A little over 30 years ago, the world of periodontal research was a very different place from the one that we inhabit now. We still had to wait over a decade before the first full genome sequence of any bacterium was solved (Fleischmann et al., 1995), several years after that before a periodontal bacterium genome appeared (Nelson et al., 2003), and another 2 decades before the human genome (Lander et al., 2001) was unfurled with much fanfare across the World Wide Web—which itself was not even a twinkle in the eye of Tim Berners-Lee in the early 1980s. While there was a growing awareness of the power of microbial genetics and the techniques of molecular biology in periodontal research, these were still in their infancy. In those bygone days, the cloning and full sequencing of a bacterial gene was considered a very respectable project for a PhD thesis. In today’s reality, this is accomplished in the blink of an eye by a computer-driven search of the cornucopia of DNA sequence information held in publically available databases or, occasionally, by a trip to your local DNA sequencing facility where, for the cost of a few dollars, you can obtain your sequence information the next morning, allowing you to press on with the biological question that drove your enquiry in the first place. Have a nice day!
However, while that epoch was clearly different from today’s informatics-driven age, it certainly did not lack innovation and excitement from a periodontal microbiology perspective. Groups of oral microbiologists around the world were using their newly acquired skills in anaerobic culture and biochemical techniques to travel deep into an unexplored world of microbial diversity encountered in supra- and subgingival plaque (e.g., Moore et al., 1983). In a seemingly endless voyage of discovery, new bacterial genera and their associated species were encountered, characterized, and named. Upon further investigation using evermore discriminatory techniques, these new bacterial types were then further subdivided into yet more species, subspecies, and, in some instances, individual clonal types—all of which added to our understanding of the richness of these microbial populations. The driving force for much of this endeavor centered on the search for the holy grail of oral microbiology at the time: the bacterial agent (or agents) of periodontal disease. With each new microbial discovery within the subgingival microbial plaque of periodontally diseased individuals, another name was added to the growing list of putative periodontal pathogens and in some instances lauded as the key to unlocking the microbial etiology of the disease. While these intrepid voyagers of the subgingival microbial world went merrily about their work, taking glee with each new finding and with each new implausibly difficult name to pronounce, other groups of investigators, also interested in the etiopathogenesis of the disease, sat darkly brooding on the sidelines.
The non-microbiological periodontal research community took a rather different, not to say disparaging, view of this cacophony of noise emanating from the microbiologists: The remarkable complexity of the periodontal microbiota may undoubtedly be the case, but the relevance of all these newly described organisms to the actual disease process was highly debatable. Indeed, possibly because it was rather difficult to fathom what was going on in oral microbiology at the time if you were not intimately involved in the science (or, possibly borne by the movement of the research spotlight away from their own endeavors), these investigators developed a rather scornful descriptor of their microbiology colleagues—“The Bug of the Month Club”—intended to convey the inappropriate promotion of this apparently bewildering array of periodontal pathogen new kids on the block. With the benefit of hindsight, this censorious attitude seems deeply ironic. Within a few short years, one of these groups of non-microbiology investigators—the oral immunologists—was embarking on their own voyage of discovery across an unchartered landscape following the breakthroughs in the field of intercellular signaling and the explosion of new molecules associated with these processes. Newly described cytokines, chemokines, and a whole host of inflammatory mediators burst onto the periodontal scene and were variously held up to be the key to the etiopathogenesis of the disease, analogous to the claims for the new periodontal pathogens a few years before (e.g., Page, 1991). That the microbiologists did not retaliate with their own disparaging terminology probably owes more to the fact that “Cell Signaling Molecule of the Month Club” does not have quite the same resonance as the earlier epithet rather than acquiescence to the new order.
A sense of microbiological order was restored, however, in large part by the hugely influential studies of Socransky and colleagues (1998), which have shaped our thinking of periodontal microbiology over the last 15 yr. The concept that complexes of specific subgingival bacteria are directly associated with the clinical periodontal status arose from large-scale analyses of subgingival plaque using DNA:DNA hybridization techniques to detect the presence of up to 40 different bacterial species—many of which were initially described by the pioneer voyages of the 1980s. In their simplest interpretation, these studies led to the description of the red and orange complexes of bacteria that appeared to be the most highly associated with destructive disease. The acceptance of this concept is evidenced by the vast number of citations to this work in the literature, the huge research focus on the microbiological properties of the 3 organisms of the red complex (Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola), and the near-universal understanding of the term red complex bacterium at dental undergraduate/postgraduate and periodontal practitioner levels across the globe. However, it is becoming increasingly clear that we need to change or at least modify this paradigm to take account of more recent microbiological findings, largely developed through the use of new technologies that overcome some of the limitations inherent in the original investigations by Socransky et al. almost 20 yr ago.
Foremost of these limitations is that the original investigations were performed with only a restricted repertoire of target bacterial species representing, at most, no more than 5% of the total number of bacteria capable of inhabiting this ecologic niche. Equally important, all the bacteria chosen for this panel are cultivatable under standard laboratory conditions, although it is clear that the non-cultivatable bacteria of the subgingival microbiota represent a significant proportion. The stupendously effective power of sequence variation in the 16S ribosomal RNA gene to discriminate among bacterial taxa was and remains at the heart of these new techniques. Supremely sensitive and culture independent, 16S rRNA sequencing has been used to define the human oral microbiome (Dewhirst et al., 2011), perform comparative analyses of periodontal health and disease (Colombo et al., 2009), and, more recently, through pyrosequencing methodologies, undertake in-depth investigation of the entire microbial population structure of periodontal samples to a level that was unthinkable only a few years ago (Griffen et al., 2012) . Almost inevitably, these studies are demonstrating that limiting the number of putative periodontal pathogens—that is, organisms whose presence and level correlate with the disease process—to a handful of bacterial species is invalid.
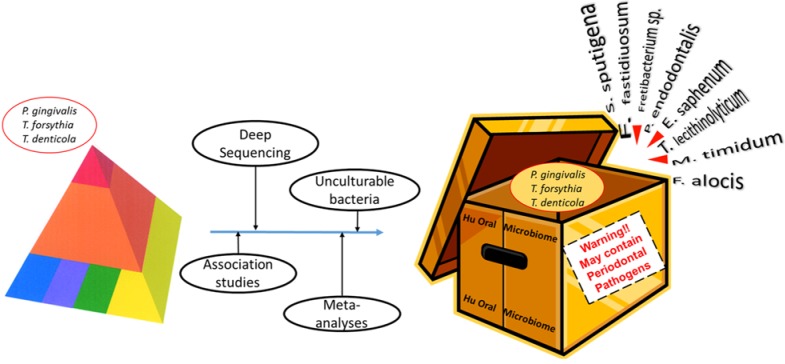
The review paper by Pérez-Chaparro et al. in this issue of the journal points a spotlight on this matter by performing a meta-analysis of 41 investigations over the last 15 yr that have demonstrated an association between periodontal disease and the presence of bacterial species outside the conventional list of presumed periodontal pathogens. The studies include some cultural and some DNA:DNA hybridization investigations, but the majority employ culture-independent polymerase chain reaction or DNA sequencing methodologies. Pérez-Chaparro et al. highlight the potential involvement of 17 new additional candidate organisms based on their reported association with periodontal disease in 3 to 5 independent investigations. These organisms include several members of bacterial phyla previously implicated but also members of the Archae domain of life, which has not previously been associated with any infectious disease of humans. Thus, in one meta-analysis stroke, the list of organisms that we should consider to be putative periodontal pathogens has more than doubled to approximately 30 separate taxa! The exploits of “The Bug of the Month Club” seem positively pedestrian by comparison (Fig).
Figure.
The application of new technologies to the analysis of the periodontal microbiome is leading to an expanded repertoire of putative periodontal pathogens.
How do we make sense of these newfound riches of putative periodontal pathogens? One approach would be to seek, through further association studies, to draw ever larger red and orange circles, which would incorporate these Johnny-come-lately bacteria within the existing framework of complexes originally described by Socransky et al. Alternatively, one could envisage a whole kaleidoscope of new complexes, each one jockeying for position as the most positively associated group of organisms with the periodontal disease process. In both cases, acknowledgment of the expanded list of putative periodontal pathogens does not lead to a paradigm shift but rather the dexterous use of a set of crayons. However, Pérez-Chaparro et al. suggest one rather different potential interpretation of their meta-analysis, in which the observed changes in the microbiology of periodontal disease represent a dysbiosis or community-wide perturbation of the entire microbial population structure of subgingival plaque. In this context, it is relatively straightforward to accommodate an expanded repertoire of organisms in the disease-associated category because this is a natural consequence of dysbiosis. One could go further—by analogy, to the current understanding of the microbial etiology of inflammatory diseases elsewhere in the gastrointestinal tract—and suggest that periodontal disease is a function of the challenge presented by this entire dysbiotic microbial community rather than necessarily by the actions of a limited number of putative periodontal pathogens. Under this paradigm, a more complete understanding of the mechanism by which this dysbiosis occurs, how a dysbiotic community structure interacts with the host, and, conversely, how a dysbiotic state may be reversed to the symbiotic state characteristic of periodontal health may prove useful avenues of research.
Acknowledgments
The author wishes to thank Juliet Ellwood for assistance in the preparation of this manuscript.
Footnotes
The author received no financial support and declares no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. (2009). Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol 80:1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. (2010). The human oral microbiome. J Bacteriol 192:5002-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Ranney RR. (1983). Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect Immun 42:510-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. (2003). Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J Bacteriol 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RC. (1991). The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodontal Res 26(3 Pt 2):230-242. [DOI] [PubMed] [Google Scholar]
- Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, et al. (2014). Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res 93:846-858. Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25:134-144. [DOI] [PubMed] [Google Scholar]