Mitochondria serve as the powerhouse of cells, respond to cellular demands and stressors, and play an essential role in cell signaling, differentiation, and survival. Aberrant mitochondria function has been linked to diverse and complex human diseases such as neurodegenerative diseases, cancers, myopathies, premature aging, and metabolic syndromes (Nunnari and Suomalainen, 2012). For example, there is clear evidence of a rapid and sustained loss of mitochondrial function following traumatic injury to the central nervous system (CNS) (Sullivan et al., 2005; McEwen et al., 2011). However, our understanding of how compromised mitochondrial activity impacts overall cellular function in the CNS is far from complete.
Recent studies indicate that a dynamic relationship exists between mitochondrial function and microRNA (miRNA) activity (Lung et al., 2006; Kren et al., 2009; Bian et al., 2010; Bandiera et al., 2011; Barrey et al., 2011; Mercer et al., 2011; Das et al., 2012; Sripada et al., 2012; Zhang et al., 2014; Shinde and Bhadra, 2015; Wang et al., 2015). MiRNAs are small (18–25 nucleotides) non-coding RNA molecules that regulate gene expression at the post-transcriptional level, and function as important mediators of many CNS biological processes including development, synaptic plasticity, and neurodegeneration (Liu and Xu, 2011). Dysregulation of certain miRNAs is associated with many human diseases including cancer, heart disease, diabetes, immune dysfunction, and neurodegenerative diseases (Calin and Croce, 2006; Nelson et al., 2008; Hata, 2013). MiRNAs have been shown to participate in important neuronal functions, for example, miR-124, a brain specific miRNA is a key player in promoting neuronal differentiation. In addition, several miRNAs (e.g., miR-107, miR-29, miR-155, miR-223, and miR-21) are known to be involved with various neurodegenerative diseases and CNS injuries. For further reading of miRNA function in the CNS, readers are referred to an excellent review by Liu and Xu (2011). MiRNAs mediate gene expression by directing their target mRNA for degradation or translational repression, and the ability of a single miRNA or miRNA family to regulate the post-transcriptional expression of hundreds of genes (Lewis et al., 2005) makes them ideal candidates for coordinating complex gene expression programs, including modifying a cell's response to stresses.
miRNA association with mitochondria: It is well established that miRNAs are transcribed in nucleus and transported into cytoplasm, which is a primary site of action. However, there is growing evidence that miRNAs also are present in or associated with other organelles or unstructured cytoplasmic foci, such as mitochondria, endoplasmic reticulum (ER), processing bodies (P-bodies), stress granules, multivesicular bodies, and exosomes (Nguyen et al., 2014). It can be suggested from these recent observations that miRNA-mediated gene regulation may be controlled within different cellular compartments, and that this miRNA-organelle crosstalk allows for more selective responses to specific cellular demands. Several studies have demonstrated the presence of miRNAs either within (Lung et al., 2006; Kren et al., 2009; Bian et al., 2010; Bandiera et al., 2011; Barrey et al., 2011; Mercer et al., 2011; Das et al., 2012; Zhang et al., 2014; Shinde and Bhadra, 2015) or associated with (Sripada et al., 2012; Wang et al., 2015) mitochondria isolated from various cell types and tissues, including the CNS (see Wang et al., 2015 for discussion). In addition, the miRNA machinery proteins, Argonaute (AGO) and Dicer have been detected in mitochondria (Bandiera et al., 2011; Das et al., 2012; Wang et al., 2015), indicating the presence of an active miRNA ribonucleoprotein complex (miRNP). The majority of these mitochondria associated miRNAs are known to be nuclear-encoded, while a few are predicted to originate from the mitochondrial genome (Lung et al., 2006; Barrey et al., 2011; Sripada et al., 2012; Shinde and Bhadra, 2015). Given the small genome of mitochondria and the presence of minimal non-coding DNA, the characterization and function of this group of intra-mitochondrial miRNA requires further investigation.
One of the presumed functions of mitochondrial miRNA is the regulation of mitochondrial gene expression. In support of this, studies performed using muscle cells and heart tissues have identified nuclear-encoded miRNAs in mitochondria that directly regulate mitochondrial proteins. For example, miR-181c is enriched in mitochondria and it targets cytochrome c oxidase subunit 1 (COX1) in rat cardiomyotubes (Das et al., 2012). Another nuclear-generated mitochondrial miRNA, miR-1 also targets COX1 and was found to increase in mitochondria during muscle cell differentiation (Zhang et al., 2014). Surprisingly, miR-1 enhanced, rather than inhibited, COX1 protein translation. These same authors determined that this unconventional action of miR-1 requires AGO2 but not GW182 (glycine-tryptophan protein of 182 kDa), which is essential for cytoplasmic miRNA gene repression but is absent in mitochondria. In a separate study, computational analysis of mitochondria enriched miRNAs isolated from human skeletal muscle cells predicted 80 putative target sites in the mitochondrial genome (Barrey et al., 2011). There is additional evidence supporting the role of miRNA in regulating mitochondrial gene expression and readers are referred to several excellent reviews (Li et al., 2012; Bienertova-Vasku et al., 2013; Duarte et al., 2014). The remainder of this Perspective focuses on the role of mitochondria in regulating cellular miRNA activities.
Mitochondria are prime candidates for regulating miRNA function: Regulation of gene expression is a tightly controlled process that involves many factors both at the transcription and post-transcriptional levels. MiRNAs broadly participate in post-transcriptional gene regulation in almost all cellular events. MiRNAs regulate gene expression by complementary binding to their target transcript, with the critical region for miRNA binding being nucleotides 2–8 from the 5′ end of the miRNA, termed the ‘seed’ sequence (Lewis et al., 2003, 2005). In addition to sequence matching, the thermodynamics of miRNA:mRNA interactions and the accessibility of the target mRNA are thought to be important factors (Kertesz et al., 2007; Wexler et al., 2007). Nevertheless, the mechanisms by which a given miRNA determines its target and the events controlling miRNA function in response to cellular demands are not well understood.
Mitochondria have been shown to interact with several cellular compartments, organelles, and cytoplasmic foci including the cytoskeleton, ER, and P-bodies (Boldogh and Pon, 2006; Huang et al., 2011; Klecker et al., 2014). These interactions create a dynamic network important for cellular energy distribution, signaling, and homeostasis. Given the rapid response capabilities of mitochondria in many cellular functions, we suggest that mitochondria are prime candidates for regulating miRNA activity and function. This is an attractive hypothesis as it allows for control of translational specificity in response to unique cellular requirements across cellular domains. In this context, mitochondria can be viewed as local rheostats that respond quickly to changes in cellular demands, both physiological and pathophysiological. As such, this hypothesis postulates that appropriate miRNA responses are determined, in part, by rapid changes in mitochondrial function due to cellular demands, stresses or perturbations. In support of this, compromising mitochondrial function with an uncoupling agent has been shown to result in delocalization of AGO proteins from P-bodies leading to a subsequent decrease in miRNA mediated RNAi efficiency (Huang et al., 2011).
Potential roles of mitochondria in regulating miRNA functions: The association of miRNAs with mitochondria raises the possibility that mitochondria regulate miRNA activities in a manner that is specific to unique cellular demands. As such, we hypothesize that mitochondria can act in several ways (Figure 1), including but not limited to: 1) Mitochondria, the warehouse; 2) Mitochondria, the vehicle; and 3) Mitochondria, the network.
Figure 1.
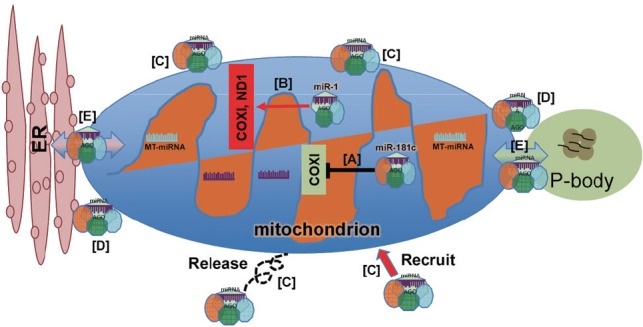
Potential roles of mitochondria in regulating miRNA activities.
The interaction of mitochondria and miRNAs may potentially extend beyond their respective functions. MiRNAs may translocate into mitochondria in order to modulate mitochondrial gene expression either by suppressing (A) or up-regulating (B) genes that are key components of mitochondrial function. In this scenario, miR-181c has been shown to inhibit COX1 expression (A), while miR-1 has been shown to increase (B) COX1 expression (see text for details). Emerging evidence also suggests that mitochondria may participate in regulating miRNA activities by serving as a storage warehouse in which miRNA or miRNA complexes may be recruited and released as needed (C), or as a vehicle to deliver miRNA and its complex to locations throughout cytoplasm (D), or as a network for distributing and exchanging miRNA and their components with other organelles and cytoplasmic foci (E). In this regard, mitochondria-associated miRNAs can utilize the existing network of mitochondrial interaction with other organelles and cellular components, as well as the sensitivity of mitochondria to make appropriate responses in gene expression based on signaling events occurring within the cellular environment. COX1: Cytochrome c oxidase subunit 1; ER: endoplasmic reticulum; ND1: NADH-ubiquinone oxidoreductase chain 1.
1) Mitochondria, the warehouse. MiRNAs may be ‘stored’ in association with mitochondria for “on demand” use as needed. The number of miRNA reported to be present within or associated with mitochondria ranges from 3 to 428 depending on tissue and cell types, with many being highly enriched in mitochondria relative to the cytoplasm. For example, miR-155 is enriched in mitochondrial fractions from mouse liver, and levels are increased following streptozotocin treatment, which induces type I diabetes and impairs mitochondrial function (Bian et al., 2010). We have recently shown that levels of several miRNAs, including miR-146a, miR-142-3p, miR-142-5p, are preferentially enriched in highly purified hippocampal mitochondria under normal physiological conditions (Wang et al., 2015). Interestingly, the mitochondrial levels of these miRNA are dramatically reduced following traumatic brain injury at time points when mitochondrial function is significantly compromised. In the same study, we observed that miR-155 and miR-223 are both elevated in mitochondria at the same post-injury time points. These observations suggest that certain miRNA, or miRNA families, associate with or dissociate from hippocampal mitochondria, possibly in response to cellular stressors impacting mitochondrial function.
2) Mitochondria, the vehicle. Mitochondria are distributed along microtubules and their shapes and positions are largely influenced by events controlling mitochondrial fusion, fission, motility and positional tethering (Lackner, 2013). These dynamic processes are highly regulated by and integrated with cellular signaling pathways and stress responses. As such, mitochondria can serve as vehicles to deliver associated miRNAs to the appropriate cellular compartments to impact a range of miRNA-mediated gene regulations. This is particularly important in the CNS where many fundamental neuronal functions, such as axonal transport and synaptic plasticity, require localized and highly regulated mRNA translation (Holt and Schuman, 2013).
3) Mitochondria, the network. In this scenario, associated miRNAs utilize the dynamic network of mitochondria interacting with other mitochondria, as well as other cellular compartments (e.g., ER and P-bodies). This allows for the delivery or exchange of ‘cargo’ miRNA, thus creating a precise control mechanism by which cellular gene expression may be regulated within the appropriate context and in a specific cellular domain.
The biological significance of mitochondria-miRNA interactions, and the mechanism(s) regulating them are largely unknown at this time. However, factors that regulate mitochondria dynamics, and/or stress-related events that alter mitochondria function may play an important role. Examples of such factors and events would include locally altered concentrations of Ca2+, the formation and/or presence of free radicals, and changes in ATP production/levels. Moreover, the degree of cellular stress and subsequent impact on mitochondrial function may determine the specific mitochondria-miRNA interaction pattern necessary to generate a response appropriate for the stress event. Therefore, cellular miRNA activity and the downstream expression of their corresponding targets would be intimately linked to mitochondrial function.
Functional importance of miRNA-mitochondria crosstalk in CNS: Various organelles arose for their distinct cellular functions. It is also true that organelles corporately engage in other non-canonical roles to orchestrate complex and compartmentalized cellular functions. This is especially important in cells of the CNS in which the spatial and temporal regulation of local protein synthesis varies across cellular compartments. It is known that neuronal mRNA transportation and local protein synthesis is crucial for many CNS events including development and plasticity. It is interesting to note that noncoding RNAs, including miRNAs, have been identified in various neuronal compartments including synaptosomes. Considering the function of miRNA in regulating mRNA stability and translation, it is conceivable that certain miRNA may have a very significant role in controlling local protein synthesis. Because many neuronal functions are dependent on mitochondria, the trafficking of these organelles to various cell compartments would allow for miRNA-mRNA-protein responses that are unique to local environmental cues.
Emerging evidence supporting the interaction and crosstalk between mitochondria and miRNA points to a highly novel dimension of gene regulation that is particularly appropriate for CNS function. We propose that miRNA-mitochondria crosstalk provides a previously undiscovered mechanism for rapid, specific, and spatially appropriate gene regulation in response to cellular cues. Further understanding these interactions will be important for advancing our knowledge of gene regulation mechanisms in the CNS.
We apologize to our colleagues whose published works were not cited or discussed due to space limitations. Supported by an endowment to JES from Cardinal Hill Rehabilitation Hospital.
References
- Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Li LM, Tang R, Hou DX, Chen X, Zhang CY, Zen K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010;20:1076–1078. doi: 10.1038/cr.2010.119. [DOI] [PubMed] [Google Scholar]
- Bienertova-Vasku J, Sana J, Slaby O. The role of microRNAs in mitochondria in cancer. Cancer Lett. 2013;336:1–7. doi: 10.1016/j.canlet.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte FV, Palmeira CM, Rolo AP. The role of microRNAs in mitochondria: small players acting wide. Genes (Basel) 2014;5:865–886. doi: 10.3390/genes5040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A. Functions of microRNAs in cardiovascular biology and disease. Annu Rev Physiol. 2013;75:69–93. doi: 10.1146/annurev-physiol-030212-183737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, Dautry F, Weil D. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem. 2011;286:24219–24230. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Klecker T, Bockler S, Westermann B. Making connections: interorganelle contacts orchestrate mitochondrial behavior. Trends Cell Biol. 2014;24:537–545. doi: 10.1016/j.tcb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL. Determining the shape and cellular distribution of mitochondria: the integration of multiple activities. Curr Opin Cell Biol. 2013;25:471–476. doi: 10.1016/j.ceb.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li P, Jiao J, Gao G, Prabhakar BS. Control of mitochondrial activity by miRNAs. J Cell Biochem. 2012;113:1104–1110. doi: 10.1002/jcb.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NK, Xu XM. MicroRNA in central nervous system trauma and degenerative disorders. Physiol Genomics. 2011;43:571–580. doi: 10.1152/physiolgenomics.00168.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen ML, Sullivan PG, Rabchevsky AG, Springer JE. Targeting mitochondrial function for the treatment of acute spinal cord injury. Neurotherapeutics. 2011;8:168–179. doi: 10.1007/s13311-011-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Brenu EW, Staines DR, Marshall-Gradisnik SM. MicroRNAs in the intracellular space, regulation of organelle specific pathways in health and disease. MicroRNA. 2014;3:98–107. doi: 10.2174/2211536604666141218154252. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde S, Bhadra U. A complex genome-microRNA interplay in human mitochondria. Biomed Res Int 2015. 2015 doi: 10.1155/2015/206382. 206382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PG, Rabchevsky AG, Waldmeier PC, Springer JE. Mitochondrial permeability transition in CNS trauma: cause or effect of neuronal cell death? J Neurosci Res. 2005;79:231–239. doi: 10.1002/jnr.20292. [DOI] [PubMed] [Google Scholar]
- Wang WX, Visavadiya NP, Pandya JD, Nelson PT, Sullivan PG, Springer JE. Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury. Exp Neurol. 2015;265:84–93. doi: 10.1016/j.expneurol.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler Y, Zilberstein C, Ziv-Ukelson M. A study of accessible motifs and RNA folding complexity. J Comput Biol. 2007;14:856–872. doi: 10.1089/cmb.2007.R020. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]


