Abstract
DNA methylation loss can produce inheritable active epialleles in plants. The mechanism involved in the stable transmission of hypomethylated epimuations is presently not clear. Here we show that maintenance of a stably hypomethylated active epiallele in rice required a CHD3 protein (CHR729) and that over-expression of an H3K4me3 demethylase (JMJ703) or H3K27me3 methyltransferase (SDG711) could stably resilence the epiallele. CHR729 and JMJ703 have antagonistic function in H3K4me3 in maintaining the active state of the epiallele, whereas SDG711-mediated H3K27me3 was sufficient to stably repress the locus. The data suggest that H3K4me3 and H3K27me3 controlled by these chromatin regulators may be involved in stable transmission/resetting of epigenetic variation in rice.
Epigenetic processes can stably alter gene transcriptional activities independently of the DNA sequence. Several biochemical mechanisms that lead to changes in DNA methylation and/or histone modifications have been discovered to be involved in the processes1. DNA methylation is important for diverse epigenetic phenomena in plants, including repression of transposable elements (TEs) and repetitive sequences and genomic imprinting.
In plants, cytosine methylation is found in three sequence contexts: CG, CHG, and CHH (in which H = A, T, C). The DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) catalyzes de novo DNA methylation in all sequence contexts and maintains asymmetric CHH methylation. Small RNAs target DRM2 to homologous genomic DNA sequences for cytosine methylation by the RNA-directed DNA methylation (RdDM) pathway. Symmetric CG methylation is maintained by METHYLTRANSFERASE1 (MET1), which recognizes hemimethylated CG sites after DNA replication. CHG methylation is catalyzed by the plant specific CHROMOMETHYLASE3 (CMT3). Other genes such as the histone deacetylase HDA6, and the SNF2/SWI2 chromatin remodeler DDM1 (decrease in DNA methylation 1) are also involved in DNA methylation2. Defects in DNA methylation genes such as met1 and ddm1 mutations lead to genome-wide hypomethylation3. DNA methylation loss from transposons in ddm1 mutants can be recovered by RdDM, when wild type DDM1 is reintroduced4. However there are many hypomethylated genomic regions that cannot be recovered, as heritable hypomethylated chromosomal segments have been propagated for many generations in so called “epiRILs” (epigenetic recombinant inbred lines)5. In ddm1 and met1 mutants hypomethylated TEs neighboring genes resulted in stable epimutations such as BONSAI and FWA (FLOWERING WAGENINGEN)3.
The rice genomic DNA is methylated also in all three cytosine contexts, with high levels of CG and CHG methylation and very low levels of CHH methylation6,7. Genes involved in DNA methylation are generally conserved in rice8,9. Besides defects in DNA methylation genes, plant tissue-culture also leads to general DNA methylation loss10,11. Loss of DNA methylation during rice callus culture was found to be associated with loss of 24 nt siRNA in callus-regenerated plants and was largely stable across generations11. However, the mechanism that maintains the transgenerationally stably hypomethylated state is not known.
Besides DNA methylation, histone modifications and chromatin remodeling also play important roles in epigenetic regulation of gene expression. For instance, trimethylation of Histone H3 lysine 4 (H3K4me3) is associated with gene activation, whereas trimethylation of Histone H3 lysine 27 (H3K27me3) marks repressed genes. Plant SET-domain genes (SDG) are involved in histone methylation that can be removed by specific histone demethylases such as Jumonji (JMJ)-C domain containing proteins identified in rice8,12.
In this work we characterized a stably hypomethylated and transcriptionally active locus in rice callus regenerated plants. We show that the stable hypomethylation and activation of the locus required the CHD3 chromatin remodeling factor CHR72913, which is involved in maintenance of H3K4me3 during cell differentiation. Removal of H3K4me3 by histone demethylase JMJ70314,15, resulted in repression and recovery of DNA methylation of the locus. Our data reveals that H3K4me3 was involved in the transmission of the active state of the epiallele and that, in addition to DNA methylation, H3K27me3 also played a role in transgenerationally stable gene repression in rice.
Results and Discussion
Identification of a callus culture-induced stable epimutation in rice
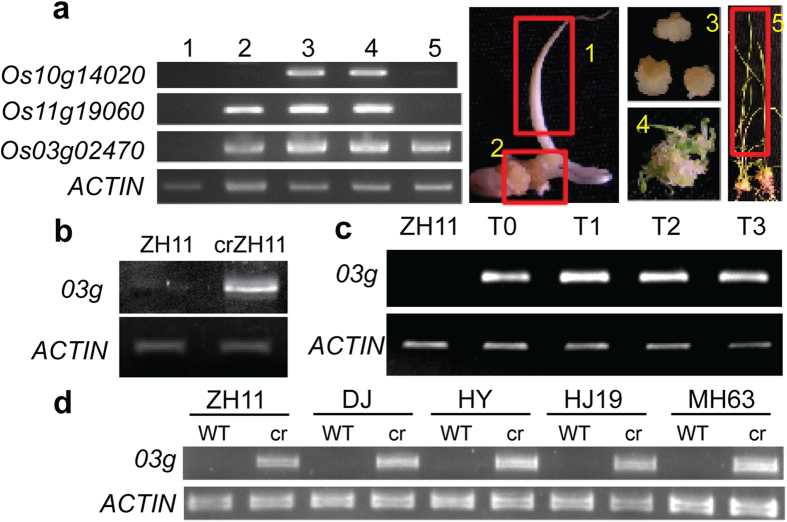
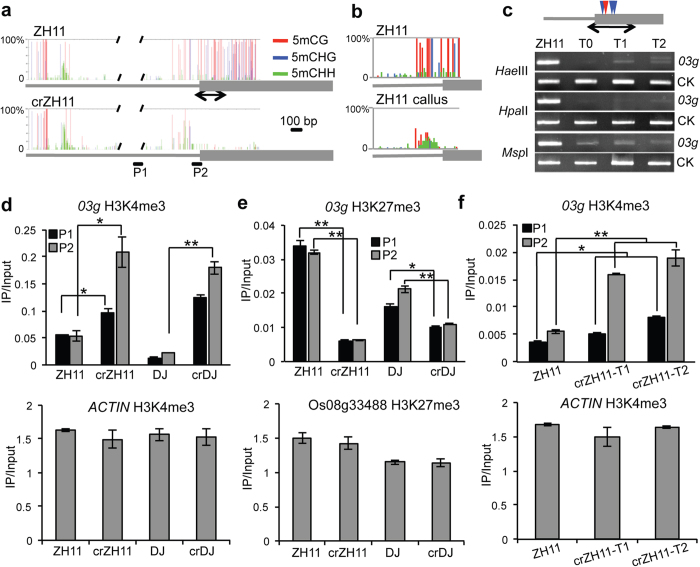
Previous data have shown that many genes are activated during callus culture16. However, most of the callus culture-induced genes are expressed in different developmental stages and/or organs/tissues and only a few genes display callus-specific expression16. Analysis by RT-PCR revealed that these genes that are expressed only in callus were resilenced during plant regeneration process, except Os03g02470 (referred as to 03g thereafter) that was not repressed in regenerated plant leaves (Fig. 1a). Transcripts of the gene could be detected in pollen and the active state of the locus was maintained in subsequent generations (Figs 1b,c). The active state of the locus was detected in all examined regenerated plants from independent callus cultures of different rice varieties (Fig. 1d), suggesting that callus culture-induced activation of the gene was not stochastic. The 03g locus encodes a protein of unknown function. In leaf tissues of normal plants, the promoter and the 5′ region of the gene displayed heavy CG (>90%) and CHG (H = A, T, or C) (>40%) methylation, which were lost in callus and regenerated shoots (Fig. 2a,b). While the other two loci, Os10g14020 and Os11g19060, were not methylated either in leaf or callus. The hypomethylation state of 03g was maintained in subsequent generations (Fig. 2c). Recent results have identified many genes that show DNA methylation loss during rice callus culture and stable hypomethylation in regenerated plants11. Hypomethylation was also found in the 03g locus in that study, but it was attributed to the downstream gene (Os03g02460), the promoter of which actually overlapped with 03g. H3K4me3 was increased, and H3K27me3 was decreased in the locus in regenerated plants (Fig. 2d,e). By comparison, H3K4me3 levels were not clearly changed in Os10g14020 and Os11g19060, although a reduction of H3K27me3 was detected in Os10g14020 (Supplementary Figure 1). In addition, the increased H3K4me3 levels in 03g were maintained in the subsequent generations (Fig. 2f). The activation of 03g was likely to be triggered by loss of DNA methylation during callus culture, as treatment of normal ZH11 seedlings with 5-azacytidine (an inhibitor of DNA methylation) led to DNA hypomethylation, gene expression, and increased H3K4me3 of the locus (Supplementary Figure 2). Moreover, the DNA hypomethylation and gene activation were maintained in the subsequent generation (Supplementary Figure 2).
Figure 1. Callus culture induced expression of Os03g02470 (03g) was stably inherited.
(a) RT-PCR detection of transcripts of three callus-specific genes during callus culture and plant regeneration processes. The stages of callus culture and regeneration are shown in the right panel. 1: seed germinated; 2: callus-like tissue in the early stage of callus induction; 3: subcultured callus; 4: mixture of callus and shoot during regeneration stage; 5: regenerated shoots/leaves. (b) 03g expression is detectable in pollens of callus-regenerated crZH11 compared to normal ZH11 pollens. (c) 03g expression was maintained in 4 subsequent generations. mRNA isolated from leaves of T0 to T3 generation plants were analyzed. (d) 03g expression was detected in all independent callus regeneration (cr) plants of different rice varieties as indicated. PCR products of different samples for each primer set were run in the same gel and cropped in the same picture.
Figure 2. Stable 03g activation was associated with DNA methylation loss, histone H3K27me3 removal and H3K4me3 deposition.
(a) 03g was heavily methylated in CG and CHG contexts at transcription start site (TSS) and 5′ UTR in leaf tissues of normal wild type ZH11, but demethylated in leaf tissues of callus regenerated ZH11 (crZH11) plants. DNA methylation was determined by bisulfite sequencing. More than 20 clones for each genotype were sequenced. Solid gray line: upstream promoter region; grey bar: transcribed region of 03g gene. Primers sets (P1, P2) used in ChIP assays are indicated. Black slashes indicate the gap, which is not sequenced. (b) DNA methylation of 03g was lost in callus. Bisulfite sequencing region is represented by the double arrows shown in a. (c) DNA demethylation was maintained in subsequent generations, shown by enzyme digestion followed by PCR amplification. PCR amplification region was the same as bisulfite sequencing region in a. CK, control DNA fragments without enzyme recognition sites. Blue triangle indicates HpaII/MspI cut sites, red triangle indicates HaeIII cut sites. PCR products of different samples for each primer set were run in the same gel and cropped in the same picture. (d,e) H3K4me3 (d) and H3K27me3 (e) at promoter (P1) and TSS region (P2) of 03g were respectively increased and decreased in callus-regenerated (cr) plants of two different rice cultivars (ZH11 and DJ). (f) Increased H3K4me3 in 03g was maintained in subsequent T1 and T2 generations. ACTIN was used as a control for H3K4me3. Os08g33488, which is methylated by H3K27me315, was used as a control for H3K27me3. Student t-tests were performed from 3 biological repeats. *p < 0.05, **p < 0.01.
To test whether the hypermethylated 03g allele in normal plants could influence the hypomethylated allele for expression, we crossed callus-regenerated ZH11 (crZH11) with normal MH63 plants, in which varieties 03g is polymorphic (Supplementary Figure 3a). 03g was found to be expressed in all tested F1 individuals and all of the sequenced transcripts were derived from the ZH11 allele (Supplementary Figure 3a). In addition, in crosses between crZH11 and normal ZH11 plants, about ¾ of the F2 segregates displayed 03g expression (Supplementary Figure 3b), suggesting that the hypomethylated epiallele was not controlled by the hypermethylated allele or vice versa.
Histone methylation regulators are involved in the maintenance of stable expression of the epiallele
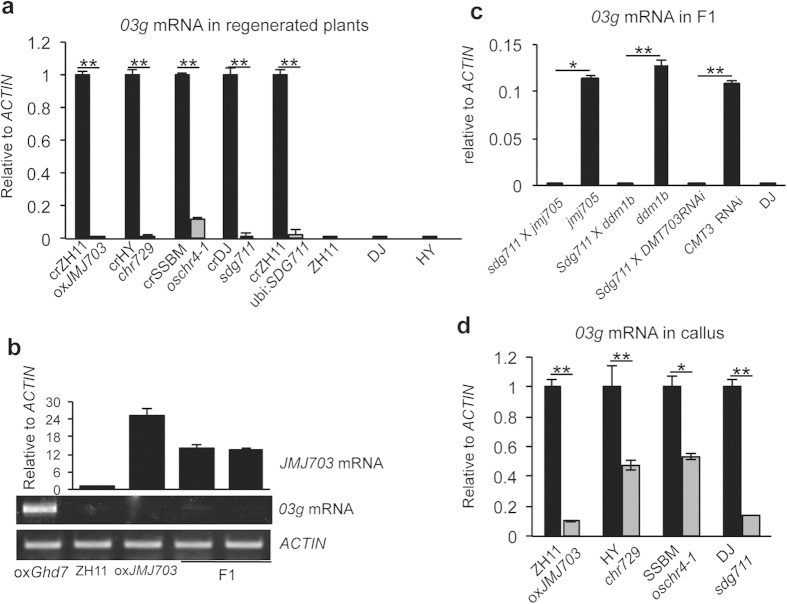
To study whether the stable activation of 03g could be affected by loss- or gain-of-function of DNA methylation, histone modification, and other chromatin regulatory genes, we analyzed 03g expression in rice T-DNA mutants, RNAi and over-expression transgenic plants of 16 chromatin protein genes encoding DNA methyltransferases (DMT702, DMT703, DMT706, and DMT707) (Supplementary Figure 4), DNA demethylase (DNG701)17, SET-domain histone methyltransferases (SDG711, SDG721, SDG723 and SDG728) (Supplementary Figure 4)14,18,19,20, Jumonji-C histone demethylases (JMJ703, JMJ705, JMJ706 and JMJ716) (Supplementary Figure 4)21,22,23,24, chromatin remodeling factors involved in DNA methylation (DDM1)25, histone modification (CHR729)13, and recognition (LHP1) (Table 1 and Supplementary Figure 4). The tissue culture-induced 03g activation was detected in most of the T-DNA mutants and transgenic plants, except in CHR729 loss-of-function T-DNA mutant (chr729)13, JMJ703 over-expression lines (oxJMJ703)21,22 and a gain-of-function T-DNA insertion mutant of SDG711 (sdg711)14,15, in which 03g was silenced as in normal wild type plants (Table 1 and Fig. 3a). The down-regulation of 03g expression in the chr729 T-DNA line was confirmed in callus-regenerated plants of an ethyl methanesulfonate (EMS)-induced mutant allele of chr729, known as oschr4-126 (Fig. 3a). In independent SDG711 over-expression lines (under the control of the maize ubiquitin gene promoter, Ubi: SDG711)14 , 03g was also repressed (Table 1 and Fig. 3a). To further confirm whether increased levels of JMJ703 repressed 03g expression, we crossed a transgenic plant (i.e. OxGhd7, which is MH63 background with overexpression of rice flowering regulator Ghd7 and showed active 03g expression)27,28 with the oxJMJ703 line and found that the 03g expression was repressed in the F1 hybrids (Fig. 3b), while remained to be expressed when crossed with normal plants (Supplementary Figure 2). Similarly, 03g expression was resilenced in the F1 hybrids of crosses between sdg711 and three different transgenic plants (Fig. 3c). These data indicated that over-expression of JMJ703 or SDG711 suppressed 03g expression. JMJ703 encodes a histone H3K4me3 demethylase21,22, and SDG711, a homolog of Enhancer of zeste, encodes a histone H3K27me3 methyltransferase14,18. Thus, in addition to DNA methylation, histone methylation homeostasis regulated by CHR729, JMJ703 and SDG711 could also control stable expression state of the gene.
Table 1. Detection of 03g expression in T-DNA mutants and transgenic plants of rice chromatin regulators.
| Name | 03gexpression | References | |
|---|---|---|---|
| DNA methyltransferase | DMT702 T-DNA mutant | Yes | * |
| DMT703 RNAi | Yes | * | |
| DMT706 over-expression | Yes | 34,* | |
| DMT707 RNAi | Yes | * | |
| DNA glycosylase | DNG701 T-DNA mutant | Yes | 17 |
| Histone methylation | SDG711 gain of function T-DNA | No | 14 |
| SDG711 over-expression | No | 14 | |
| SDG711 RNAi | Yes | 14 | |
| SDG721 RNAi | Yes | * | |
| SDG723 RNAi | Yes | * | |
| SDG728 over-expression | Yes | 20 | |
| Histone demethylase | JMJ703 T-DNA mutant | Yes | 21,22 |
| JMJ703 over-expression | No | 21,22 | |
| JMJ705 T-DNA mutant | Yes | 24 | |
| JMJ705 over-expression | Yes | 24 | |
| JMJ706 T-DNA mutant | Yes | 23 | |
| JMJ716 T-DNA mutant | Yes | * | |
| Chromatin remodeler | DDM1b T-DNA mutant | Yes | 25 |
| CHR729 T-DNA mutant | No | 13 | |
| LHP1 RNAi | Yes | * | |
| LHP1 over-expression | Yes | * |
*characterization of the mutant, over-expression or RNAi plants are shown in Supplementary Figure 4.
Figure 3. Callus culture-induced 03g expression was repressed by over-expression of JMJ703 and SDG711 and mutations of CHR729.
(a) 03g expression was repressed in JMJ703 over-expression (oxJMJ703), SDG711 gain-of-function T-DNA mutant (sdg711) and over-expression (ubi: SDG711), and CHR729 T-DNA (chr729) and EMS-induced (oschr4) mutant plants compared to the respective regenerated wild plants (crZH11, crHY, crDJ, and crSSBM). The expression levels are presented as relative to regenerated wild type plants (set at 1). (b) 03g expression was inhibited in the F1 hybrids of crossed between oxJMJ703 and a 03g-expressing transgenic plant (oxGhd7). (c) 03g expression was inhibited in the F1 hybrids of crosses between sdg711 and other three 03g active regenerated plants (i.e. T-DNA mutants of jmj705 and ddm1b, and DMT703 RNAi). 03g expression levels were normalized with ACTIN. (d) 03g expression was detected in callus of oxJMJ703, chr729, oschr4, sdg711, and ubi: SDG711 plants. But the expression levels were reduced compared to the respective wild types (set at 1). Student t-tests were performed from 3 biological repeats. *p < 0.05, **p < 0.01.
To check whether the repression of 03g expression in the above mentioned mutants and transgenic plants was initiated early during callus induction or shoot regeneration process, we produced callus from chr729, oxJMJ703, and sdg711 plants and found that unlike in the respective regenerated plants where 03g was almost totally silent, the transcripts of 03g could be detected in callus derived from these plants especially from chr729 and oschr4-1 mutants (Fig. 3d), suggesting that repression of 03g by CHR729 mutation or SDG711 and JMJ703 over-expression initiated during tissue culture was reinforced during plant regeneration.
Antagonistic function of CHR729 and JMJ703 is involved in regulating the 03g expression state
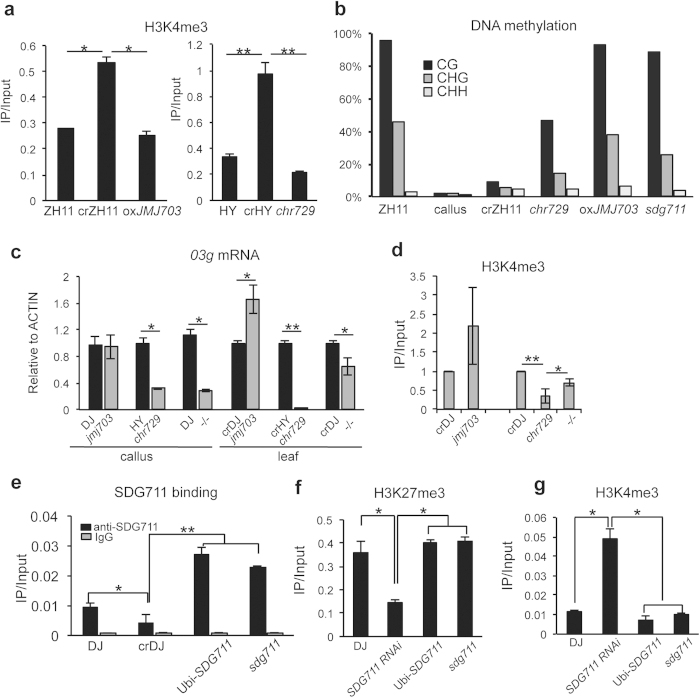
CHR729 was shown to be preferentially involved in regulating H3K4me3 levels of under-expressed genes13. The requirement of CHR729 for the maintenance of 03g expression during plant regeneration and the opposite effect of JMJ703 over-expression suggests that H3K4me3 levels may be important for the stabilization of the callus-induced epigenetic expression state of the gene. Analysis of chromatin modifications on the 03g locus revealed a decrease of H3K4me3 (Fig. 4a) and restoration of DNA methylation in chd729 T-DNA mutant and JMJ703 over-expression plants compared to regenerated wild type (Fig. 4b). However, in jmj703 loss-of-function mutant21,22, 03g expression in callus was not affected, while some increase of 03g expression was observed in jmj703 leaves (Fig. 4c), suggesting that on the 03g locus JMJ703 might be more active in differentiated cells than in callus. In jmj703/chr729 double mutant leaves, 03g expression was increased back to the level of regenerated wild type plants, but there was no clear difference for 03g expression in callus between chr729 and the double mutant (Fig. 4c,d). This observation supported the hypothesis that JMJ703 preferentially functioned in differentiated tissues and suggested that higher H3K4me3 in jmj703 mutant might compensate the loss of H3K4me3 for 03g expression in chr729 plants. CHR729 and JMJ703 might be functional antagonists for H3K4me3 in a subset of genes, as jmj703 mutation resulted in elevated H3K4me3 in many loci22, while loss-of-function of CHR729 leads to decrease of H3K4me3 preferentially from under-expressed genes13. Comparison of the two sets of data revealed that half of the genes that showed higher H3K4me3 in jmj703 were among those with decreased H3K4me3 in chr729 (Supplementary Table 1). We tested some of the loci and found that the decrease of H3K4me3 in chr729 mutant was partially or fully recovered in the jmj703/chr729 double mutant (Supplementary Figure 4). This suggests that the two proteins may have antagonistic function in H3K4me3 of a subset of loci in the genome.
Figure 4. H3K4me3 and H3K27me3 had antagonistic functions in stabilizing the expression and DNA methylation levels on 03g locus.
(a) H3K4me3 in 03g was reduced in oxJMJ703 and chr729 mutant respectively compared to in wild type regenerated plants. (b) DNA methylation of 03g locus was partially or fully recovered in chr729 mutant, oxJMJ703, and sdg711 plants. DNA methylation was determined by bisulfite sequencing. More than 20 clones for each genotype were sequenced. (c) Comparison of 03g expression in jmj703, chr729, and jmj703/chr729 double mutant (−/−) callus and leaves compared to wild type (DJ, HY) or regenerated wild type leaves (crDJ, crHY). The expression levels were normalized with ACTIN and then set as 1 in wild types. (d) H3K4me3 levels of 03g in jmj703, chr729, and jmj703/chr729 double mutant (−/−). (e) SDG711 protein was enriched in 03g. ChIP analysis was performed with anti-SDG71114 and with preimmune serum (IgG) as controls. (f,g) H3K4me3 (f) and H3K27me3 (g) levels of 03g in SDG711 RNAi, over-expression (ubi: SDG711), and gain-of-function T-DNA line (sdg711) compared to wild type (DJ). Because H3K4me3 and H3K27me3 are enriched in the 5′ end of pant genes, P2 primer set shown in Fig. 2a was used for the ChIP analysis. Primers used for ChIP qPCR were P2 indicated in Fig. 2a. Student t-tests were performed from 3 biological repeats. *p < 0.05, **p < 0.01.
SDG711-mediated H3K27me3 is sufficient to stably repress the epiallele
ChIP assay with anti-SDG711, prepared with E. coli-produced SDG711 protein and previously tested14, revealed a direct association of SDG711 to the 03g locus (Fig. 4e). A lower level of SDG711-binding detected in regenerated plants (crDJ) might be due to 03g activation that might consequently inhibit SDG711 association. H3K27me3 levels on 03g were maintained in sdg711 and Ubi: SDG711 and were reduced in SDG711 RNAi plants compared to wild type (Fig. 4f). Likely, SDG711-mediated maintenance of H3K27me3 was sufficient to mediate 03g stable repression. Meanwhile, in sdg711 plants 03g displayed similar levels of DNA methylation as in wild type (Fig. 4b), suggesting that either the maintenance of H3K27me3 or the association of SDG711 to the locus prevented DNA methylation loss from the locus in regenerated plant. Consistent with the repressive state of 03g, the H3K4me3 levels in sdg711 and Ubi: SDG711 plants were similar to that in wild type (Fig. 4g).
The mechanism that causes DNA hypomethylation in regenerated plants which occurs preferentially at promoter regions is not known11. As the T-DNA mutant line of DNG70117, which encodes a rice DNA demethylase, showed 03g expression (Table 1), it is unlikely that this demethylase activity was involved in the DNA methylation loss from the locus during callus culture. Previous results have indicated that siRNAs play an important role in the recovery of DNA methylation loss from a majority of genomic regions over generations in Arabidopsis4, and that stable DNA hypomethylation in regenerated rice plants may be due to loss of relevant siRNA11. The present data suggest that deposition of H3K4me3 during callus-induced gene activation also plays a role in maintaining stable DNA hypomethylation and expression in regenerated plants. This is consistent with previous results showing that H3K4me3 can prevent DNA remethylation29,30,31,32. The recovery of 03g DNA methylation by over-expression of H3K4me3 demethylase gene JMJ703 corroborated the observations that mutation of Arabidopsis H3K4 demethylase genes reduces DNA methylation in genomic regions that show coincidental increases in H3K4me2/me330. The function of CHR729 in maintaining 03g H3K4me3 likely contributed to the maintenance of active state of the locus. Therefore, CHR729/JMJ703-regulated H3K4me3 homeostasis may provide a mechanism to stabilize active epialleles that could be generated by epigenomic variations in plants, although it is not excluded that an independent function of CHR729 might be involved. In addition, our data show that stable 03g silencing was not only mediated by DNA methylation but also by SDG711-mediated H3K27me3, suggesting that SDG711 may be involved in transgenerational silencing of gene expression. Taken together, the data in this work suggest that variation in expression levels of histone methylation regulatory genes may contribute to stable transmission or resetting of epigenetic variation of gene expression in crop plants.
Methods
Rice materials
Rice (Oryza sativa) cultivated varieties ZH11 (Zhonghua11), MH63 (Minghui63), DJ (Dongjin), HY (Hwayoung), SSBM (Shi Shou Bai Mao) were used in this study. Callus culture and plant regeneration were performed as described previously33. Briefly, rice seeds were sterilized and placed on callus induction media for 30 days, then primary callus was transferred to subculture media. After two weeks, callus was used for transformation or transferred to regeneration media. Primary plants regenerated from callus were transferred to rooting media and transferred to field or greenhouse after 1 week. T-DNA mutants, RNAi and over-expression plants were generated by Agrobacterium-mediated transformation using standard callus culture, transformation, selection, and regeneration procedures33.
Gene expression analysis
RNA was extracted from rice leaves or callus with TRIzol reagent (15596-026, Life Technology) and reverse-transcribed with SSIII (18080-044, Life Technology). Synthesized first strand cDNA was used as template for regular PCR amplification or real-time PCR (ABI7500).
5-azacytidine treatment
Sterilized rice seeds were placed on 1/2 MS medium containing 50 mg/L 5-aza-2′-deoxycytidine (A3656, Sigma) and grown under dark condition for 1 month. Treated seedlings (aerial parts) were then harvested and pooled for further analysis or transferred to normal condition for propagation.
DNA methylation assay
Genomic DNA was extracted from rice leaf and callus. For bisulfite sequencing, 1 μg DNA was treated with bisulfite salt and recovered using EpiTect Bisulfite Kit (Qiagen, 59104). PCR fragments were cloned into T Easy Vector (Promega, A1360) for sequencing. Sequences were analyzed using online software (http://katahdin.mssm.edu/kismeth/revpage.pl). At least 20 clones were sequenced for each. For Enzyme Digestion PCR analysis, 500 ng DNA was treated with 20U HaeIII, HpaII and MspI for 4 hours in a 20 μl reaction volume, respectively. Digested DNA (1 μl) was used as PCR template. DNA fragments without recognition sites of those enzymes were amplified as DNA loading control.
ChIP assay
ChIP analysis was performed as previous described23. Briefly, chromatin isolated from 2 g rice leaf or 1 g callus was incubated with antibody coated beads (Life technology, 10001D) overnight. After wash and elution, products were reverse crosslinked. Then the products were treated with protease K (Takara, 9034), recovered, and used as template for real-time PCR with primers listed in Supplementary Table 2. Antibodies for histone modifications are anti-H3K4me3 (ab8580, Abcam), anti-H3K27me3 (07–449, Millipore) and anti-FLAG (F3165, Sigma) antibodies respectively. Antibody of SDG711 was produced by immunizing rabbits with E. coli produced full-length SDG711 protein14.
Accession Numbers
DMT702 (Os03g58400), DMT703 (Os05g13790), DMT706 (Os03g02010), and DMT707 (Os07g08500), DNG701 (Os05g37350), SDG711 (Os06g16390), SDG721 (Os01g11950), SDG723 (Os09g04890) and SDG728 (Os05g41170), JMJ703 (Os05g10770), JMJ705 (Os01g67970), JMJ706 (Os10g42690), JMJ716 (Os03g22540), CHR729 (Os07g31450), OsDDM1 (Os09g27060), OsLHP1 (Os10g17770).
Additional Information
How to cite this article: Chen, X. et al. Histone H3K4me3 and H3K27me3 regulatory genes control stable transmission of an epimutation in rice. Sci. Rep. 5, 13251; doi: 10.1038/srep13251 (2015).
Supplementary Material
Acknowledgments
We thank Qianwen Sun, Ning Zhu, Yiran Ye for assistance in experiments; Wenhao Yan and Feng Tan for providing help on rice hybridization. This work was supported by grants from the National Science Foundation of China (31371241), the 863 Key Project “Rice Functional Genomics” of the Chinese Ministry of Science and Technology (2012AA10A304), and the French Agence Nationale de la Recherche (ANR 12-BSV6-0010).
Footnotes
Author Contributions X.C., X.L. and Y.Z. performed the experiments. X.C. and D.X.Z. analyzed the data and prepared the figures. D.X.Z. wrote the main manuscript text. All authors reviewed the manuscript.
References
- Law J. A. & Jacobsen S. E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11, 204–220, 10.1038/nrg2719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun C. et al. Use of forward genetic screens to identify genes required for RNA-directed DNA methylation in Arabidopsis thaliana. Cold Spring Harb Symp Quant Biol 77, 195–204, 10.1101/sqb.2012.77.015099 (2012). [DOI] [PubMed] [Google Scholar]
- Slotkin R. K. & Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–285, 10.1038/nrg2072 (2007). [DOI] [PubMed] [Google Scholar]
- Teixeira F. K. et al. A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604, 10.1126/science.1165313 (2009). [DOI] [PubMed] [Google Scholar]
- Johannes F. et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5, e1000530, 10.1371/journal.pgen.1000530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Jacobsen S. E. & Reik W. Epigenetic reprogramming in plant and animal development. Science 330, 622–627, 10.1126/science.1190614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A. et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107, 18729–18734, 10.1073/pnas.1009695107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D. X. & Hu Y. F. Regulatory Function of Histone Modifications in Controlling Rice Gene Expression and Plant Growth. Rice 3, 103–111, 10.1007/S12284-010-9045-8 (2010). [DOI] [Google Scholar]
- Chen X. & Zhou D. X. Rice epigenomics and epigenetics: challenges and opportunities. Curr Opin Plant Biol 16, 164–169, 10.1016/j.pbi.2013.03.004 (2013). [DOI] [PubMed] [Google Scholar]
- Tanurdzic M. et al. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6, 2880–2895, 10.1371/journal.pbio.0060302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H. et al. Plants regenerated from tissue culture contain stable epigenome changes in rice. Elife 2, e00354, 10.7554/eLife.00354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hu Y. & Zhou D. X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim Biophys Acta 1809, 421–426, 10.1016/j.bbagrm.2011.03.004 (2011). [DOI] [PubMed] [Google Scholar]
- Hu Y. et al. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc Natl Acad Sci USA 109, 5773–5778, 10.1073/pnas.1203148109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. et al. The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front Plant Sci 5, 591, 10.3389/fpls.2014.00591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. et al. Regulation of Histone Methylation and Reprogramming of Gene Expression in the Rice Inflorescence Meristem. Plant Cell 10.1105/tpc.15.00201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J 61, 752–766, 10.1111/j.1365-313X.2009.04100.x (2010). [DOI] [PubMed] [Google Scholar]
- La H. et al. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc Natl Acad Sci USA 108, 15498–15503, 10.1073/pnas.1112704108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Platten D., Chaudhury A., Peacock W. J. & Dennis E. S. Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2, 711–723, 10.1093/mp/ssp036 (2009). [DOI] [PubMed] [Google Scholar]
- Choi S. C. et al. Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol 164, 1326–1337, 10.1104/pp.113.228049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F. J., Sun Q. W., Huang L. M., Chen X. S. & Zhou D. X. Rice SUVH histone methyltransferase genes display specific functions in chromatin modification and retrotransposon repression. Mol Plant 3, 773–782, 10.1093/mp/ssq030 (2010). [DOI] [PubMed] [Google Scholar]
- Chen Q. et al. Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet 9, e1003239, 10.1371/journal.pgen.1003239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. et al. Control of transposon activity by a histone H3K4 demethylase in rice. Proc Natl Acad Sci USA 110, 1953–1958, 10.1073/pnas.1217020110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q. & Zhou D. X. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105, 13679–13684, 10.1073/pnas.0805901105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. et al. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736, 10.1105/tpc.113.118802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo H. et al. DDM1 (decrease in DNA methylation) genes in rice (Oryza sativa). Mol Genet Genomics 287, 785–792, 10.1007/s00438-012-0717-5 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao C. et al. Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta 236, 1165–1176, 10.1007/s00425-012-1667-1 (2012). [DOI] [PubMed] [Google Scholar]
- Xue W. et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40, 761–767, 10.1038/ng.143 (2008). [DOI] [PubMed] [Google Scholar]
- Weng X. et al. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol 164, 735–747, 10.1104/pp.113.231308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A. et al. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep 11, 950–955, 10.1038/embor.2010.158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. V. et al. Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 9, e1003946, 10.1371/journal.pgen.1003946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. L. et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci USA 106, 22187–22192, 10.1073/pnas.0905767106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I. R., Pontes O., Melnyk C. W., Smith L. M. & Baulcombe D. C. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24, 986–991, 10.1101/gad.579910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. J. & Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23, 540–547, 10.1007/s00299-004-0843-6 (2005). [DOI] [PubMed] [Google Scholar]
- Dangwal M., Malik G., Kapoor S. & Kapoor M. De novo methyltransferase, OsDRM2, interacts with the ATP-dependent RNA helicase, OseIF4A, in rice. J Mol Biol 425, 2853–2866, 10.1016/j.jmb.2013.05.021 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.