig. 1b) were separated by liquid chromatography and by the masses of their precursor ions (Fig. 1c and Table S1). Since the most intense signals in our system were associated with histone H4 tails with a charge of +4, we analyzed +4 peaks here. Three biological replicates were independently analyzed with a standard label-free semi-quantification and integrated by normalizing to histone H4 C-terminal peptides DVVYALKRQGRTLYGFGG (+3 and +4) (Table S1). To adjust the dynamic range rigorously, the injection volume was fine-tuned to adjust the ion current for the normalization of H4 C-terminal peptides. Resulting dynamic range was 6×105, for the largest peak abundance was 1.1×107 [Histone H4, DVVYALKRQGRTLYGFGG, +3, 4 h; CV=2.5% (normalized), 3.4% (raw)] and the smallest was 19.2 (Histone H4, SGRGKGGKGLGKGGAKRHRKVLR, 5+, 14 h; CV=120%). The median CV was 29% for identified peptides and 25% for histone H4 tails (N=3 peaks in Table S1). Each dataset contained quantitative data for about 90,000 peaks, of which about 600 had possible peptide identifications (MASCOT expect score<0.05, Fig. 1c). For H4 tail peptide of +4, we obtained 41, 39, and 46 separated peaks in the three datasets, respectively (Figs. 1c, 1d). All ion abundances obtained were given in Fig. S4. Most probable identification for the peaks were determined by the number of MS/MS spectra obtained in the retention time window of the peak (Table S2, Fig. S4). Best MS/MS spectra for the most probable identification of each peak were shown in Fig. S5. Note that some peaks had small number of assigned MS/MS spectra for the representative identification because of mixed MS/MS spectra (Table S2, Fig. S4). Some isobaric species were separated chromatographically to be quantified as individual peaks (Fig. S2). MASCOT search results contained a larger number of combinatorial PTMs (at least 53 in Table S3, Fig. 1d) than the number of chromatographically separated peaks because of co-eluted inseparable isobaric species in the measurement.
Fig. 1. Workflow to identify histone modifications.
(a) Workflow. (b) Histone H4 tail and common PTMs. Ac (acetylation), Me (methylation), Ph (phosphorylation). Amino acids with number on the top include the sites PTM were considered in our MASCOT search. (c) LC-MS map of the histone H4 tail (+4). Peaks are separated by their parent mass and LC retention time depending on their acetylation, methylation, and phosphorylation statuses. Representative results (Dataset 3) are shown. All maps for each dataset are given in Fig. S3. (d) Summary of the analysis. *: Obtained from Table S3.
Fig. 2. Label-free semi-quantification of modification on histone H4 tail during the cell cycle.
(a) Cell cycle synchronization: HeLa S3 cells were synchronized by double thymidine blocking and collected at two-hour intervals after release. Cell cycle synchrony was confirmed by flow cytometry. (b) Hierarchical clustering of temporal changes in abundance of modifications during the cell cycle: Ion abundances of +4 ion peaks were summed and normalized to the maximum abundance (shown in columns) of each species. Representative modifications for the peaks are selected as annotation from Table S2. Peaks whose PTM assignment showed conflicts between datasets were written in italics. Peak number in bold red indicates S1ph containing species.
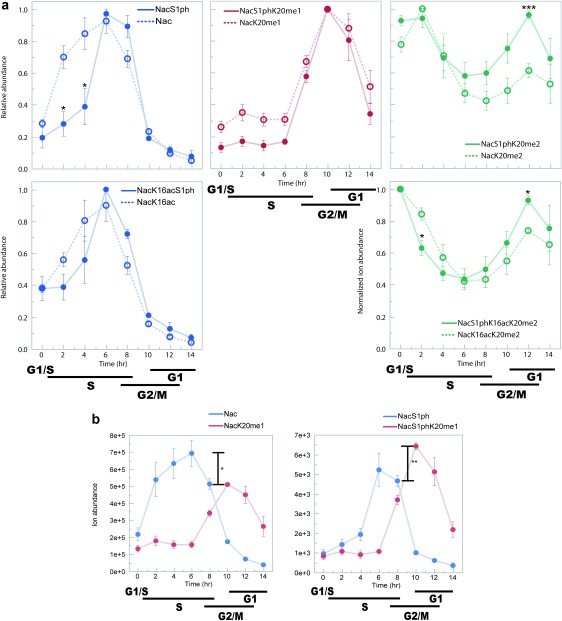
To compare the temporal changes among the modification species with various abundances, we normalized abundances to the maximum abundance of each species to obtain relative abundances. By hierarchical clustering, profiles of the relative abundances were clustered by K20 methylation state and were distinguished by the presence of S1ph (Fig. 2b). The methylation state of K20 was observed to change from unmodified in the S-phase through monomethylated in the G2-phase to dimethylated in the M-phase (Figs. 2b, 3a), as previously shown.9,29–31) K20me2/3, R3me1/2 (peaks 2, 5, 6, 7+8, 11+12, 13) increased not only in M-phase but also in early S-phase, which can be interpreted as hypoacetylation of histone tails in early-S and M phase. Accordingly, increase in acetylation is observed on G1/S and then deacetylated in early S-phase. Comparing profiles between with and without S1ph revealed histones with K20me0 increased in early to mid S-phase were not phosphorylated and phosphorylated in late S-phase and then methylated to be K20me1 in G2-phase and then to K20me2 in M-phase in parallel to those without S1ph (Fig. 3a). Comparing profiles between K20me0 and K20me1 revealed histones with S1ph increased even after the conversion to K20me1 (Fig. 3b). Then K20me2 and S1ph co-occurred in M-phase. Therefore, phosphorylation is likely to occur from late S-phase to M-phase on histones with K20me0. Whereas, phosphorylation onto histones with K20me2 increased in early S-phase and M-phase like K20me2/3 and R3me1/2 did. Co-existence of K16ac showed a slight effect on their temporal changes such as quick decrease from G1/S to early S-phase, like other acetylated species did.
Fig. 3. H4S1ph profiles.
(a) Temporal changes of K20 methylation state with (closed circle) and without (open circle) S1ph: With respect to accompanying modifications (K16ac or unmodified, and K20me0, 1, 2), relative abundances are plotted. Average of the relative abundances among the datasets. Error bars represent standard errors. Differences between with and without S1ph are tested by a pooled t-test. (b) Comparison between K20me0 and K20me1 abundances with (right panel) or without (left panel) S1ph: Average ion abundances among the datasets. Error bars represent standard errors. Differences indicated in the bars are tested by a pooled t-test. * p<0.05; ** p<0.01; *** p<0.005.
The combinatorial patterns for acetylation were largely consistent with previous works29) (Table S3); monoacetylation was frequent on K16 and K12; diacetylation was frequent on K16/12 and K12/8; and triacetylation was frequent on K16/12/8 and K12/8/5. Nascent histone (represented by unmodified K20) preferentially showed different combinatorial acetylations, such as K5/12 diacetylation.8)
The LC-chromatogram and MS spectra of histone H4 are shown in Fig. S6. ETD spectra, compared with CID, showed high-quality fragmentation and no neutral loss of phosphoryl groups (Fig. S6). The ETD fragments were well assigned to the theoretical fragment masses, whereas no identification was possible from the CID spectra (p<0.05; the ETD spectral assignment is shown in Fig. S6).
Identification of potential interaction partners for H4S1ph
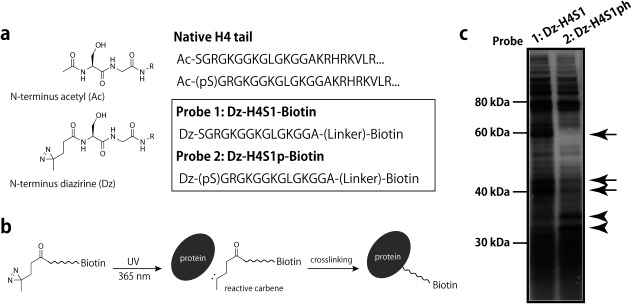
There have been several immunocytological and genomic studies on H4S1ph-related cellular events and the genomic distributions of the PTM,32–34) but the nature of the effector protein that directly recognizes and mediates such processes is largely unknown.35,36) Phosphorylation has some negative effects on binding of histone methyltransferase to the H4 tail37,38); however, to our knowledge, no binder for H4S1ph has been positively identified. Here, we synthesized and tested several photoaffinity probes for the direct capture of H4S1/H4S1ph-interacting proteins (see Fig. 4a for probes 1 and 2, and Fig. S8 for probes 3–6). Photoreactive moieties capable of being activated by UV irradiation were incorporated in three positions. A benzophenone moiety was incorporated into the H4G7 (probes 3–4) or H4G11 (probes 5–6) side-chains, whereas a diazirine group was incorporated into the H4 tail N-terminus just beside H4S1/S1ph (probes 1 and 2). Because binding sites should not be sterically hindered by the bulky benzophenone moiety, we avoided placing Bpa too close to H4S1. By using these probes, we conducted crosslink-assisted protein pull-down from HeLa S3 lysate. Proteins which are crosslinked to those probes are bonded to biotin through the bait peptides, so they can be detected with high sensitivity by using SA-HRP (Fig. 4b). In the cases of probes 3–6 (G7Bpa and G11Bpa substitution), no differential crosslinking product between H4S1 and H4S1ph was observed (Fig. S8). Therefore, only probes 1 and 2 showed an obviously differential landscape of interacting proteins, distinguishing between H4S1 and H4S1ph. Figure 4c shows evidence for the existence of differentially interacting proteins of the H4S1/S1ph N-termini. H4S1-specific bands are present in the 60 kDa and 40 kDa regions, and H4S1ph-specific bands are present in the 30 kDa regions.
Fig. 4. Detection of H4S1/S1ph interacting proteins.
(a) Design of photoreactive peptide probes 1 (Dz-H4S1) and 2 (Dz-H4S1ph). The N-terminal acetyl groups were replaced with a diazirine-containing moiety. (b) Scheme for the crosslinking reaction (carbene insertion). (c) Crosslinking pull-down experiment using probes 1 and 2. Photoaffinity-captured proteins were enriched by biotin-streptavidin affinity purification and detected by streptavidin–HRP. H4S1-specific (arrows) and H4S1ph-specific bands (arrowheads) are indicated.
Interaction between H4S1ph and 14-3-3
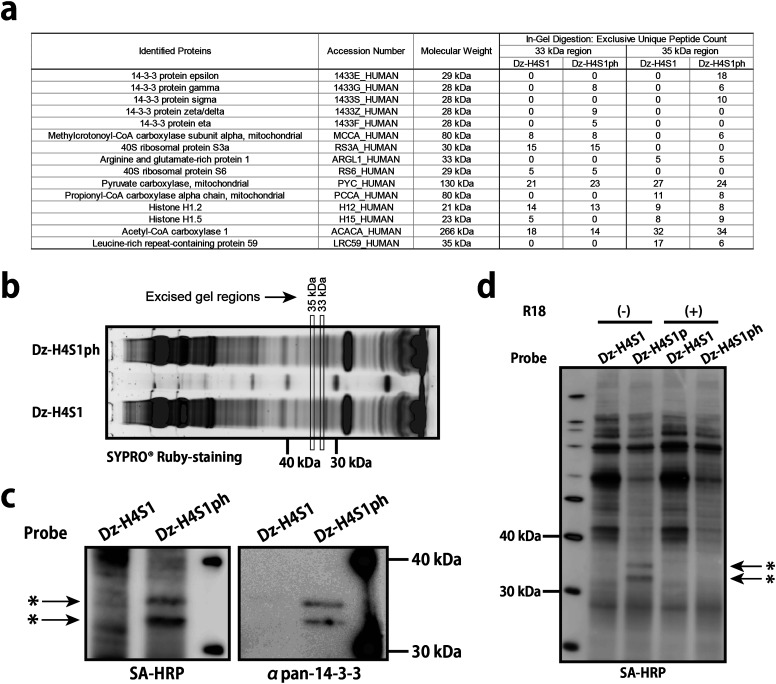
Having performed the pull-down experiment, we identified the Dz-H4S1ph-specific 30 kDa bands as 14-3-3 proteins by means of mass spectrometry (Figs. 5a, b, Table S4); this identification was validated by immunodetection using anti-pan-14-3-3 antibody (Fig. 5c). This interaction was further confirmed by an inhibition experiment with 14-3-3 binding peptide R18 (PHCVPRDLSWLDLEANMCLP; known to antagonize phospho-S/T binding groove of 14-3-3 at KD≈80 nM),39) in which Dz-H4S1ph-specific 14-3-3 bands specifically disappeared (Fig. 5d).
Fig. 5. Identification and verification of H4S1p interacting proteins.
(a) Identification of H4S1ph interacting protein by LC-MS/MS. Pull-down product were separated by gel electrophoresis, SYPRO Ruby-stained, and excised as shown in (b) for in-gel tryptic digestion. The bands of interest were not recognizable on the SYPRO® Ruby-stained gel, so excision of gels were performed by estimating the position from molecular weight markers. Identified proteins are shown in the table. (c) Immunodetection of crosslinked product using pan-14-3-3 antibody. Asterisks indicates crosslink-product of 14-3-3 proteins and Dz-H4S1ph-biotin. (d) Specific inhibition of the interaction between H4S1ph and 14-3-3 by R18 peptide (10 μM).
DISCUSSION
We have defined PTM dynamics on histone H4 during cell cycle using label-free semi-quantification. Among the PTMs we quantified, we found S1ph elevated from late S-phase to M-phase on histones with K20me0. It is notable that histones with K20me0 were not phosphorylated in early S-phase on contrary to histones with K20me2. This observation was acquired by measurement of K20me state and S1ph simultaneously by our middle-down procedure and by our sensitive quantification of less abundant H4S1ph species otherwise undetectable by antibody-based immunodetection because antibodies typically recognize only one single PTM at a time. Since histones synthesized in S-phase is largely unmodified at K20,30,40) and S1ph was reported as a mark on nascent histone,41) histones with S1phK20me0 would mostly be nascent histone. If histones with S1phK20me0 are nascent histone, this characteristic profile of S1ph on nascent histone might be associated with replication of late-replicating region. Previous observations such as cdk1 activation to replicate the late-replicating region42) and DNA concentration–dependent H4S1ph observed during Xenopus development43) might be associated phenomena. Since H4S1ph is involved in double strand break repair33) and late S-phase is a vulnerable phase for DNA stability,44) H4S1ph in late-S phase might also be associated with the double strand break repair.
Our pull-down assay has defined 14-3-3 proteins as S1ph interaction partners, which is a feasible result as 14-3-3 recognizes most of phosphorylated proteins.45,46) Actually, 14-3-3 proteins are known to bind H3S10ph in the nucleus47) whereas most fraction of 14-3-3 proteins are located in cytoplasm, which implies the possibility that 14-3-3 can bind both nuclear H4S1ph and cytosolic nascent H4. Some additional PTMs may modify the 14-3-3 binding affinity with or without other interjacent proteins48) but we cannot tell in advance which PTM acts as such an affinity modifier, herein photocrosslinking is helpful in finding a clue for PTM reader discovery. The phosphorylated H4 N-terminal sequence (Ac-pSGRGKGG...) is not a conventional 14-3-3 recognition sequence,49) which in turn reflects our technical improvement in the screening of weak protein–protein interactions. Indeed, without cross-linking, we could not capture any H4S1ph-specific binding proteins by pull-down experiments. Our next challenge will be to identify PTM-related proteins with functional relevance using photo-reactive probes. In addition to the challenges to good PTM quantification, the identification of binding partners of PTMs is not an easy process because of the variability of the binding affinities of histone PTM readers. Here, we tried direct incorporation of photoreactive moieties into pull-down baits, and we realized that the bait design is of critical importance for successful identification of interacting proteins. The implications are that (i) the crosslinker moiety should not disrupt PTM–protein interactions sterically, (ii) that bioisosteric design is desirable, and (iii) the crosslinker moiety should be located as near as possible to the PTM site. Diazirine is compact and less disruptive to intermolecular interaction than benzophenone, which facilitates ideal molecular mimicry.50,51) In the case of the H4 tail, almost every N-terminus is physiologically acetylated, so we substituted the N-terminal acetyl group with a diazirine-containing moiety just beside the H4S1 residue. Irreversible N2 removal from diazirine by photoirradiation and the high reactivity of the resulting carbene intermediate52,53) resulted in the efficient capture of proteins weakly binding with the target amino acid, otherwise unsuccessful by benzophenone crosslinking. However, incorporation of diazirine is sometimes difficult depending on the amino acid sequence of interest. Therefore, to capture a PTM reader robustly, we have to consider the structural traits of the PTM itself on a case-by-case basis.
Overall, here we have developed a mass spectrometric method for more-comprehensive histone code analysis. However, not only H4, but also H3 and H2A/H2B carry epigenetic information in cis- or trans-histone manners. For further elucidation of the code, extension of the target repertoire and more-versatile analysis are essential.
Acknowledgments
We thank Drs. Andrea Sinz, Junko Ohkanda, and Yuzo Yamazaki for helpful comments, and Aya Nakayama and Haruna Saito and Yuko Nakamura for technical assistance.
This study was supported by a grant for the New Energy and Industrial Technology Development Organization (NEDO), Japan and by a grant for Translational Systems Biology and Medicine Initiative (TSBMI) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Mass Spectrom (Tokyo) 2015; 4(1): A0039
- ac
acetylation
- AGC
automatic gain control
- BSA
bovine serum albumin
- DIEA
N,N-diisopropylethylamine
- DTT
dithiothreitol
- Dz
diazirine
- ETD
electron transfer dissociation
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- HBTU
(1H-benzotriazol-1-yloxy)(dimethylamino)-N,N-dimethylmethaniminium hexafluorophosphate
- HOBt
1-hydroxybenzotriazole
- HRP
horseradish peroxidase
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- me
methylation
- NMM
N-methylmorpholine
- NSI
nanospray ionization
- PBS
phosphate-buffered saline
- PEG
poly(ethylene glycol)
- ph
phosphorylation
- PTM
post-translational modification
- PVDF
poly(vinylidene difluoride)
- PyBOP
(benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate
- SA
streptavidin
- SDS
sodium dodecylsulfate
- TCEP
tris(2-carboxyethyl)phosphine
- TFA
trifluoroacetic acid
- TIPS
triisopropylsilane
Supporting Information
References
- 1) T. Jenuwein, C. D. Allis. Translating the histone code. Science 293: 1074–1080, 2001. [DOI] [PubMed] [Google Scholar]
- 2) A. M. Arnaudo, B. A. Garcia. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin 6: 24, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3) Z. Tian, N. Tolic, R. Zhao, R. J. Moore, S. M. Hengel, E. W. Robinson, D. L. Stenoien, S. Wu, R. D. Smith, L. Pasa-Tolic. Enhanced top-down characterization of histone post-translational modifications. Genome Biol. 13: R86, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4) D. Phanstiel, J. Brumbaugh, W. T. Berggren, K. Conard, X. Feng, M. E. Levenstein, G. C. McAlister, J. A. Thomson, J. J. Coon. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105: 4093–4098, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5) N. L. Young, P. A. DiMaggio, M. D. Plazas-Mayorca, R. C. Baliban, C. A. Floudas, B. A. Garcia. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 8: 2266–2284, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6) R. Zhao, S. J. Ding, Y. Shen, D. G. Camp 2nd, E. A. Livesay, H. Udseth, R. D. Smith. Automated metal-free multiple-column nanoLC for improved phosphopeptide analysis sensitivity and throughput. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 663–670, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7) M. Sandin, A. Ali, K. Hansson, O. Mansson, E. Andreasson, S. Resjo, F. Levander. An adaptive alignment algorithm for quality-controlled label-free LC-MS. Mol. Cell. Proteomics 12: 1407–1420, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8) R. E. Sobel, R. G. Cook, C. A. Perry, A. T. Annunziato, C. D. Allis. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U.S.A. 92: 1237–1241, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9) J. C. Rice, K. Nishioka, K. Sarma, R. Steward, D. Reinberg, C. D. Allis. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 16: 2225–2230, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10) B. T. Seet, I. Dikic, M. M. Zhou, T. Pawson. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7: 473–483, 2006. [DOI] [PubMed] [Google Scholar]
- 11) M. Mann, O. N. Jensen. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21: 255–261, 2003. [DOI] [PubMed] [Google Scholar]
- 12) H. C. Eberl, C. G. Spruijt, C. D. Kelstrup, M. Vermeulen, M. Mann. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell 49: 368–378, 2013. [DOI] [PubMed] [Google Scholar]
- 13) M. Vermeulen, H. C. Eberl, F. Matarese, H. Marks, S. Denissov, F. Butter, K. K. Lee, J. V. Olsen, A. A. Hyman, H. G. Stunnenberg, M. Mann. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980, 2010. [DOI] [PubMed] [Google Scholar]
- 14) J. Wysocka. Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods 40: 339–343, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15) D. J. Patel, Z. Wang. Readout of epigenetic modifications. Annu. Rev. Biochem. 82: 81–118, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16) P. W. Lewis, M. M. Muller, M. S. Koletsky, F. Cordero, S. Lin, L. A. Banaszynski, B. A. Garcia, T. W. Muir, O. J. Becher, C. D. Allis. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17) X. Li, E. A. Foley, S. A. Kawashima, K. R. Molloy, Y. Li, B. T. Chait, T. M. Kapoor. Examining post-translational modification-mediated protein–protein interactions using a chemical proteomics approach. Protein Sci. 22: 287–295, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18) X. Li, E. A. Foley, K. R. Molloy, Y. Li, B. T. Chait, T. M. Kapoor. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein–protein interactions. J. Am. Chem. Soc. 134: 1982–1985, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19) X. Li, T. M. Kapoor. Approach to profile proteins that recognize post-translationally modified histone “tails”. J. Am. Chem. Soc. 132: 2504–2505, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20) Z. Liu, L. C. Myers. Med5(Nut1) and Med17(Srb4) are direct targets of mediator histone H4 tail interactions. PLoS ONE 7: e38416, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21) S. H. Yu, M. Boyce, A. M. Wands, M. R. Bond, C. R. Bertozzi, J. J. Kohler. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc. Natl. Acad. Sci. U.S.A. 109: 4834–4839, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22) M. Knehr, M. Poppe, M. Enulescu, W. Eickelbaum, M. Stoehr, D. Schroeter, N. Paweletz. A critical appraisal of synchronization methods applied to achieve maximal enrichment of HeLa cells in specific cell cycle phases. Exp. Cell Res. 217: 546–553, 1995. [DOI] [PubMed] [Google Scholar]
- 23) M. R. Bond, H. Zhang, P. D. Vu, J. J. Kohler. Photocrosslinking of glycoconjugates using metabolically incorporated diazirine-containing sugars. Nat. Protoc. 4: 1044–1063, 2009. [DOI] [PubMed] [Google Scholar]
- 24) M. Fujinoki, T. Kawamura, T. Toda, H. Ohtake, T. Ishimoda-Takagi, N. Shimizu, S. Yamaoka, M. Okuno. Identification of 36-kDa flagellar phosphoproteins associated with hamster sperm motility. J. Biochem. 133: 361–369, 2003. [DOI] [PubMed] [Google Scholar]
- 25) K. Daigo, T. Kawamura, Y. Ohta, R. Ohashi, S. Katayose, T. Tanaka, H. Aburatani, M. Naito, T. Kodama, S. Ihara, T. Hamakubo. Proteomic analysis of native hepatocyte nuclear factor-4alpha (HNF4alpha) isoforms, phosphorylation status, and interactive cofactors. J. Biol. Chem. 286: 674–686, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26) K. Daigo, N. Yamaguchi, T. Kawamura, K. Matsubara, S. Jiang, R. Ohashi, Y. Sudou, T. Kodama, M. Naito, K. Inoue, T. Hamakubo. The proteomic profile of circulating pentraxin 3 (PTX3) complex in sepsis demonstrates the interaction with azurocidin 1 and other components of neutrophil extracellular traps. Mol. Cell. Proteomics 11: M111.015073, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27) P. Rodriguez-Collazo, S. H. Leuba, J. Zlatanova. Robust methods for purification of histones from cultured mammalian cells with the preservation of their native modifications. Nucleic Acids Res. 37: e81, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28) H. Imamura, M. Wakabayashi, Y. Ishihama. Analytical strategies for shotgun phosphoproteomics: Status and prospects. Semin. Cell Dev. Biol. 23: 836–842, 2012. [DOI] [PubMed] [Google Scholar]
- 29) J. J. Pesavento, C. R. Bullock, R. D. LeDuc, C. A. Mizzen, N. L. Kelleher. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 283: 14927–14937, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30) J. J. Pesavento, H. Yang, N. L. Kelleher, C. A. Mizzen. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 28: 468–486, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31) R. Y. Tweedie-Cullen, A. M. Brunner, J. Grossmann, S. Mohanna, D. Sichau, P. Nanni, C. Panse, I. M. Mansuy. Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PLoS ONE 7: e36980, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32) C. M. Barber, F. B. Turner, Y. Wang, K. Hagstrom, S. D. Taverna, S. Mollah, B. Ueberheide, B. J. Meyer, D. F. Hunt, P. Cheung, C. D. Allis. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 112: 360–371, 2004. [DOI] [PubMed] [Google Scholar]
- 33) W. L. Cheung, F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, C. D. Allis. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15: 656–660, 2005. [DOI] [PubMed] [Google Scholar]
- 34) J. Govin, J. Schug, T. Krishnamoorthy, J. Dorsey, S. Khochbin, S. L. Berger. Genome-wide mapping of histone H4 serine-1 phosphorylation during sporulation in Saccharomyces cerevisiae. Nucleic Acids Res. 38: 4599–4606, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35) T. Banerjee, D. Chakravarti. A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 31: 4858–4873, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36) D. Rossetto, N. Avvakumov, J. Cote. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 7: 1098–1108, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37) M. C. Ho, C. Wilczek, J. B. Bonanno, L. Xing, J. Seznec, T. Matsui, L. G. Carter, T. Onikubo, P. R. Kumar, M. K. Chan, M. Brenowitz, R. H. Cheng, U. Reimer, S. C. Almo, D. Shechter. Correction: Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS ONE 8(8): 10.1371, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38) M. C. Ho, C. Wilczek, J. B. Bonanno, L. Xing, J. Seznec, T. Matsui, L. G. Carter, T. Onikubo, P. R. Kumar, M. K. Chan, M. Brenowitz, R. H. Cheng, U. Reimer, S. C. Almo, D. Shechter. Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS ONE 8: e57008, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39) B. Wang, H. Yang, Y. C. Liu, T. Jelinek, L. Zhang, E. Ruoslahti, H. Fu. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38: 12499–12504, 1999. [DOI] [PubMed] [Google Scholar]
- 40) B. M. Zee, L. M. Britton, D. Wolle, D. M. Haberman, B. A. Garcia. Origins and formation of histone methylation across the human cell cycle. Mol. Cell. Biol. 32: 2503–2514, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41) A. Ruiz-Carrillo, L. J. Wangh, V. G. Allfrey. Processing of newly synthesized histone molecules. Science 190: 117–128, 1975. [DOI] [PubMed] [Google Scholar]
- 42) J. A. Farrell, A. W. Shermoen, K. Yuan, P. H. O’Farrell. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev. 26: 714–725, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43) W. L. Wang, L. C. Anderson, J. J. Nicklay, H. Chen, M. J. Gamble, J. Shabanowitz, D. F. Hunt, D. Shechter. Phosphorylation and arginine methylation mark histone H2A prior to deposition during Xenopus laevis development. Epigenetics Chromatin 7: 22, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44) N. Donley, M. J. Thayer. DNA replication timing, genome stability and cancer: Late and/or delayed DNA replication timing is associated with increased genomic instability. Semin. Cancer Biol. 23: 80–89, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45) B. C. Collins, L. C. Gillet, G. Rosenberger, H. L. Rost, A. Vichalkovski, M. Gstaiger, R. Aebersold. Quantifying protein interaction dynamics by SWATH mass spectrometry: Application to the 14-3-3 system. Nat. Methods 10: 1246–1253, 2013. [DOI] [PubMed] [Google Scholar]
- 46) K. Kakiuchi, Y. Yamauchi, M. Taoka, M. Iwago, T. Fujita, T. Ito, S. Y. Song, A. Sakai, T. Isobe, T. Ichimura. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46: 7781–7792, 2007. [DOI] [PubMed] [Google Scholar]
- 47) S. Winter, E. Simboeck, W. Fischle, G. Zupkovitz, I. Dohnal, K. Mechtler, G. Ammerer, C. Seiser. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 27: 88–99, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48) A. Zippo, R. Serafini, M. Rocchigiani, S. Pennacchini, A. Krepelova, S. Oliviero. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138: 1122–1136, 2009. [DOI] [PubMed] [Google Scholar]
- 49) M. B. Yaffe, K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, L. C. Cantley. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91: 961–971, 1997. [DOI] [PubMed] [Google Scholar]
- 50) A. Sinz. Investigation of protein–protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal. Bioanal. Chem. 397: 3433–3440, 2010. [DOI] [PubMed] [Google Scholar]
- 51) G. W. Preston, A. J. Wilson. Photo-induced covalent cross-linking for the analysis of biomolecular interactions. Chem. Soc. Rev. 42: 3289–3301, 2013. [DOI] [PubMed] [Google Scholar]
- 52) A. F. Gomes, F. C. Gozzo. Chemical cross-linking with a diazirine photoactivatable cross-linker investigated by MALDI- and ESI-MS/MS. J. Mass Spectrom. 45: 892–899, 2010. [DOI] [PubMed] [Google Scholar]
- 53) B. Albertoni, J. S. Hannam, D. Ackermann, A. Schmitz, M. Famulok. A trifluoromethylphenyl diazirine-based SecinH3 photoaffinity probe. Chem. Commun. (Camb.) 48: 1272–1274, 2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.