Abstract
The repeated failures reported in cultivating some microbial lineages are a major challenge in microbial ecology and probably linked, in the case of Frankia microsymbionts to atypical patterns of auxotrophy. Comparative genomics of the so far uncultured cluster-2 Candidatus Frankia datiscae Dg1, with cultivated Frankiae has revealed genome reduction, but no obvious physiological impairments. A direct physiological assay on nodule tissues from Coriaria myrtifolia infected with a closely-related strain permitted the identification of a requirement for alkaline conditions. A high pH growth medium permitted the recovery of a slow-growing actinobacterium. The strain obtained, called BMG5.1, has short hyphae, produced diazovesicles in nitrogen-free media, and fulfilled Koch’s postulates by inducing effective nodules on axenically grown Coriaria spp. and Datisca glomerata. Analysis of the draft genome confirmed its close proximity to the Candidatus Frankia datiscae Dg1 genome with the absence of 38 genes (trehalose synthase, fumarylacetoacetase, etc) in BMG5.1 and the presence of 77 other genes (CRISPR, lanthionine synthase, glutathione synthetase, catalase, Na+/H+ antiporter, etc) not found in Dg1. A multi-gene phylogeny placed the two cluster-2 strains together at the root of the Frankia radiation.
Ever since Robert Koch first proposed a series of postulates to establish a causal link between a microbe and a disease, a basic tenet of microbiology has been the isolation of the causative microbes in pure culture1. Because many microbes such as fungi (Gigaspora, Rhizophagus) or bacteria (Mycobacterium leprae, Wolbachia, etc), cannot be cultivated in pure culture, it is now widely accepted these postulates should not be strictly considered2. Nevertheless, growth in pure culture remains an irreplaceable tool to study the physiology of an organism. Reasons behind these failures are varied including fragility of cells buffered within host tissues, cumulative damages by reactive compounds to cellular constituents such as DNA and membranes, or loss of genes resulting in auxotrophy3.
Frankia is a slow-growing actinobacterium. First described in 1866 as the causative agent of nodules found on the roots of Alnus and Elaeagnus4, it resisted cultivation attempts for a century. Because of its importance in ecological successions, studies were made using crushed nodules. However, the lack of isolates caused erroneous conclusions on plant host range and phylogenetic affiliation of the microbes present5. In 1978, the first Frankia isolation was reported6 using an enzymatic maceration of host tissues and a complex growth medium, and has led to many subsequent successful reports on the isolation of strains from the different host plants. Molecular phylogenetic approaches using both cultured and uncultured bacteria have grouped Frankia into 4 clusters7 and 3 of these clusters now routinely yield isolates from most provenances. However in spite of repeated attempts in numerous laboratories, cluster-2 strains that nodulate Rosaceae, Coriariaceae, Datiscaceae and Ceanothus have consistently resisted isolation. Nevertheless, the genome of a cluster-2 strain was determined by directly sequencing DNA isolated from nodule tissues8 and a comprehensive study of its metabolic pathways permitted to conclude there was no missing function that would impede cluster-2 strains to grow in pure culture9.
One of the aims of the cultivation of a microbe is to provide knowledge on its physiological properties, but this information is also necessary to tailor the growth media for isolation of this given microbe. This circular logic is usually broken through trials and errors. The Biolog (Hayward CA) assay, which allows the assessment of physiological properties of cells, does not depend on growth, but rather on the ability of cells to reduce an indicator dye, tetrazolium to the purple insoluble formazan10. We decided to use this system to study the physiology of cluster-2 Frankia strains and thus adapt our isolation conditions to fit the endophytic cells.
The genome of Candidatus Frankia datiscae Dg1, an uncultured cluster-2 strain, was analyzed to provide information on these symbionts for their isolation. Although no metabolic pathways appeared to be incomplete, the genome contained a reduced number of genes involved in stress responses8. With this information, a high-throughput microplate assay was used to first assess the viability of the microsymbiont. These live/dead assays were performed under different pH conditions, with various carbon sources and in the presence or absence of oxygen scavengers and osmolytes. Strikingly, the viability of these endophytic cell clusters was highly sensitive to pH. Viable endophytic cells were recoverable only from PM10 microplate wells with a pH higher than 8.5 for up to two months as shown by the live/dead assay. Therefore, buffers used to release vesicle clusters from root nodules and to inoculate Biolog microplates were adjusted to pH 9 for all subsequent experiments, which permitted a substantial extension of cell endophyte viability in numerous wells in PM1 and PM2A microplates. Pyruvate, propionate, succinate, and acetate are all generally used to cultivate other Frankia strains11,12,13, and were identified as potential carbon and energy sources for the isolation medium.
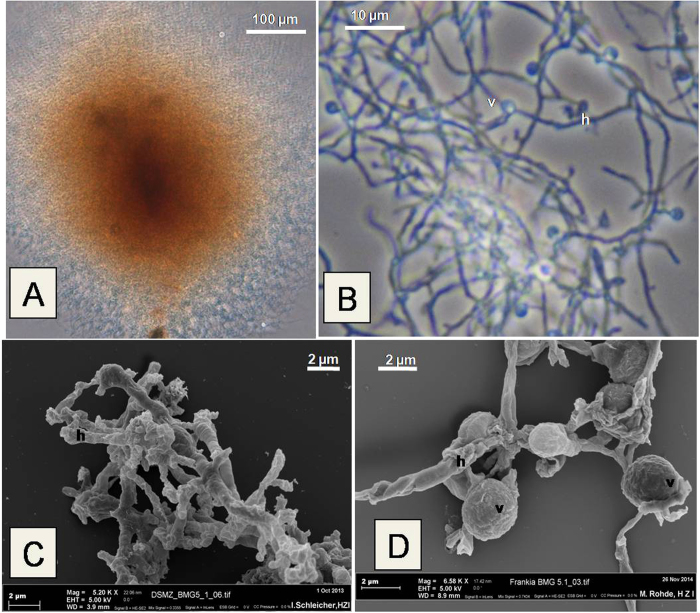
Based on this information, a modified BAP medium13,14 buffered to pH 9 and supplemented with several organic acids to replace the usual propionate as carbon source and several oxygen scavengers was used, and yielded very slow-growing brownish colonies. These colonies required a 14 month incubation period to yield sufficient material for subculturing. One colony, termed BMG 5.1, was selected, and gently fragmented for propagation. The isolate was maintained in pure culture and found to be free of contaminants after plating a sample of it on solid LB medium. The isolate grew slowly, forming hard-to-fragment colonies in liquid medium (Fig. 1A). Under phase contrast microscopy, the isolate was observed to consist of a mycelial mat containing short irregular filaments (Fig. 1B) with thick elbow-like regions that could be due to discontinuous growth (Fig. 1C). Globular diazovesicles, the site of nitrogen fixation in Frankia under symbiotic and pure-culture conditions15, were produced in nitrogen-deficient (BAP medium without NH4Cl) medium (Fig. 1D). No sporangia were observed under the conditions tested. The culture grown on nitrogen-deficient medium was also able to reduce acetylene (135 +/−11 nmoles h-1 mg-1 protein), a proxy for nitrogen-reduction, as soon as vesicles were formed. The results from the Biolog assays on isolate BMG5.1 grown in pure culture (Table S1) showed a similar pattern of physiological properties as predicted from the viability screening of microsymbiont clusters. Furthermore, isolate BMG5.1 showed an optimum growth yield at pH 9.5 (Fig. 2). In contrast, the other Frankia strains showed optimum growth yield around pH 7-7.5. Isolate BMG5.1 failed to grow in medium at pH 6.5 that was usually used for other Frankia isolates14.
Figure 1. Frankia strain BMG5.1 in pure culture.
(A) a micro-colony grown on BAP medium at pH 9. (B) a higher magnification with short hyphae (h) and spherical nitrogen-fixing vesicles (v). (C) and (D) scanning electron micrographs showing short thick hyphae and vesicles, respectively.
Figure 2.
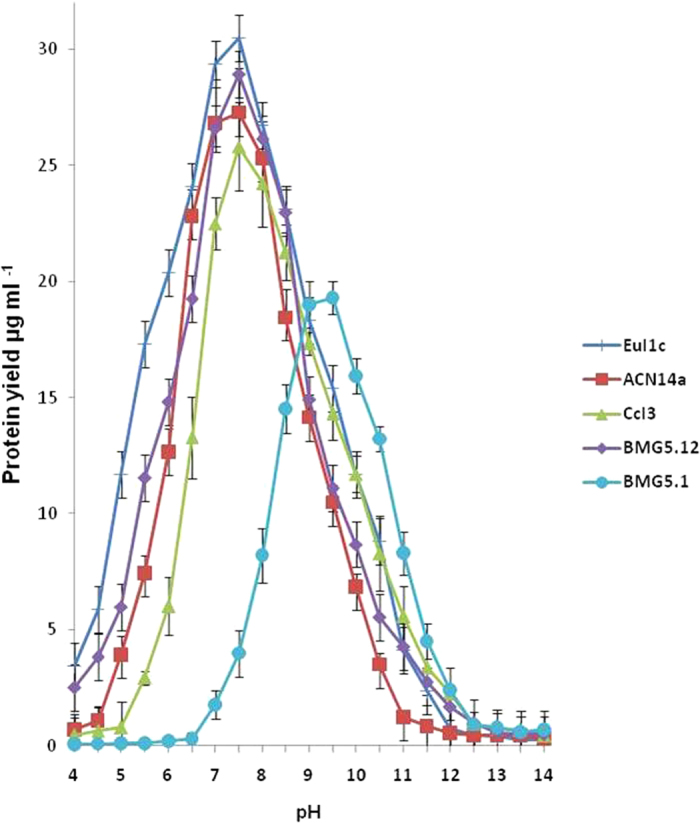
Protein yield of Frankia sp. strain BMG5.1 compared to Frankia sp. strains EuI1c (cluster-4), ACN14a (cluster-1a), CcI3 (cluster-1c) and BMG5.12 (cluster-3) grown in BAP medium with 5mM NH4Cl, buffered at different pH values.
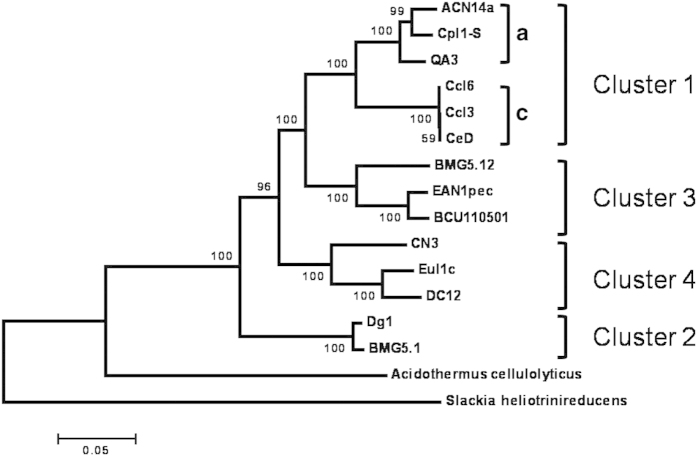
Phylogenetic analysis shows that BMG5.1 strain belongs to Frankia cluster-2 and shares 99.6%, 98.16%, 98.76% and 99.4% identity with the Candidatus Frankia datiscae Dg1, respectively on the basis of 16S rRNA, nifH, gyrB and glnA gene sequences. A multigene analysis using concatenated atpI, gyrA, ftsZ, secA and dnaK genes confirms the close proximity of the new isolate with Candidatus Frankia datiscae Dg1 at the root of the genus radiation (Fig. 3).
Figure 3. Maximum Likelihood phylogenetic tree estimated from concatenation of atp1, ftsZ, dnaK, gyrA and secA genes.
The tree was rooted using Acidothermus cellulolyticus and the deeply branching actinobacterium Slackia helionitrireducens and bootstrap values based on 1,000 replicates shown as percentages at branching points. Frankia clusters are designated as described previously7.
The draft genome of Frankia sp. strain BMG5.1 was determined and found to consist of 5,795,263 bp with a 71% G+C and 5,245 predicted protein-coding genes, characteristics which are very close to those of the Candidatus Frankia datiscae Dg1 genome (Table 1). All of the generated 500000 reads were found to be Frankia sequences indicating the absence of potential contaminants. Comparative analysis showed that the two genomes had an average nucleotide identity (ANI) of 96.93%, which would correspond to the same species16 and ANI scores of 82.32, 82.97, 83.05 and 83.68% with Frankia sp. strains EuI1c, ACN14a, EAN1pec and CcI3 genomes, respectively17. Several genes were found to be present in BMG5.1, but were absent in Dg1 (Table S2) including catalase, choline dehydrogenase and glutathione synthase that would correspond to a higher level of resistance to in vitro culture conditions for BMG5.1. Interestingly, the Frankia sp. BMG5.1 genome contains multiple Na+/H+ antiporter genes (Fig. S1).
Table 1. Summary of genome characteristics of isolated Frankia strain BMG5.1 and the Candidatus Frankia datiscae Dg1.
| BMG5.1 | Dg1 | |
|---|---|---|
| Status | Draft (116 scaffolds) | Finished (1 contig) |
| Size in bp | 5,795,263 | 5,323,186 |
| Proteins | 5,245 | 4,527 |
| Pseudogenes | 375 | 355 |
| rRNA | 3 | 6 |
| tRNA | 47 | 44 |
| %G+C | 70.16 | 70.04 |
| Transposons and IS elements | 29 | 180 |
| Horizontally Transferred Genes | 44 | 1 |
| Accession | JWIO01000000 | CP002801.1 |
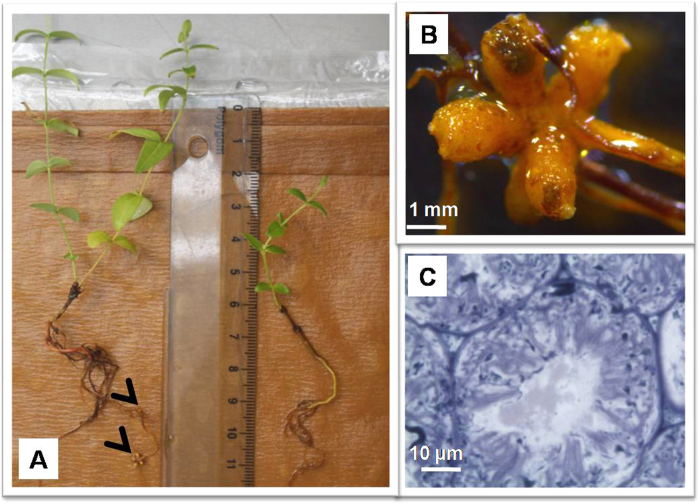
To fulfill Koch’s postulates, plant infectivity experiments were performed on seedlings of Coriaria japonica, Coriaria myrtifolia and Datisca glomerata, all of which are hosts of cluster-2 Frankia. The seedlings were grown axenically and transferred to growth pouches prior to inoculation with the isolate. After inoculation with isolate BMG5.1, the plants produced root nodules within 30–35 days. These nodules were actively fixing nitrogen as determined by the better growth of inoculated seedlings in N-free medium relative to non-inoculated controls (Fig. 4) and by the result of the acetylene reduction assay18 (211 + 3 nmoles h−1 mg−1 of nodule). The presence of BMG5.1 was confirmed by extracting DNA from the plant root nodules, amplifying and sequencing of the four DNA markers (16S rRNA, nifH, gyrB and glnA), which yielded identical sequences to the BMG5.1 isolate. Furthermore, BMG5.1 was reisolated from these nodules using the same isolation methods, grown in pure culture, and shown by PCR-sequencing to have identical markers (16S rRNA, nifH, gyrB and glnA).
Figure 4. Coriaria myrtifolia seedlings grown in growth pouches.
(A) seedlings were inoculated (left) with a pure culture of Frankia strain BMG5.1 or not inoculated (right). (B) a multi-lobed nodule. (C) a section through a cortical cell with peripheral nitrogen-fixing Frankia vesicles.
It is a stressful situation for a bacterium buffered by host tissues and under oxygen levels kept low by neighboring mitochondria19 to grow on synthetic medium. Down-regulation of many stress proteins in symbiotic Frankia alni20 also indicates that symbiosis may be less stressful than saprotrophic growth. In the case of alders, defense peptides have been shown to increase Frankia porosity21 and would render symbiotic cells particularly fragile. Thus, if in vitro growth conditions greatly differed from those found in the host tissues, isolation would be extremely difficult and would probably not occur. This is the reason why it is necessary to modify growth media to make them as comparable to the physiological conditions existing inside host tissues as possible. In the case of cluster-2 Frankia, these changes to the growth medium included the addition of oxygen scavengers and the use of an alkaline pH. Physiological assessment of uncultivated cells can thus provide reliable physiological data, leading to testable hypotheses for pure culture conditions.
This new Frankia isolate may form, together with Candidatus Frankia datiscae Dg1 strain, a different species that is mildly alkaliphilic. A number of alkaliphiles have been found among various lineages, many of which are actinobacteria in arid soils22. Adaptation to an alkaline biotope occurs through either local acidification of the medium or modification of the wall. Among the genes present in the BMG5.1 genome but absent in Dg1 were the Na+/H+ antiporters, which have been described as permitting cells to adjust to high pH22. A pure culture of Frankia BMG5.1 should be helpful in experiments directed toward our understanding this requirement for these unusual physiological conditions.
Clearly, the use of bioinformatics analysis of the genome and physiological bioassays has resulted in the isolation of a previously recalcitrant microbe. This dual approach could be extended to other microbes considered as yet uncultivable such as Mycobacterium leprae or Rhizophagus intraradices, which should markedly facilitate their study and biotechnological uses.
Methods
Vesicle clusters viability and growth requirement screening
Root nodules obtained from Coria-ria japonica23 growing in Japan (Tosa district) were used to inoculate Coriaria myrtifolia seedlings grown on sterile sand and maintained in a greenhouse. Root nodules were harvested, surface disinfected (Na hypochlorite 1.05% for 30 min) and washed several times with sterile distilled water. Preparation of vesicle clusters was performed as described previously24. Lobes were crushed in 0.05 M sodium phosphate buffer, pH 7.2 using a Broeck tissue homogenizer and incubated with the following scavengers for hydrogen peroxide (10 mM N,N’-dimethylthiourea) hydroxyl radical (0.28 M dimethylsulfoxide), for singlet oxygen (0.02 M L-histidine), for superoxide anion (10 mM 4,5-dihydroxybenzene-1,3-disulfonate), for peroxyl radical (100 μM α-tocopherol) and for peroxynitrite anion (100 μM uric acid). Released microsymbiont vesicle clusters were retained by gravity filtration on the 52-μm nylon screen and used to assess cells viability using PM1 and PM2A microplates (Biolog) for carbon sources, as well as PM9 and PM10 microplates (Biolog) for pH and NaCl tolerance, respectively. Viability of endophytic cell clusters was weekly assessed in a subset of PM1, PM2A, PM9 and PM10 microplates using only the live/dead assay BacLight Bacterial Viability Kit for microscopy (Life Technologies, Paisley, UK).
Growing Coriaria endophyte
Vesicle clusters of Coriaria endophyte were prepared as described above except that sodium phosphate buffer was adjusted to pH9. BAP medium14 containing in mM: K2HPO4, 4; KH2PO4, 7; MgSO4, 0.1; CaCl2, 0.07; FeNaEDTA, 10 mg/l and Hoagland’s microelements25 with 5 mM NH4Cl (BAP+) was supplemented with selected carbon sources, each added to a final concentration of 10 mM; pyruvate, propionate, succinate and acetate and with a mixed vitamin stock solution contained the following (per liter): 4-aminobenzoic, acid, 40 mg; D-(+)-biotin, 10 mg; nicotinic acid, 100 mg; calcium D(+) −pantothenate, 50 mg; pyridoxamine dihydrochloride, 100 mg; and thiamine dihydrochloride, 100 mg was inoculated with clumps of microsymbiont vesicle clusters in 10 ml glass tubes containing 3 ml liquid medium adjusted to pH9 using TRIS (tris-hydroxy-methyl-aminomethane) and incubated in the dark at 25 °C without agitation. Subcultures of isolated bacteria were characterized by 16S rRNA PCR amplification and sequencing using primers S-D-Bact0008-a-S-20 and S-D-Bact-1495-a-A-2026.
Host plant infectivity and molecular identification
Surface sterilized seeds of C. japonica, C. myrtifolia and D. glomerata were germinated on moistened filter paper in Petri dishes, at 20 °C under daylight illumination and transferred to sterile moist sand. Three week old seedlings were inoculated by applying the equivalent to 10 μg of protein of washed cultures of the isolate previously syringed with a 21G sterile needle. Plantlets were checked for nodules every week. After 9 weeks, nodules were harvested and used for endophyte reisolation and direct DNA extraction. DNA was extracted from root nodules and isolates using Plant DNeasy kits (Qiagen, Hilden, Germany) and characterized by PCR/sequencing of 16S rRNA , nifH, gyrB and glnA genes using respectively S-D-Bact0008-a-S-20 and S-D-Bact-1495-a-A-2026, nifHF and nifHR27, FragyrBF and FragyrBR28 and DB41 and DB4429. The generated sequences were then analyzed by BLASTN30 and compared to those obtained for BMG5.1.
Acetylene reduction activity
C. japonica and D. glomerata seedlings with and without nodules were assayed for nitrogenase activity by monitoring acetylene reduction18 . About 1g of nodules was harvested and placed in sterile Vacutainer (Beckton-Dickinson, Franklin Lakes, NJ) tubes which had 10% of the gas volume replaced with acetylene. After a 60 min incubation at 30 °C, 1 mL of the gas volume was injected into a Girdel gas chromatograph (Suresnes, France) equipped with a flame ionization detector.
A similar assay was made for a BMG5.1 culture in BAP medium without ammonium (BAP-). Vacutainer tubes (10 mL) were inoculated with 5 ml of the liquid cultures and after 1 week, 10% (vol/vol) of the remaining gas volume was replaced with acetylene and incubated for 60 min at 30 °C.
Biolog and growth at different pH
Frankia BMG5.1 cells were harvested by centrifugation (9000 x g, 15 min), washed three time and incubated four days in BAP- mineral solution (no carbon source) to deplete nutritional reserves. The cells were homogenized by repeated passages through a 25G needle. Biolog microplates were inoculated with cells at 10% transmittance suspended in IF0 buffer (supplied by the manufacturer) adjusted to pH 9 for PM1, PM2A and PM9 microplates and supplemented with pyruvate (10 mM final) for PM9 and PM10 microplates and incubated at 28 °C for 20 days. The new isolate BMG5.1 and strains representing the three other Frankia clusters; BMG5.12 (cluster-3), ACN14a (cluster-1a), CcI3 (cluster-1b) and EuI1c (cluster-4) were grown in BAP+ media adjusted to different pH. Organic buffers used to reach the desired pH were MES (2-N-morpholinoethanesulfonicacid) for pH 6.5, and MOPS (3-(N-morpholino)-propanesulfonic acid) for the pH range 6.5–7 and TRIS (tris-hydroxy-methyl-aminomethane) for pH above 7. After 3 weeks of growth, growth yield was determined by measuring total cellular protein. After harvesting, bacterial cell samples were suspended in 1.0 N NaOH and solubilized by incubating for 15 min at 90 °C. Total cellular protein levels were measured using the BCA method31.
Genome sequencing of the BMG5.1 strain
Approximately 500 mg of cells obtained from 2 L of 2 months old cultures were washed and their DNA extracted using a Plant DNeasy kit (Qiagen). Highly pure genomic DNA from strain BMG5.1 was used to generate a 300-bp library for paired-end 2 × 100 Illumina sequencing (Imagif, Gif-sur-Yvette, France) from 500000 reads. Sequences assembly was performed using the Velvet de novo assembler32. The assembled Frankia sp. strain BMG5.1 genome was analyzed via the Integrated Microbial Genomes (IMGer) platform of the Joint Genome Institute, Walnut Creek, CA. This whole genome shotgun sequence has been deposited at DDBJ/EMBL/GenBank under accession number JWIO00000000. The version described in this paper is JWIO01000000.
The draft genome was compared to that of Candidatus Frankia datiscae to identify those genes different between the two genomes using the default values of 30% amino acid identity over 80% of the length of the shortest sequence on the IMGer platform33.
Phylogenetic analysis
Selected MLSA phylogenetic markers34 were downloaded from the JGI-IMG (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) and used for Maximum Likelihood35 phylogenetic inference using MEGA 5.036.
Additional Information
How to cite this article: Gtari, M. et al. Cultivating the uncultured: growing the recalcitrant cluster-2 Frankia strains. Sci. Rep. 5, 13112; doi: 10.1038/srep13112 (2015).
Supplementary Material
Acknowledgments
This research was supported by The Ministere de l’Enseignement Superieur et de la Recherche Scientifique, Tunisie project LR03ES03. We would like to thank Prof. Manfred Rohde (Helmholz Centre for Infection Biology, Braunschweig, Germany) for the scanning Electron Microscopy. PN acknowledges funding from the French ANR (BugsInACell ANR-13-BSV7-0013-03). Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution Number 2617. This work was partially supported by the USDA National Institute of Food and Agriculture Hatch Project 022821.
Footnotes
Author Contributions M.G. conceived the study and designed experiments. M.G., F.G.G. and I.N. performed Frankia isolation and cultivation; F.G.G. performed Biolog and plant infectivity experiments; A.A., A.K. and W.M. performed pH experiment; I.N., I.S., K.H. and A.K. performed plant experiment. T.Y. provided Coriaria root nodules and seeds. A.B. advised in data analysis; M.G. and P.N. performed the comparative genomics; M.G., L.S.T. and P.N. wrote the paper. All authors read the final version of the manuscript.
References
- Koch R. Die Aetiologie der Tuberculose. Berliner klinische Wochenschrift 19, 221–230 (1882). [Google Scholar]
- Relman D. A. The identification of uncultured microbial pathogens. The Journal of infectious diseases 168, 1–8 (1993). [DOI] [PubMed] [Google Scholar]
- Stewart E. J. Growing unculturable bacteria. Journal of bacteriology 194, 4151–4160, 10.1128/JB.00345-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woronin M. S. Uber die bei der Schwarzerle (Alnus glutinosa) und bei der gewöhnlichen Garten-Lupine (Lupinus mutabilis) auftretenden Wurzelanschwellungen. Mem Acad Imp Sci St.Petersbourg VII Series. 10, 1–13 (1866). [Google Scholar]
- Becking J. H. Frankiaceae fam. nov. (Actinomycetales) with one new combination and six new species of the genus Frankia Brunchorst 1886, 174. Int J Syst Bacteriol 20, 201–220 (1970). [Google Scholar]
- Callaham D., Del Tredici P. & Torrey J. G. Isolation and cultivation in vitro of the actinomycete causing root nodulation in Comptonia. Science 199, 899–902 (1978). [DOI] [PubMed] [Google Scholar]
- Normand P. et al. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol 46, 1–9 (1996). [DOI] [PubMed] [Google Scholar]
- Persson T. et al. Genome sequence of “Candidatus Frankia datiscae” Dg1, the uncultured microsymbiont from nitrogen-fixing root nodules of the dicot Datisca glomerata. J Bacteriol 193, 7017–7018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson T. et al. Candidatus Frankia datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS ONE 10, e0127630, 10.1371/journal.pone.0127630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. M. & Rhoden D. L. Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J Clin Microbiol 29, 1143–1147 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. & O’Keefe D. A modified sucrose fractionation procedure for the isolation of frankiae from actinorhizal root nodules and soil samples. Plant Soil 78, 23–28 (1984). [Google Scholar]
- Murry M., Fontaine M. & Torrey J. Growth kinetics and nitrogenase induction in Frankia sp. HFPArI3 grown in batch culture. Plant Soil 78, 61–78 (1984). [Google Scholar]
- Tisa L., McBride M. & Ensign J. C. Studies on growth and morphology of Frankia strains EAN1pec, EuI1c, CpI1 and ACN1AG. Can. J. Bot. 61, 2768–2773 (1983). [Google Scholar]
- Murry M., Fontaine M. & Torrey J. Growth kinetics and nitrogenase induction in Frankia sp. HFPArI3 grown in batch culture. Plant & Soil 78, 61–78 (1984). [Google Scholar]
- Tjepkema J. D., Ormerod W. & Torrey J. G. Vesicle formation and acetylene reduction activity in Frankia sp. CpI1 cultured in defined nutrient media. Nature 287, 633–635 (1980). [Google Scholar]
- Goris J. et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57, 81–91, 10.1099/ijs.0.64483-0 (2007). [DOI] [PubMed] [Google Scholar]
- Normand P. et al. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res 17, 7–15 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart W., Fitzgerald & Burris, R. In situ studies on N2 fixation using the acetylene reduction technique. Proc Nat Acad Sci (USA) 58, 2071–2078 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvester W. B., Langenstein B. & Berg R. H. Do mitochondria provide the oxygen diffusion barrier in root nodules of Coriaria and Datisca? Can. J. Bot. 77, 1358–1366 (1999). [Google Scholar]
- Alloisio N. et al. The Frankia alni symbiotic transcriptome. Mol Plant Microbe Interact 23, 593–607 (2010). [DOI] [PubMed] [Google Scholar]
- Carro L. et al. Alnus peptides modify membrane porosity and induce the release of nitrogen-rich metabolites from nitrogen-fixing Frankia. The ISME journal 10.1038/ismej. 2014.257, 10.1038/ismej.2014.257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. F. et al. Nesterenkonia rhizosphaerae sp. nov., a novel alkaliphilic actinobacterium isolated from rhizosphere soil of Reaumuria soongorica in saline-alkaline desert. Int J Syst Evol Microbiol, 10.1099/ijs.0.066894-0 (2014). [DOI] [PubMed] [Google Scholar]
- Nouioui I. et al. Absence of cospeciation between the uncultured Frankia microsymbionts and the disjunct actinorhizal Coriaria species. BioMed research international 2014, 924235, 10.1155/2014/924235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. Isolation of Frankia strains from alder actinorhizal root nodules. Applied and Environmental Microbiology 44, 461–465 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. R. & Arnon D. I. The Water Culture Method for Growing Plants Without Soil., 142 (Circular 347, California Agricultural Experiment Station, 1950). [Google Scholar]
- Daffonchio D., Borin S., Frova G., Manachini P. L. & Sorlini C. PCR fingerprinting of whole genomes: the spacers between the 16S and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis [corrected]. Int J Syst Bacteriol 48, 107–116 (1998). [DOI] [PubMed] [Google Scholar]
- Gtari M. et al. Genetic diversity among Elaeagnus compatible Frankia strains and sympatric-related nitrogen-fixing actinobacteria revealed by nifH sequence analysis. Soil Biol. Biochem. 39, 372–377 (2007). [Google Scholar]
- Nouioui I., Ghodhbane-Gtari F., Beauchemin N. J., Tisa L. S. & Gtari M. Phylogeny of members of the Frankia genus based on gyrB, nifH and glnII sequences. Antonie Van Leeuwenhoek 100, 579–587, 10.1007/s10482-011-9613-y (2011). [DOI] [PubMed] [Google Scholar]
- Clawson M. L., Bourret A. & Benson D. R. Assessing the phylogeny of Frankia-actinorhizal plant nitrogen-fixing root nodule symbioses with Frankia 16S rRNA and glutamine synthetase gene sequences. Mol Phylogenet Evol 31, 131–138 (2004). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Molec Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Smith P. K. et al. Measurement of protein using bicinchoninic acid. Analytical biochemistry 150, 76–85 (1985). [DOI] [PubMed] [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18, 821–829, 10.1101/gr.074492.107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V. M. et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res 34, D344–348, 10.1093/nar/gkj024 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A. et al. Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders ‘Frankiales’ and Micrococcales should be split into coherent entities: proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov. and Nakamurellales ord. nov. Int J Syst Evol Microbiol 64, 3821–3832, 10.1099/ijs.0.063966-0 (2014). [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729, 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.