Abstract
MicroRNA-124 (miR-124) is abundantly expressed in neurons in the mammalian central nervous system, and plays critical roles in the regulation of gene expression during embryonic neurogenesis and postnatal neural differentiation. However, the expression profile of miR-124 after spinal cord injury and the underlying regulatory mechanisms are not well understood. In the present study, we examined the expression of miR-124 in mouse brain and spinal cord after spinal cord injury using in situ hybridization. Furthermore, the expression of miR-124 was examined with quantitative RT-PCR at 1, 3 and 7 days after spinal cord injury. The miR-124 expression in neurons at the site of injury was evaluated by in situ hybridization combined with NeuN immunohistochemical staining. The miR-124 was mainly expressed in neurons throughout the brain and spinal cord. The expression of miR-124 in neurons significantly decreased within 7 days after spinal cord injury. Some of the neurons in the peri-lesion area were NeuN+/miR-124−. Moreover, the neurons distal to the peri-lesion site were NeuN+/miR-124+. These findings indicate that miR-124 expression in neurons is reduced after spinal cord injury, and may reflect the severity of spinal cord injury.
Keywords: nerve regeneration, spinal cord injury, microRNA, spinal cord, in situ hybridization, immunohistochemistry, digoxin, NeuN protein, brain, neural plasticity, repair, apoptosis, NSFC grants, neural regeneration
Introduction
Spinal cord injury is a traumatic event with a poor prognosis affecting mobility and resulting in early mortality. Current understanding of the mechanisms underlying spinal cord injury is limited, and traditional therapeutic methods lack a molecular approach to prevent the loss of sensory function and paralysis (Strickland et al., 2011). Investigators have previously reported that altered gene expression plays a major role in the pathogenesis of secondary spinal cord injury (Bareyre et al., 2003; De et al., 2005), but there is limited knowledge of the regulatory networks that control gene expression.
A novel class of tiny RNA molecules, microRNAs, are involved in the regulation of mRNA stability and translation, heterochromatin formation, genome rearrangement and gene regulation (Gu et al., 2014). MicroRNA-124 (miR-124) is the most abundant microRNA in the adult mammalian brain, accounting for 25-48% of all brain microRNAs (Doeppner et al., 2013). The sequence of mature miR-124 is highly conserved from worm to human, and the number of predicted potential target genes for miR-124 exceed 1,000 (Lewis et al., 2005). miR-124 is present at undetectable or very low levels in neural progenitors, but is highly expressed in differentiating and mature neurons (Deo et al., 2006).
In this study, we examined the expression of miR-124 in mature neurons throughout the central nervous system by in situ hybridization (ISH). We found that miR-124 expression significantly decreased after spinal cord injury in the crush model using quantitative RT-PCR. ISH combined with NeuN immunohistochemistry (IHC) revealed that miR-124 was significantly downregulated in neurons in the peri-lesion area, but not in the distal grey matter of the spinal cord. These results suggest that the absence of miR-124 in neurons might reflect cellular damage after spinal cord injury.
Materials and Methods
Experimental animals
A total of 45 adult male C57BL/6 mice, aged 6–8 weeks, weighing 22–29 g (Shanghai Laboratory Animal Center, Shanghai, China) were used in this study. All mice were housed in a specific pathogen-free environment at the animal facility of the Institute of Neurosciences, the Fourth Military Medical University of Chinese PLA (Xi’an, Shaanxi Province, China; animal license No. 130003). All procedures were approved by the Animal Care and Use Committee of the Fourth Military Medical University of Chinese PLA in China.
Animal grouping
The 45 mice were randomly divided into three groups. Three mice comprised the ISH group for miR-124 ISH staining and six comprised the spinal cord injury ISH/IHC group for miR-124 ISH in combination with NeuN IHC on day 7 after spinal cord injury. The remaining 36 mice with spinal cord injury constituted the PCR group for assessing miR-124 expression. These mice were randomly divided into sham operation and 1, 3 and 7 days after injury groups, with nine mice in each group.
Spinal cord crush injury
All animal surgical procedures were performed under sodium pentobarbital anesthesia (50 mg/kg, intraperitoneally; Sigma Aldrich, St. Louis, MO, USA) using a Leica M651 MSD surgical microscope (Leica, Wetzlar, Germany). A graded crush spinal cord injury model was established with forceps (Plemel et al., 2008). Briefly, laminectomy was carried out at the T8–10 vertebral level, leaving the dura intact. Each mouse was subjected to 20-second spinal cord crush at the level of T9 by compressing the cord laterally from both sides with #5 forceps (66 Vision Medical Apparatus Co., Ltd., Suzhou, Jiangsu Province, China). The spinal cord was compressed at an interval of 0.25 mm to produce severe injury (Plemel et al., 2008). A successful procedure resulted in paralysis of the lower limbs. The surgical procedure was performed in the ISH/IHC and PCR groups after spinal cord injury. The mice were sacrificed at 1, 3 and 7 days after spinal cord injury (Nakanishi et al., 2010). The sham operation group received an identical treatment, including exposure, laminectomy and placement of the forceps around the spinal cord, but no crush injury was inflicted.
RNA extraction and quantitative RT-PCR
Mice were sacrificed at 1, 3 and 7 days after spinal cord injury with an overdose of 1% pentobarbital sodium intraperitoneal injection. An 8-mm spinal cord segment (including the site of injury and 4 mm on either side) was removed quickly for RNA isolation. Mice in the sham operation group were sacrificed 1 day after operation.
Spinal cord total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The concentration and quality of RNA were evaluated by measuring UV optical density at 260 and 280 nm (260/280) and checked by gel electrophoresis. Equal amounts of RNA were used for quantitative RT-PCR.
Quantitative RT-PCR was performed using the NCode™ EXPRESS SYBR GreenER™ microRNA quantitative RT-PCR Kit (Invitrogen). Poly(A) tailing and RT reactions consisted of 4 μL 5 × reaction mix, 2 μL 10 × SuperScript enzyme mix and 200 ng total RNA in a final volume of 20 μL. 1 μL of the RT product was transferred into a PCR reaction mixture consisting of 10 μL Express SYBR green quantitative RT-PCR SuperMix, 0.4 μL microRNA-specific forward primer (10 μM) and 0.4 μL universal quantitative PCR primer (10 μM) in a final volume of 20 μL. PCR cycling parameters were 95°C for 2 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, performed in a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA). The miR-124 forward primer sequence of 18 bp was 5′-TAA GGC ACG CGG TGA ATG-3′ (Ho et al., 2014). U6 was used as the internal reference standard. Normalized expression was calculated using the comparative Ct method, and fold change was derived from the equation 2–ΔΔCt for each microRNA (Kim et al., 2012).
Preparation of digoxigenin-labeled RNA probes
A mirVana miRNA Probe Construction Kit (Ambion, Austin, Texas, USA) and a digoxigenin RNA Labeling Kit (SP6/T7; Roche, Basel, Switzerland) were used to construct digoxigenin-UTP-labeled RNA probes. The antisense probe for miR-124 was 5′-TAA GGC ACG CGG TGA ATG CCC CTG TCT C-3′ and the sense probe for miR-124 was 5′-CCG TAA GTG GCG CAC GGA ATC CTG TCT C-3′. Briefly, dsDNA transcription was carried out by hybridizing the DNA oligonucleotide to the T7 promoter primer at 70°C for 5 minutes, and then at room temperature for 5 minutes. Extension was performed by incubating with exo-Klenow DNA polymerase and dNTP Mix at 37°C for 30 minutes. The templates were mixed with T7 RNA polymerase and 10 × NTP labeling mixture, and then incubated at 37°C for 2 hours. Then, 2 μL DNase I was added and incubated at 37°C for 30 minutes to remove the DNA templates. The digoxigenin-labeled RNA transcript was stored at −20°C.
Tissue preparation and ISH
The mice were anesthetized and perfused transcardially with cold 4% paraformaldehyde in PBS for 10 minutes. Brain and spinal cord from normal adult C57BL/6 mice were removed and post-fixed in the same fixative for 4 hours at 4°C. The damaged spinal cord segment taken 7 days post-injury was carefully dissected out and post-fixed in the same fixative for 4 hours at 4°C. Then, all the samples were immersed in 20% sucrose-PBS at 4°C until they sank to the bottom of the container. Tissues were prepared for cryostat sectioning, with three animals from each group used for analysis.
For ISH, sections were washed twice in PBS for 5 minutes each and treated with 100 mM glycine-PBS twice for 5 minutes each. After Triton X-100 treatment (0.3%, v/v) for 15 minutes, sections were washed twice in PBS for 5 minutes each. Then sections were treated with 1 μg/mL proteinase K for 30 minutes at 37°C and re-fixed in 4% paraformaldehyde for 5 minutes. After washing in PBS for 10 minutes, sections were acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine hydrochloride for 10 minutes. Then, sections were prehybridized for 30 minutes at 37°C in prehybridization buffer (50% formamide and 4 × SSC). Hybridization was performed at 42°C overnight in 200 μL hybridization buffer containing 50 ng digoxigenin-labeled probe. The next day, sections were washed twice in 2 × SSC at 37°C for 15 minutes each and then washed twice with 1 × SSC at 37°C for 15 minutes each on a bath shaker. For signal development, sections were incubated with blocking solution (Roche, Basel, Switzerland) for 30 minutes at room temperature, and then incubated with alkaline phosphatase-conjugated Fab anti-digoxigenin antibody (Roche) for 30 minutes. NBT/BCIP solution (Roche) was incubated with the sections in the dark for 16 hours, and reactions were stopped with PBS washing.
ISH combined with IHC
After ISH for miR-124, the sections were washed with PBS three times and treated with blocking buffer (1% bovine serum albumin + 0.3% Triton X-100) for 1 hour at room temperature. Then, biotinylated mouse anti-NeuN monoclonal antibody (1:1,000; Chemicon, Billerica, MA, USA) was incubated with the sections at 4°C overnight. The next day, sections were washed with PBS and incubated with avidin-peroxidase (1:500; Sigma-Aldrich) for 1 hour at room temperature. Finally, the signals were developed with 3,3′-diaminobenzidene tetrahydrochloride until the dark brown reaction product was visible under the B51 microscope (Olympus, Tokyo, Japan).
Statistical analysis
Data were analyzed using SPSS 10.0 software (SPSS, Chicago, IL, USA) and were expressed as the mean ± SEM. Data were compared by one-way analysis of variance and Scheffe's post hoc test. P < 0.05 was considered to indicate a significant difference.
Results
Expression pattern of miR-124 in adult mouse brain and spinal cord
ISH with an RNA oligonucleotide probe complementary to miR-124 (antisense probe) was performed to assess expression levels of miR-124 throughout the central nervous system of adult mice (Figure 1A–E, G). miR-124 was mainly distributed in the grey matter, where neurons are located, such as the olfactory bulbs (Figure 1A), cerebral cortex (Figure 1B, C), basal nuclei, hippocampal pyramidal cell layer (Figure 1C), and cerebellar cortex (Figure 1D) and the grey matter of the spinal cord (Figure 1E, G). Cells in white matter regions, such as the corpus callosum and white matter of the spinal cord, were negative for miR-124 by ISH (Figure 1B–D). Under the same hybridization conditions, the sense probe for miR-124 generated no signal (Figure 1F, H), indicating that the antisense probe is specific for miR-124.
Figure 1.
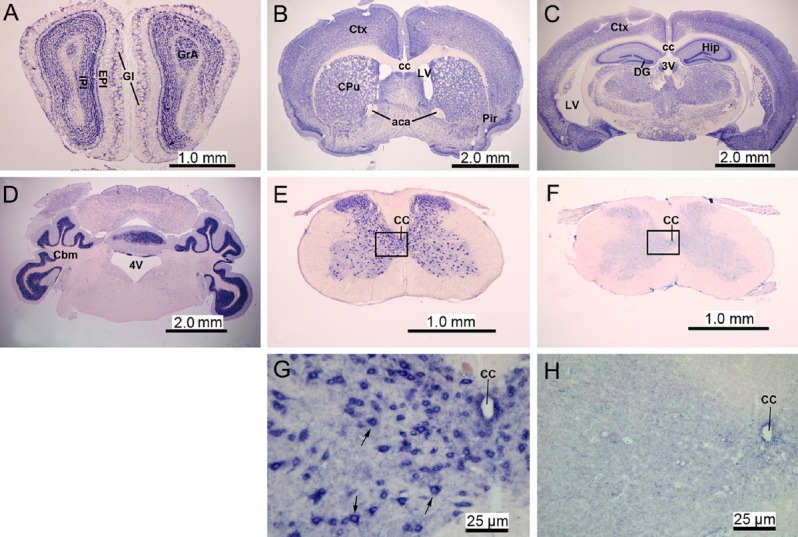
MicroRNA-124 (miR-124) expression in the central nervous system of adult mice (in situ hybridization, B51 microscope).
The miR-124 was mainly distributed in grey matter regions where neurons are located, such as the olfactory bulbs, cerebral cortex, basal nuclei, hippocampal pyramidal cell layer, and cerebellar cortex. (A–E) The expression of miR-124 in the olfactory bulbs (A), basal ganglia and basal forebrain (B), hippocampal region (C), cerebellum (D), and spinal cord (E). Panels A–E are representative sections hybridized with the antisense oligonucleotide probe against miR-124. (F) Spinal cord sections were hybridized with the sense oligonucleotide probe for miR-124 as negative control. (G) Higher magnification of panel E showing miR-124 signal (blue color) located in the cytoplasm of neurons (arrows) in the grey matter of the spinal cord. (H) Higher magnification of panel F showing lack of signal using the sense probe for miR-124 in the grey matter of the spinal cord. GI: Glomerular layer; EPI: external plexiform layer of the olfactory bulb; IPI: internal plexiform layer of the olfactory bulb; GrA: granule cell layer of the accessory olfactory bulb; Ctx: cerebral cortex; Cpu: caudate putamen; Pir: piriform cortex; cc: corpus callosum; aca: anterior commissure, anterior; LV: lateral ventricle; Hip: hippocampus; DG: dentate gyrus; 3V: 3rd ventricle; Cbm: cerebellum; 4V: 4th ventricle; CC: central canal.
Temporal expression profile of miR-124 after spinal cord injury
We examined the expression of miR-124 at 1, 3 and 7 days after spinal cord injury using quantitative RT-PCR analysis with SYBR Green probes. In the PCR group, the expression of miR-124 was significantly decreased at 1 day compared with the sham operation group (P < 0.05; Figure 2A). The expression of miR-124 was also significantly decreased at 3 and 7 days compared with the sham operation group (P < 0.01; Figure 2A). The expression of miR-21, used as a positive control (Liu et al., 2009), was significantly increased between 1 and 7 days post-injury compared with the sham operation group (P < 0.01; Figure 2B).
Figure 2.
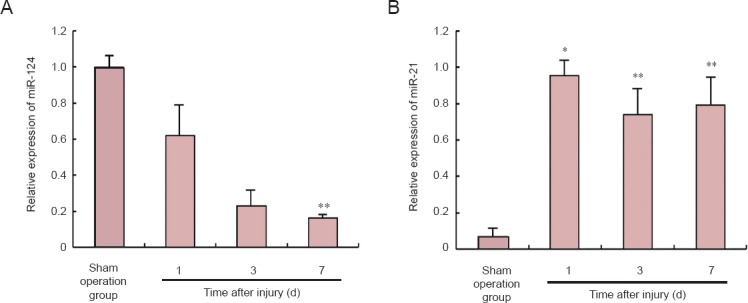
Quantitative RT-PCR analysis of the relative expression levels of microRNA-124 (miR-124) and microRNA-21 (miR-21) after spinal cord injury.
The expression of miR-124 was significantly decreased within the first 7 days after spinal cord injury. In comparison, the expression of miR-21 was significantly increased after spinal cord injury. (A) The relative expression of miR-124 on days 1, 3 and 7 after spinal cord injury. (B) The relative expression of miR-21 on days 1, 3 and 7 after spinal cord injury. U6 was used as the internal reference standard. Data are expressed as the mean ± SEM. The differences were compared with one-way analysis of variance and Scheffe's post hoc test. *P < 0.05; **P < 0.01, vs. sham operation group. d: Day(s).
Loss of miR-124 expression in neurons in the peri-lesion area after spinal cord injury
Although previous ISH studies have examined the expression pattern of miR-124 after spinal cord injury (Nakanishi et al., 2010), the expression of miR-124 in neurons after spinal cord injury remains unclear. To clarify the changes in expression of miR-124 in neurons after spinal cord injury, ISH was combined with NeuN IHC 7 days post-injury in the ISH/IHC group. The ISH signal for miR-124, visible as small blue dots, was distributed in the cytoplasm (Figure 3A), and the brown NeuN IHC signal was localized in the cytoplasm (Figure 3B). When the miR-124 and NeuN labeling was combined, double-positive cells appeared dark brown, and the dark blue dots could still be detected under an oil-immersion lens (Figure 3C).
Figure 3.
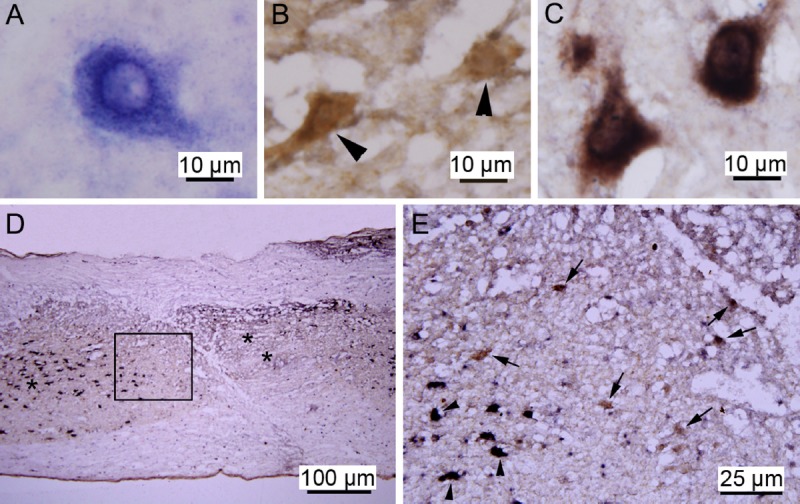
MicroRNA-124 (miR-124) expression in the damaged spinal cord (in situ hybridization (ISH)/immunohistochemistry).
The ISH miR-124-positive signal was distributed in the neuronal cytoplasm. In the peri-lesion areas, there were many neurons positive for NeuN, but negative for miR-124. (A) Representative images of miR-124-positive cells in the spinal cord as shown by ISH. (B) Representative images of NeuN-positive neurons in the spinal cord (arrowheads) as shown by immunohistochemistry. (C) Representative images of NeuN-positive neurons (dark brown) showing the miR-124 ISH signal. (D) miR-124 and NeuN displayed distinct expression patterns in the damaged spinal cord section. “*” Area distal to the focal site of injury; “**” the focal site of injury. (E) Higher magnification of the boxed area in panel D showing NeuN-positive neurons positive for miR-124 (arrowheads, black) or negative for miR-124 (arrows, brown).
At the center of the site of injury, miR-124 and NeuN signals were undetectable (Figure 3D). In areas distal to the peri-lesion area, all neurons were miR-124 and NeuN double-positive, in which blue and brown colors overlapped to result in a dark brown color (Figure 3C, E). In the peri-lesion areas that were adjacent to the center of the site of injury, there were many neurons positive for NeuN, but negative for miR-124 (Figure 3D, E). This suggests that there is a loss of neuronal miR-124 expression 7 days after spinal cord injury.
Discussion
MicroRNAs are ~22-nucleotide noncoding RNAs, derived from genome-encoded stem-loop precursors (Thum, 2014), and they play critical roles in the posttranscriptional regulation of gene expression by repressing translation and/or by destabilizing target mRNAs (Bartel, 2004; Leopold et al., 2014). An individual microRNA can down-regulate hundreds of mRNA targets by interacting with partially complementary sequences within their 3′ untranslated regions (Vella et al., 2005), and comparative genomics analyses have predicted that a third of all human genes are potential microRNA targets (Lim et al., 2005).
Several brain-specific/enriched microRNAs have been reported to play important roles in physiological and pathological processes in the brain (Cao et al., 2006; Courts et al., 2013). Among these, miR-124 is the most abundantly expressed in the adult mammalian brain (Doeppner et al., 2013). The expression of miR-124 increases during brain development (Krichevsky et al., 2006; Xu et al., 2012), and it promotes a neural-like phenotype by decreasing the levels of hundreds of non-neuronal transcripts (Lim et al., 2005). In non-neural cells and neural progenitors, a transcriptional repressor, RE1 silencing transcription factor, inhibits miR-124 expression (Conaco et al., 2006; Bitel et al., 2010). As progenitors differentiate into mature neurons, RE1 silencing transcription factor no longer represses miR-124 expression (Conaco et al., 2006). miR-124 also directly targets small C-terminal domain phosphatase 1 to induce neurogenesis during embryonic development of the central nervous system (Visvanathan et al., 2007). In adult mouse brain, miR-124 enhances neural maturation by targeting the SRY-box transcription factor Sox9 in the subventricular zone (Cheng et al., 2009). Moreover, miR-124 directly represses polypyrimidine tract binding protein 1, an RNA binding protein, which leads to increased expression of polypyrimidine tract binding protein 2 and correct alternative mRNA splicing that is necessary for the transition from a non-neuronal to a neuronal phenotype (Makeyev et al., 2007).
Recently, a number of studies have shown that the expression of several microRNAs related to neural plasticity, repair, apoptosis and neuroinflammation change after spinal cord injury (Liu et al., 2010; Izumi et al., 2011). miR-124 was demonstrated by microarray assay to be significantly down-regulated in the site of injury in a mouse spinal cord contusion model, and quantitative RT-PCR analysis revealed that miR-124 expression decreased 7–14 days after spinal cord injury (Liu et al., 2009; Nakanishi et al., 2010). Using ISH, Nakanishi and colleagues observed miR-124 expression only in the control group. Expression was absent at the site of injury due to tissue loss in the spinal cord injury group (Nakanishi et al., 2010).
In the present study, miR-124 was found to be continuously down-regulated after spinal cord injury in mice by quantitative RT-PCR. We combined ISH with IHC to specifically evaluate miR-124 expression in neurons. We showed that there was a lack of miR-124 and NeuN staining in the peri-lesion area. Distal to the peri-lesion area, all neurons were miR-124/NeuN double positive. These results are consistent with previous studies showing that miR-124 is neural specific and is mainly expressed in differentiated neurons (Lagos-Quintana et al., 2002; Deo et al., 2006). In the peri-lesion areas, there were numerous neurons positive for NeuN, but negative for miR-124. This indicates that NeuN-positive neurons in the peri-lesion area lose miR-124 expression after spinal cord injury. In our previous study, we found that there was a pale ischemic zone (peri-lesion area) surrounding the center of the site of injury in the contused spinal cord, in which a few remaining neurons have weakly-stained or absent Nissl bodies (Shen et al., 2009). The cellular impact of neuronal loss of miR-124 expression after spinal cord injury remains unknown, but the loss of expression appears to be a response to the tissue damage. We hypothesize that the neuronal loss of miR-124 expression may reflect the state of tissue damage after spinal cord injury.
Acknowledgments
We are grateful to Ms Jian-yong Qiu and Ling-ling Fei from Institute of Neurosciences, the Fourth Military Medical University for their assistances.
Footnotes
Funding: This work was supported by the National Natural Science Foundation of China, No. 81371364.
Conflicts of interest: None declared.
Copyedited by Patel B, Norman C, Wang J, Yang Y, Li CH, Song LP, Zhao M
References
- Bareyre FM, Schwab ME. Inflammation, degeneration and regeneration in the injured spinal cord: insights from DNA microarrays. Trends Neurosci. 2003;26:555–563. doi: 10.1016/j.tins.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomices, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bitel CL, Perrone-Bizzozero NI, Frederikse PH. HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens. Mol Vis. 2010;16:2301–2316. [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cheng L, Pastrana E, Taazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han J, Mandel G. Reciprocal actions of REST and microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courts C, Grabmüller M, Madea B. Dysregulation of heart and brain specific micro-RNA in sudden infant death syndrome. Forensic Sci Int. 2013;228:70–74. doi: 10.1016/j.forsciint.2013.02.032. [DOI] [PubMed] [Google Scholar]
- De Biase A, Knoblach SM, Di Givoanni S, Fan C, Molon A, HoffmanEP, Faden AI. Gene expression profiling of experimental traumatic spinal cord injury as a function of distance from impact site and injury severity. Physiol Genomics. 2005;22:368–381. doi: 10.1152/physiolgenomics.00081.2005. [DOI] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Müller B, Koch JC, Bähr M, Hermann DM, Michel U. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol. 2013;126:251–265. doi: 10.1007/s00401-013-1142-5. [DOI] [PubMed] [Google Scholar]
- Gu W, Xu Y, Xie X, Wang T, Ko JH, Zhou T. The role of RNA structure at 5’ untranslated region in microRNA-mediated generegulation. RNA. 2014;20:1369–1375. doi: 10.1261/rna.044792.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho VM, Dallalzadeh LO, Karathanasis N, Keles MF, Vangala S, Grogan T, Poirazi P, Martin KC. GluA2 mRNA distribution and regulation by miR-124 in hippocampal neurons. Mol Cell Neurosci. 2014;61:1–12. doi: 10.1016/j.mcn.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi B, Nakasa T, Tanaka N, Nakanishi K, Kamei N, Yamamoto R, Nakamae T, Ohta R, Fujioka Y, Yamasaki K, Ochi M. MicroRNA-223 expression in neutrophils in the early phase of secondary damage after spinal cord injury. Neurosci Lett. 2011;492:114–118. doi: 10.1016/j.neulet.2011.01.068. [DOI] [PubMed] [Google Scholar]
- Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Stem Cells. 2006;24:857–864. [Google Scholar]
- Leopold JA. MicroRNAs regulate vascular medial calcification. Cells. 2014;3:963–980. doi: 10.3390/cells3040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu N, Wang X, Lu Q, Xu X. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Keeler BE, Zhukareva V, Houlé JD. Cycling exercise affects the expression of apoptosis-associated microRNAs after spinal cord injury in rats. Exp Neurol. 2010;226:200–206. doi: 10.1016/j.expneurol.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Nakasa T, Ishikawa M, Yamada K, Ymasaki K, Kamei N, Izumi B, Adachi N, Miyaki S, Asahara H, Ochi M. Responses of microRNAs 124a and 223 following spinal cord injury in mice. Spinal Cord. 2010;48:192–196. doi: 10.1038/sc.2009.89. [DOI] [PubMed] [Google Scholar]
- Plemel JR, Duncan G, Chen KK, Shannon C, Park S, Sparling JS, Tetzlaff W. A graded forceps crush spinal cord injury model in mice. J Neurotrauma. 2008;25:350–370. doi: 10.1089/neu.2007.0426. [DOI] [PubMed] [Google Scholar]
- Shen X, Zhao Y, Zhang Y, Jia L, Ju G. A modified ferric tannate method for visualizing a blood vessel and its usage in the study of spinal cord injury. Spinal Cord. 2009;47:852–856. doi: 10.1038/sc.2009.30. [DOI] [PubMed] [Google Scholar]
- Strickland ER, Hook MA, Balaraman S, Huie JR, Grau JW, Miranda RC. MicroRNA dysregulation following spinal cord contusion: implication for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol. 2014;11:655–663. doi: 10.1038/nrcardio.2014.125. [DOI] [PubMed] [Google Scholar]
- Vella MC, Slack FJ. C. elegans microRNAs. WormBook. 2005;21:1–9. doi: 10.1895/wormbook.1.26.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Li P, Qin K, Wang X, Jiang X. miR-124 regulates neural stem cells in the treatment of spinal cord injury. Neurosci Lett. 2012;529:12–17. doi: 10.1016/j.neulet.2012.09.025. [DOI] [PubMed] [Google Scholar]


