Abstract
Recurrent aphthous stomatitis (RAS) is the most common disease affecting oral mucosae. Etiology is unknown, but several factors have been implicated, all of which influence the composition of microbiota residing on oral mucosae, which in turn modulates immunity and thereby affects disease progression. Although no individual pathogens have been conclusively shown to be causative agents of RAS, imbalanced composition of the oral microbiota may play a key role. In this study, we sought to determine composition profiles of bacterial microbiota in the oral mucosa associated with RAS. Using high-throughput 16S rRNA gene sequencing, we characterized the most abundant bacterial populations residing on healthy and ulcerated mucosae in patients with RAS (recruited using highly stringent criteria) and no associated medical conditions; we also compared these to the bacterial microbiota of healthy controls (HCs). Phylum-level diversity comparisons revealed decreased Firmicutes and increased Proteobacteria in ulcerated sites, as compared with healthy sites in RAS patients, and no differences between RAS patients with healthy sites and HCs. Genus-level analysis demonstrated higher abundance of total Bacteroidales in RAS patients with healthy sites over HCs. Porphyromonadaceae comprising species associated with periodontal disease and Veillonellaceae predominated in ulcerated sites over HCs, while no quantitative differences of these families were observed between healthy sites in RAS patients and HCs. Streptococcaceae comprising species associated with oral health predominated in HCs over ulcerated sites but not in HCs over healthy sites in RAS patients. This study demonstrates that mucosal microbiome changes in patients with idiopathic RAS—namely, increased Bacteroidales species in mucosae of RAS patients not affected by active ulceration. While these changes suggest a microbial role in initiation of RAS, this study does not provide data on causality. Within this limitation, the study contributes to the understanding of the potential role of mucosal microbiome changes in oral mucosal disease.
Keywords: oral ulcer, oral mucosa, chronic disease, microbiota, host-pathogen relations, high-throughput DNA sequencing
Introduction
Recurrent aphthous stomatitis (RAS) is the most common oral mucosal disease in the general population (5% to 60% in different study groups). RAS is characterized by multiple recurrent round or ovoid inflammatory ulcerations with circumscribed margins, erythematous haloes, and yellow or gray floors (Jurge et al. 2006). RAS causes considerable pain, can interfere with oral functions (eating, speech, toothbrushing), and can thereby have a negative impact on quality of life (Al-Omiri et al. 2014). Current treatment is based on topical corticosteroids and systemic immunosuppressants depending on severity but is still palliative, as it only reduces the severity of the ulceration and does not stop recurrence (Altenburg et al. 2007; Sheikh et al. 2013). Understanding the etiology and pathogenesis of RAS, currently unknown, will aid the development of more effective therapeutic strategies.
Host genetics, nutritional deficiencies, as well as a number of systemic conditions (including chronic inflammatory disorders and immunodeficiencies) have been recognized as systemic modulating factors of RAS (Jurge et al. 2006; Slebioda et al. 2013; Sun et al. 2014). While not directly involved in the etiology of RAS, all these factors influence the composition of the community of microorganisms that colonize the oral mucosa (oral microbiota), which can in turn modulate inflammatory responses and thus affect progression of RAS. There is an increasing body of evidence suggesting that perturbations of mucosal microbiota—at the intestinal mucosa in particular—can modulate innate and adaptive immune responses, with inflammation arising upon reduction of the number of symbiont microorganisms and/or increase in the number of pathobiont microorganisms (commensal bacteria with pathogenic potential). This status is referred to as microbial dysbiosis (Petersen and Round 2014). Several immune mechanisms of disease suppression by symbiont bacteria have been hypothesized, including induction of IL-10, suppression of TNF-α and IL-8, and modulation of Toll-like receptors (Round and Mazmanian 2009). These mechanisms have all been implicated in remission of RAS (Taylor et al. 1992; Miyamoto et al. 2008; Gallo et al. 2012).
An association between Helicobacter pylori and RAS has been shown but remains controversial (Riggio et al. 2000; Mansour-Ghanaei et al. 2005). Oral streptococci have also been implicated (Hasan et al. 1995). So far, however, no bacterial species have been conclusively shown to be the causative agents of RAS. Nonetheless, the oral microbiota of patients with RAS was found to differ from that of healthy controls (HCs; Marchini et al. 2007). Indeed, pathogen-associated molecular patterns shared by different microbial species have been implicated in the etiopathogenesis of RAS (Hietanen et al. 2012). As suggested for periodontal disease (Kumar et al. 2005), targets of early stages of inflammation in RAS may be pathobionts that are overrepresented during global perturbation of the normal oral microbiota. The role of Porphyromonas gingivalis in periodontal disease has recently been emphasized by demonstration of its ability to promote dysbiosis of oral microbiota (Maekawa et al. 2014).
Identification of protective symbionts and pathobionts that contribute to disease progression would be crucial to the development of effective treatment for RAS. In this study, we used high-throughput sequencing of 16S ribosomal genes to characterize the oral microbiota residing on healthy mucosae and ulcerated mucosae in patients with RAS. We also compared each of these profiles with the bacterial populations from corresponding oral mucosal sites of healthy individuals.
Methods
Ethics Statement
Ethical approval for involvement of human participants in this study was granted by North of Scotland Research Ethics Service (REC reference 12/NS/0006). Written informed consent was obtained from all subjects according to the World Health Organization guidelines for good clinical practice.
Patient Recruitment Method
Participants were recruited at the Maxillofacial Unit, Aberdeen Royal Infirmary. Idiopathic RAS cases were diagnosed through accepted clinical criteria, and the severity of the condition was defined via a recently described ulcer severity score (Tappuni et al. 2013). A group of HCs matched by sex and age (± 5 y) was also recruited. Patient assessment included a full history and neck and oral mucosae examination. As part of a comprehensive oral health assessment, we also carried out tooth charting, periodontal health assessment, measurement of plaque index, and whole salivary flow rate and candida culture. To exclude the presence of underlying medical conditions, serology investigations were carried out (full blood count, liver function test, renal profile, red cell folate, ferritin, and vitamin B12). Exclusion criteria included the following: hematologic and biochemical tests outside the normal range, whole salivary flow rate <1 mL/min, candida count >1,000 colony-forming unit/mL, smoking, use of antibiotics in the preceding 3 mo, having commenced any therapy for RAS in the preceding 6 mo, presence of any known medical conditions, presence of other oral mucosal diseases (including trauma-related injury), periodontal disease, high-sugar diet, D3MFT >2, plaque index >30%. To reduce influences by dietary and oral hygiene habits, participants were asked to avoid antibacterial mouthwashes, dental floss, and consumption of sugary food and drinks for 72 h prior to the visit. Patients were asked to avoid the foods and drinks defined as potentially cariogenic by the Department of Health and the British Association for the Study of Community Dentistry (2009). Recruits were also asked to brush their teeth twice a day using only toothpaste provided by the clinic. Toothbrushes were not provided by the clinic, but we ensured that all patients used manual toothbrushes. Optimal toothbrushing technique was discussed with all patients as part of routine clinical practice. The full recruitment method is illustrated in the Appendix Figure.
Sample Collection
Samples for microbiota analyses were all taken in the morning before the participants ate breakfast. Direct swabs of the following mucosal areas were taken: healthy site (lower buccal sulcus) in RAS patients, ulcerated area in RAS patients, and lower buccal sulcus in HCs. In RAS patients where the buccal sulcus was affected by ulceration, healthy sites were sampled from the contralateral lower buccal sulcus. In RAS patients with multiple lesions, only 1 ulcer was sampled, with the following order of site preference: buccal sulcus, buccal mucosae, labial mucosa, and ventral and lateral surface of the tongue. Additional details of mucosal sites sampled from RAS patients are reported in Appendix Table 1 along with demographic and clinical details of RAS patients recruited.
Microbiota Analyses
Swabs were immersed directly in DNA extraction buffer (300 μL), and the suspension was stored at 4oC until DNA extraction was performed, which was a maximum of 24 h postcollection. DNA extraction was conducted with the commercially available QIAamp Mini Kit (Qiagen, Crawley, UK), with minor amendments as described previously (Thomson et al. 2011; full details are reported in the Appendix).
Community polymerase chain reaction (PCR) for denaturing gradient gel electrophoresis (DGGE) analysis was conducted with universal primers U968 + GC clamp and L1401r targeting the variable V8/9 region of the 16S rRNA gene corresponding to Escherichia coli positions 968 to 1401 (Zoetendal et al. 1998). These samples were then assessed by DGGE through a 40% to 55% gradient as described previously (Hold et al. 2001).
Preparation of Samples for High-throughput Sequencing
Bacterial DNA was quantified by spectrophotometry (NanoDrop 1000, Labtech, East Sussex, UK) prior to analysis. Initial PCR amplification was undertaken with FastStart High Fidelity PCR reagents (Roche Diagnostics, West Sussex, UK) as described previously (Hansen et al. 2012). The 16S rDNA primers spanned the V3 to V6 region of the 16S rRNA gene, providing a base pair (bp) product of ~726. Full details are provided in the Appendix.
Bioinformatic and Statistical Analysis
Data analysis of the 454 sequence data was performed with the QIIME 1.1.0 workflow (Caporaso et al. 2010). Full details are provided in the Appendix.
For analysis of diversity within samples (alpha diversity), operational taxonomic unit (OTU) tables were rarefied at 3,000 reads, and diversity indices Chao 1, Shannon, Simpson, observed species, and phylogenetic diversity (PD_whole_tree) were determined. To assess differences between microbial communities (beta diversity), weighted Unifrac analysis (Lozupone et al. 2006) on rarefied data with 97% (equivalent to species level) and 95% (equivalent to genus level) OTU clustering was performed, followed by principal components analysis. Statistical analysis of pyrosequencing phylum and genus data was undertaken between pairs of phenotypic groups with the analysis of variance (ANOVA) test. Additional sample comparisons were done between groups with t tests and the Mann-Whitney U test where appropriate. All statistical tests were undertaken through PASW Statistics 22.
Results
Details of Participants
RAS patients recruited in the study comprised 12 RAS patients from whom both healthy and ulcerated tissue was swabbed and 6 additional RAS patients presenting with a history of RAS and active ulceration at the time of recruitment into the study but no active ulceration at the clinic for sample collection. In the latter group of patients, a single sample was taken from the healthy lower buccal sulcus. The range of ulcer severity scores was 17 to 39. Demographic and clinical details of RAS patients as well as sites sampled are reported in Appendix Table 1. We also recruited 17 HCs who each provided a single sample from the lower buccal sulcus (HCs were matched to the 18 RAS patients, but 1 dropped out of the study).
As shown in the Appendix Figure, 32 patients (16 RAS patients and 16 matching HCs) from a total of 35 initially recruited met the criteria for inclusion in high-throughput sequence analysis.
Bacterial Diversity Analysis
Denaturing Gradient Gel Electrophoresis
All samples yielded DNA upon extraction, which was amplifiable through universal bacterial primers, with a median DNA concentration of 48.6 ng/µL (interquartile range, 23.65 to 89.45). Bacterial DNA was amplifiable from all samples via conventional 16S rRNA gene primers for DGGE analysis. DGGE analysis was used to provide a visual representation of bacterial diversity and was not intended to provide quantitative data. DGGE analysis showed diversity across all samples with individuals each harboring a unique collection of bacteria, depicted by differing DGGE profiles (Fig. 1). In RAS cases where ulcers sampled were >2 mm in diameter, intrapatient differences between the bacterial profiles generated from healthy and ulcerated mucosal sites were dramatic and not confined to differences in band intensity, suggesting that different bacterial communities were present in healthy sites when compared with ulcerated sites in the same individual (Fig. 1, lanes 8 and 9 vs. lanes 10 and 11).
Figure 1.
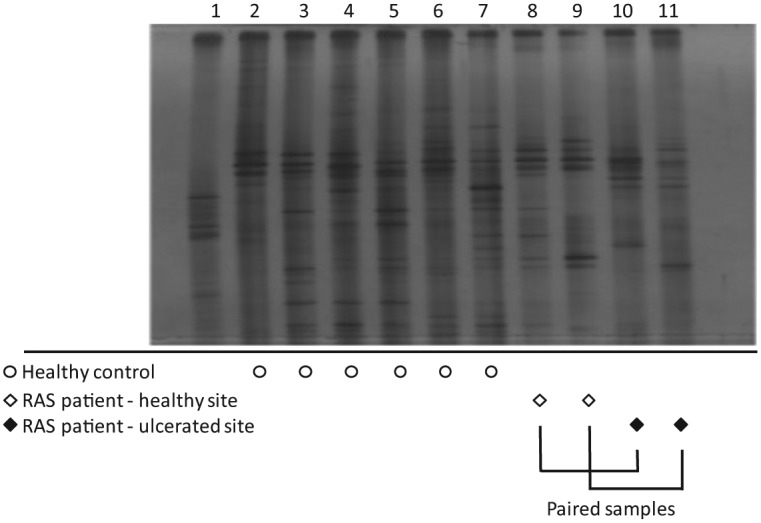
Denaturing gradient gel electrophoresis (DGGE) analysis of samples from recurrent aphthous stomatitis (RAS) patients and healthy controls. DGGE profiles of samples from healthy controls MU5 to MU10 (lanes 2 to 7, indicated as open circles), samples from healthy sites (lanes 8 and 9, indicated as open diamonds), and ulcerated sites (lanes 10 and 11, indicated as solid diamonds) in RAS patients MU11 and MU12. Lane 1: DGGE ladder. Unique profiles are apparent for individuals, but significant differences are also seen between healthy and ulcerated sites from the same individual (lane 8 vs. lane 10 for MU11; lane 9 vs. lane 11 for MU12).
High-throughtput Sequence Analysis
High-throughput sequencing generated 389,194 individual sequencing reads before and 307,940 after initial quality control analysis, with a mean yield of 6,415 reads per sample after bioinformatic processing but before rarefaction (set at 3,000 reads). Out of 47 samples, 4 did not meet the 3,000-read threshold (HC MU7, both ulcerated and healthy sites samples for MU15, and healthy site sample for MU25), and the respective participants were excluded from analysis. Thus, 32 subjects were ultimately represented by 3,000 rarefied reads after bioinformatic processing, allowing inter- and intrasubject comparisons to be made.
Bacterial richness was assessed by 5 distinct indices (Shannon, Simpson, Chao 1, observed species, and PD whole tree). No significant differences were seen in bacterial diversity when comparisons were made between (1) HC and ulcerated sites, (2) HC and healthy sites in RAS subjects, and (3) ulcerated and healthy sites in RAS subjects (Appendix Table 2).
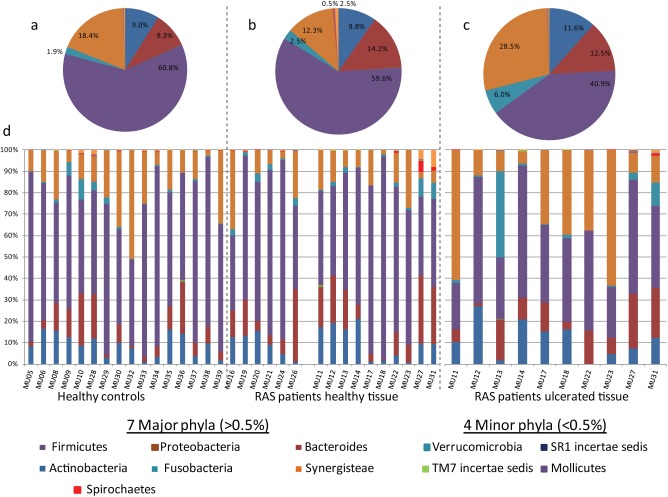
Phylum-level diversity comparisons across the 12 phyla of high-throughput sequencing findings revealed statistically significant differences for Firmicutes and Proteobacteria between ulcerated and healthy sites in RAS patients with decreased levels of Firmicutes detected in ulcerated sites, which coincided with an increase in Proteobacteria (P = 0.034 and 0.032 respectively; Fig. 2, Table 1). There were no statistically significant phylum-level differences between the HC group and healthy sites in RAS patients (P = 0.14 to 0.804, ANOVA; Fig. 2, Table 1).
Figure 2.
Phylum-level diversity assessment. Pie charts comprised all patient-rarefied reads (3,000 reads per patient) to represent overall diversity of each sample type; phyla constituting <0.5% have been removed for clarity: (a) healthy controls composed of 16 participants, (b) healthy tissue in 16 recurrent aphthous stomatitis (RAS) patients, (c) ulcerated tissue in 10 RAS patients. (d) Individual patient sample diversity at phylum level as stacked bars.
Table 1.
Phylum-level Statistical Comparisons for High-throughput Sequencing Data
| Statistical Comparison by ANOVA |
||||||||
|---|---|---|---|---|---|---|---|---|
| Percentage of 3,000 Patient Reads, Mean ± SD |
HC vs RAS-HS |
RAS-HS vs RAS-US |
||||||
| Phyla | HCs | RAS-HS | RAS-US | RAS-HS (No Ulcers) a | F | P | F | P |
| Actinobacteria | 9.0 ± 5.3 | 10.0 ± 7.9 | 11.6 ± 8.5 | 9.4 ± 5.6 | 0.071 | 0.791 | 0.27 | 0.610 |
| Bacteroides | 9.3 ± 7.9 | 14.8 ± 10.3 | 12.5 ± 8.3 | 13.2 ± 11.3 | 2.133 | 0.157 | 0.277 | 0.605 |
| Cyanobacteria | 0.1 ± 0.3 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.126 | 0.726 | 0.799 | 0.383 |
| Firmicutes | 60.8 ± 14.5 | 58.7 ± 18.7 | 40.9 ± 14.0 | 61.2 ± 19.9 | 0.227 | 0.638 | 5.272 | 0.034 b |
| Fusobacteria | 1.9 ± 2.6 | 2.6 ± 3.1 | 6.0 ± 12.5 | 2.5 ± 1.2 | 0.303 | 0.587 | 0.695 | 0.415 |
| Proteobacteria | 18.4 ± 12.9 | 11.6 ± 7.9 | 28.5 ± 21.8 | 13.4 ± 13.7 | 2.335 | 0.140 | 5.387 | 0.032 b |
| SR1 incertae sedis | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.2 | 0.0 ± 0.0 | 0.883 | 0.357 | 2.257 | 0.150 |
| Spirochaetes | 0.2 ± 0.2 | 0.8 ± 1.8 | 0.2 ± 0.4 | 0.1 ± 0.1 | 1.782 | 0.194 | 1.103 | 0.308 |
| TM7 incertae sedis | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.2 | 0.0 ± 0.0 | 0.063 | 0.804 | 0.305 | 0.587 |
| Mollicutes | 0.0 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.01 ± 0.1 | 2.336 | 0.139 | 0.389 | 0.541 |
| Verrucomicrobia | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.170 | 0.290 | ||
| Synergistia | 0.3 ± 0.6 | 1.3 ± 2.7 | 0.2 ± 0.5 | 0.1 ± 0.2 | 2.151 | 0.155 | 1.698 | 0.209 |
F, Firmicutes; HC, healthy control; HS, healthy site; P, Proteobacteria; RAS, recurrent aphthous stomatitis; US, ulcerated site.
RAS patients who presented with active ulceration at the time of recruitment but no ulcers at the visit for sample collection.
P < 0.05.
An additional ANOVA comparison was made between each of the 3 pairings of sample groups against the top 99.9% of genus-level matches (118 genera). Of a possible 411 comparisons, 9 achieved statistical significance, although some were numerically small genera. Statistically significant results are summarized in Table 2. Most notably, Bacteroidetes and Bacteroidales were increased in healthy sites of RAS patients when compared with HCs (mean ± SD, 4.1 ± 5.6 vs. 0.9 ± 1.6 reads, P = 0.041; 61.9 ± 88.6 vs. 12.9 ± 17.8 reads, P = 0.040, respectively). Ulcerated sites showed increased Porphyromonadaceae and Veillonellaceae when compared with HCs (17.5 ± 20.6 vs. 3.6 ± 4.3 reads, P = 0.014; 3.8 ± 7.3 vs. 1.9 ± 3.4 reads, P = 0.048, respectively), along with decreased Streptococcaceae (5.7 ± 4.0 vs. 2.2 ± 1.4 reads, P = 0.01). No statistically significant differences of these 3 families were noted between healthy sites in RAS patients and HCs.
Table 2.
Genus-level Statistical Comparisons That Reached Statistical Significance
| Percentage of 3,000 Patient Reads, Mean ± SD |
||||
|---|---|---|---|---|
| Genus | HC | RAS-HS | RAS-US | P Valuea |
| Bacteroidalesb | 12.9 ± 17.8 | 61.9 ± 88.6 | 0.04 | |
| Bacteroidetesb | 0.9 ± 1.6 | 4.1 ± 5.6 | 0.041 | |
| Caulobacter | 0.0 ± 0.0 | 0.1 ± 0.3 | 0.018 | |
| Akkermansia | 0.2 ± 0.5 | 0.0 ± 0.0 | 0.031 | |
| Porphyromonadaceaeb | 3.6 ± 4.3 | 17.5 ± 20.6 | 0.014 | |
| Bacillib | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.035 | |
| Streptococcaceaeb | 5.7 ± 4.0 | 2.2 ± 1.4 | 0.01 | |
| Veillonellaceaeb | 3.8 ± 7.3 | 1.9 ± 3.4 | 0.048 | |
| Clostridiab | 0.3 ± 0.8 | 0.1 ± 0.3 | 0.003 | |
HC, healthy control; HS, healthy site; RAS, recurrent aphthous stomatitis; US, ulcerated site.
Statistical comparison by analysis of variance. False discovery rate correction was not applied to the 411 comparisons.
Unclassified.
To explore clustering within patient sample groups, principal component analysis was undertaken on weighted Unifrac distances (Fig. 3). No distinct group clustering was observed within any sample group.
Figure 3.
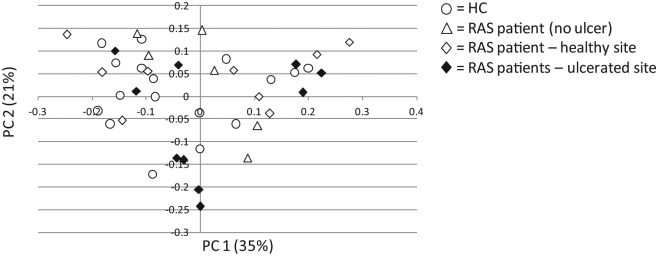
Bacterial beta diversity from high-throughput sequencing. Principal components analysis of weighted Unifrac distances for denoised, chimera-checked sequences for patient sample groups. Sample distribution is shown by 2 major principal components (PC1 and PC2) plotted in 2 dimensions. Healthy controls (HCs) are indicated as open circles; healthy sites from recurrent aphthous stomatitis (RAS) patients with active ulceration at the time of recruitment—but no ulcers at the visit for sample collection—are indicated as open triangles; healthy sites in RAS patients presenting with active ulceration are indicated as open diamonds; ulcerated sites in RAS patients are indicated as solid diamonds.
Discussion
This study is the first to show an association between RAS and imbalances of the oral mucosal microbiome. Numerous reviews have appraised the robustness of studies attributing an infectious basis (viral and bacterial) to the etiology of RAS and agreed that the evidence is contradictory (Jurge et al. 2006). This study suggests that imbalances of the oral mucosal microbiome, rather than individual infectious pathogens as suggested previously (Mansour-Ghanaei et al. 2005), may be implicated in the etiopathogenesis of RAS. Recent evidence suggests that mucosal microbiome changes may play a role in the etiology of chronic mucosal inflammatory conditions historically associated with individual infectious pathogens (Petersen and Round 2014).
We recruited RAS patients and matched HCs using highly stringent criteria to minimize known influences by oral and extraoral factors on the composition of oral mucosal microbiota. We also asked all recruits to comply with strict dietary and oral hygiene instructions for 72 h prior to sample collection. We then described the differences in composition of bacterial microbiota among healthy and ulcerated oral mucoase in RAS patients and compared the 2 with the microbiota of HC. The study is a novel exploratory investigation aimed to determine whether changes in the oral microbiota could be associated with RAS, a condition with poorly characterized mucosal microbiota data. Our approach was hypothesis generating, and findings will require replication and validation in larger cohorts.
In RAS patients presenting with ulcers >2 mm at the time of the visit for sample collection, DGGE analyses showed major qualitative differences in banding profiles, indicating that different bacterial communities were present in healthy versus ulcerated mucosal sites. This was in agreement with a previous report where the oral microbiota retrieved from ulcerated areas in RAS patients was found to differ from that of HCs (Marchini et al. 2007). Equally, phylum-level diversity comparisons of high-throughput sequencing findings revealed statistically significant differences between healthy and ulcerated mucosal sites of RAS patients in Firmicutes and Proteobacteria, 2 of the 6 major phyla containing around 96% of the taxa in the oral microbiome (Dewhirst et al. 2010). No statistically significant phylum-level differences were observed between HCs and healthy sites in RAS patients.
Diversity comparisons of lower level taxonomic groups revealed statistically significant differences among the 3 groups of samples. Total genera from the order Bacteroidales comprising several Gram-negative microorganisms of the normal colonic microbiota as well as oral opportunistic pathogens (Colombo et al. 2013) predominated in healthy sites from RAS patients over HCs. Porphyromonadaceae comprising species specifically associated with periodontal disease (Mysak et al. 2014) and Veillonellaceae for which diverse roles in oral health and disease are reported predominated only in ulcerated sites over HC. No quantitative differences of these 2 families were observed between healthy sites in RAS patients and HCs. Total Clostridia, to which the family of Veillonellaceae belongs, showed the reverse trend, with statistically significant higher abundance in HC over ulcerated sites. Equally, Streptococcaceae comprising commensal species associated with oral health (Liu et al. 2012) were more abundant in HCs over ulcerated sites but not in HCs over healthy sites in RAS patients. Consistently, the overall Bacilli population, which includes Streptococcaceae, showed higher abundance in HCs over ulcerated sites. The biological relevance of statistically significant changes associated to Caulobacter and Akkermansia was difficult to interpret, particularly in view of the low numerical abundance.
This study demonstrates mucosal microbiome changes in patients with idiopathic RAS—namely, increased abundance of species from the Bacteroidales order in mucosae of RAS patients not affected by active ulceration. Importantly, the observed increase of Bacteroidales was independent of the presence of medical conditions, other oral diseases, smoking, and reduced salivary flow rate. Potential influences by dietary and oral hygiene habits were also minimized. Oral hygiene instructions included suspension of flossing for 3 participants (1 HC and 2 RAS) who used floss as part of their routine oral hygiene. We acknowledge that suspension of flossing may influence the composition of oral mucosal microbiota in healthy individuals.
Increased Porphyromonadaceae and Veillonellaceae species were seen in ulcerated sites only, suggesting that these changes are unlikely to be involved in initiation of RAS. Yet, recent studies have shown the ability of P. gingivalis to inhibit phagocytosis of otherwise susceptible bacteria and hence promote dysbiosis itself (Maekawa et al. 2014). The coinciding decrease of Streptococcaceae species was equally observed in active ulcers only. Streptococci-based products to repopulate the oral environment with “beneficial species” have been marketed for control of halitosis. The potential of these products for controlling oral inflammatory conditions has been suggested, although randomized clinical trials to test their efficacy have not been carried out (Pradeep et al. 2014). The work presented here shows no evidence of increased abundance of Streptococcaceae species in healthy subjects over healthy mucosae in RAS patients and therefore does not support the use of streptococci-based products to prevent recurrence of RAS. This, however, does not exclude a role for these products in aiding restoration of oral health following acute exacerbations.
While the observed increase of Bacteroidales in RAS patients (including those who presented with no active ulceration at the time of sample collection) suggests a microbial role in initiation of the disease, this study does not provide clear data on causality as yet. A comparison of the mucosal microbiome in patients with idiopathic RAS and those with recurrent oral ulceration associated with similar immune pathogenesis but different etiology (e.g., underlying systemic conditions such as iron deficiency; Challacombe et al. 1983) may provide data on causality. Future studies should also investigate the role of the oral mycobiome (Ghannoum et al. 2010) in etiopathogenesis of RAS. Metagenomic studies of the oral microbiome in RAS patients, as recently reported for healthy individuals (Jorth et al. 2014), will define conserved changes in metabolic and virulence gene expression of bacterial communities associated with RAS.
Author Contributions
K. Hijazi, contributed to conception, design, and data interpretation, drafted and critically revised the manuscript; T. Lowe, J. Foley, contributed to design, critically revised the manuscript; C. Meharg, contributed to data acquisition and analysis, drafted the manuscript; S.H. Berry, contributed to data acquisition and analysis, critically revised the manuscript; G.L. Hold, contributed to data acquisition and analysis, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We thank all patients and healthy volunteers for contributing to this study, as well as Moira Munro, Angela Reid, and Haleigh Scott for valuable assistance in patient recruitment.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This work was funded by Tenovus Scotland (registered charity SC009675, Glasgow, UK), award G12/09.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Al-Omiri MK, Karasneh J, Alhijawi MM, Zwiri AM, Scully C, Lynch E. 2014. Recurrent aphthous stomatitis (RAS): a preliminary within-subject study of quality of life, oral health impacts and personality profiles. J Oral Pathol Med [epub ahead of print 26 August 2014] in press. doi: 10.1111/jop.12232 [DOI] [PubMed] [Google Scholar]
- Altenburg A, Abdel-Naser MB, Seeber H, Abdallah M, Zouboulis CC. 2007. Practical aspects of management of recurrent aphthous stomatitis. J Eur Acad Dermatol Venereol. 21(8):1019–1026. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challacombe SJ, Scully C, Keevil B, Lehner T. 1983. Serum ferritin in recurrent oral ulceration. J Oral Pathol. 12(4):290–299. [DOI] [PubMed] [Google Scholar]
- Colombo AV, Barbosa GM, Higashi D, di Micheli G, Rodrigues PH, Simionato MR. 2013. Quantitative detection of Staphylococcus aureus, Enterococcus faecalis and Pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol. 62(Pt 10):1592–1600. [DOI] [PubMed] [Google Scholar]
- Department of Health and the British Association for the Study of Community Dentistry. 2009. Delivering better oral health: an evidence-based toolkit for prevention [accessed 2014 December 3]. http://www.oralhealthplatform.eu/sites/default/files/field/document/NHS_Delivering%20Better%20Oral%20health.pdf. 2nd ed. Sec 4.
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C, Barros F, Sugaya N, Nunes F, Borra R. 2012. Differential expression of toll-like receptor mRNAs in recurrent aphthous ulceration. J Oral Pathol Med. 41(1):80–85. [DOI] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6(1):e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R, Russell RK, Reiff C, Louis P, McIntosh F, Berry SH, Mukhopadhya I, Bisset WM, Barclay AR, Bishop J, et al. 2012. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn’s but not in ulcerative colitis. Am J Gastroenterol. 107(12):1913–1922. [DOI] [PubMed] [Google Scholar]
- Hasan A, Childerstone A, Pervin K, Shinnick T, Mizushima Y, Van der Zee R, Vaughan R, Lehner T. 1995. Recognition of a unique peptide epitope of the mycobacterial and human heat shock protein 65–60 antigen by T cells of patients with recurrent oral ulcers. Clin Exp Immunol. 99(3):392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietanen J, Hayrinen-Immonen R, Al-Samadi A, Trokovic N, Koskenpato K, Konttinen YT. 2012. Recurrent aphthous ulcers: a Toll-like receptor-mediated disease? J Oral Pathol Med. 41(2):158–164. [DOI] [PubMed] [Google Scholar]
- Hold GL, Smith EA, Rappé MS, Maas EW, Moore ER, Stroempl C, Stephen JR, Prosser JI, Birkbeck TH, Gallacher S. 2001. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiology Ecology. 37(2):161–173. [Google Scholar]
- Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 5(2):e01012–e01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurge S, Kuffer R, Scully C, Porter SR. 2006. Mucosal disease series: number VI. Recurrent aphthous stomatitis. Oral Dis. 12(1):1–21. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 43(8):3944–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, et al. 2012. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 7(6):e37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, Hashim A, Hoch S, Curtis MA, Nussbaum G, et al. 2014. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 15(6):768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour-Ghanaei F, Asmar M, Bagherzadeh AH, Ekbataninezhad S. 2005. Helicobacter pylori infection in oral lesions of patients with recurrent aphthous stomatitis. Med Sci Monit. 11(12):CR576–CR579. [PubMed] [Google Scholar]
- Marchini L, Campos MS, Silva AM, Paulino LC, Nobrega FG. 2007. Bacterial diversity in aphthous ulcers. Oral Microbiol Immunol. 22(4):225–231. [DOI] [PubMed] [Google Scholar]
- Miyamoto NT, Jr, Borra RC, Abreu M, Weckx LL, Franco M. 2008. Immune-expression of HSP27 and IL-10 in recurrent aphthous ulceration. J Oral Pathol Med. 37(8):462–467. [DOI] [PubMed] [Google Scholar]
- Mysak J, Podzimek S, Sommerova P, Lyuya-Mi Y, Bartova J, Janatova T, Prochazkova J, Duskova J. 2014. Porphyromonas gingivalis: major periodontopathic pathogen overview. J Immunol Res 2014:476068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Round JL. 2014. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 16(7):1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep K, Kuttappa MA, Prasana KR. 2014. Probiotics and oral health: an update. SADJ. 69(1):20–24. [PubMed] [Google Scholar]
- Riggio MP, Lennon A, Wray D. 2000. Detection of Helicobacter pylori DNA in recurrent aphthous stomatitis tissue by PCR. J Oral Pathol Med. 29(10):507–513. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 9(5):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh S, Gupta D, Pallagatti S, Singla I, Gupta R, Goel V. 2013. Role of topical drugs in treatment of oral mucosal diseases: a literature review. N Y State Dent J. 79(6):58–64. [PubMed] [Google Scholar]
- Slebioda Z, Szponar E, Kowalska A. 2013. Recurrent aphthous stomatitis: genetic aspects of etiology. Postepy Dermatol Alergol. 30(2):96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Chen HM, Cheng SJ, Wang YP, Chang JY, Wu YC, Chiang CP. 2014. Significant association of deficiencies of hemoglobin, iron, vitamin B12, and folic acid and high homocysteine level with recurrent aphthous stomatitis. J Oral Pathol Med [epub ahead of print 22 July 2014] in press. doi: 10.1111/jop.12241 [DOI] [PubMed] [Google Scholar]
- Tappuni AR, Kovacevic T, Shirlaw PJ, Challacombe SJ. 2013. Clinical assessment of disease severity in recurrent aphthous stomatitis. J Oral Pathol Med. 42(8):635–641. [DOI] [PubMed] [Google Scholar]
- Taylor LJ, Bagg J, Walker DM, Peters TJ. 1992. Increased production of tumour necrosis factor by peripheral blood leukocytes in patients with recurrent oral aphthous ulceration. J Oral Pathol Med. 21(1):21–25. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Hansen R, Berry SH, Hope ME, Murray GI, Mukhopadhya I, McLean MH, Shen Z, Fox JG, El-Omar E, et al. 2011. Enterohepatic helicobacter in ulcerative colitis: potential pathogenic entities? PLoS One. 6(2):e17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans AD, De Vos WM. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 64(10):3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.