Abstract
The cell cycle is important for growth, genome replication, and development in all cells. In bacteria, studies of the cell cycle have focused largely on unsynchronized cells making it difficult to order the temporal events required for cell cycle progression, genome replication, and division. Caulobacter crescentus provides an excellent model system for the bacterial cell cycle whereby cells can be rapidly synchronized in a G0 state by density centrifugation. Cell cycle synchronization experiments have been used to establish the molecular events governing chromosome replication and segregation, to map a genetic regulatory network controlling cell cycle progression, and to identify the establishment of polar signaling complexes required for asymmetric cell division. Here we provide a detailed protocol for the rapid synchronization of Caulobacter NA1000 cells. Synchronization can be performed in a large-scale format for gene expression profiling and western blot assays, as well as a small-scale format for microscopy or FACS assays. The rapid synchronizability and high cell yields of Caulobacter make this organism a powerful model system for studies of the bacterial cell cycle.
Keywords: Cellular Biology, Issue 98, cell cycle, cell biology, systems biology, synchronization, Caulobacter, asymmetric cell division
Introduction
The bacterial cell cycle controls both the replication of the genome and the division of daughter cells. Importantly, as antibiotic resistance is a growing threat to public health, the bacterial cell cycle presents an untapped target for antibiotic development.
In the bacterium Caulobacter crescentus, each cell cycle leads to an asymmetric division, yielding two daughter cells of different fates (Figure 1A) 1,2. One daughter cell inherits a flagellum and is motile while the other daughter inherits a stalk and is sessile. An integrated genetic circuit controls cell cycle progression and cell fate by transcriptional regulation, phospho-signaling, and regulated proteolysis 3. In addition, chromosome replication and concurrent segregation yield daughter cells that contain exactly one copy of the chromosome 4. Importantly, these two cell types can be rapidly separated by colloidal silica particle density centrifugation in the synchronizable NA1000 strain 5-7 allowing the isolation of the swarmer cells from the rest of the population with high yields (Figure 1B). Isolated swarmer cells then proceed synchronously through asymmetric cell division. Here, we detail the protocol used for synchronizing Caulobacter strain NA1000. We provide protocols and common troubleshooting tips for both large- and small-scale synchronizations. This experimental procedure provides a powerful tool to interrogate the spatiotemporal control of the Caulobacter cell cycle and cell fate.
Protocol
1. Large-scale Synchrony - Optimal for Western Blot, Microarray/RNA-Seq, and Other Material Intensive Assays
From a freezer stock or a plate, grow a 5 ml O/N culture of strain NA1000 by shaking at 28 °C in PYE medium.
Inoculate 0.5 ml of the cells from step 1 in 25 ml of M2G (Tables 1-2) and shake at 28 °C until the culture reaches an OD600 between 0.5 and 0.6.
Inoculate the cells into 1 L of M2G and shake at 28 °C.
Once OD600 reaches 0.5 to 0.6, confirm the presence of swarmer cells using liquid mounted phase microscopy. Spot 1 µl of cells on a glass slide, cover with a cover slip, and image by phase microscopy. Confirm the presence of swarmer cells by visualizing rapidly swimming cells in the population.
Spin cells for 15 min at 7 k x g at 4 °C in a JA-10 rotor.
Discard the supernatant and add 180 ml of cold M2 (Tables 1-2) and gently resuspend all the cells using a serological pipet. Discard loosely pelleted cells; they are predivisional and stalked cells.
Add 60 ml of cold Colloidal silica solution (be sure to mix the Colloidal silica suspension well before adding to the cells) and mix the cell suspension well.
Pour the cell suspension into eight 30 ml tubes and spin for 30 min at 6.4 k x g at 4 °C in a JA-20 rotor. NOTE: One should see two distinct bands; the swarmer band is the lower band while stalked/predivisional cells are in the top band (Figure 1B).
Carefully aspirate the top band off and remove the liquid to ~1 cm above the swarmer band (the lower band).
(Critical) Using a Pasteur pipet, carefully remove the swarmer band and place into a clean tube. To wash away the Colloidal silica, top the tube off with cold M2 and spin for 10 min at 6.4 k x g at 4 °C in a JA-20 rotor.
Carefully discard the supernatant and resuspend the cells in 20 ml of cold M2 and spin for 10 min at 6.4 k x g at 4 °C in a JA-20 rotor.
Resuspend all the pellets into 30 ml of cold M2 and measure the OD600 using a spectrophotometer and blank using cold M2 medium. Save 1 µl for phase imaging to check for swarmer cells; 90-95% of cells should be swarmers.
Spin down the cells for 5 min at 6.4 k x g at 4 °C in a JA-20 rotor. Resuspend cells in 28 °C M2G medium so that the A600 is ~0.3-0.4 and begin shaking at 28 °C. NOTE: Typical yields are between 30 and 60mL of swarmer cell culture from 1L of unsynchronized cells.
Begin taking time points (wild type culture will take approximately 135-140 minutes to divide) 8,9. At each time point, measure the OD600. Check that the OD600 after division is approximately 2X the initial OD600.
For western blot or gene expression assays, remove 1mL aliquots of the culture at the desired time points, spin down at max speed in a tabletop centrifuge for 30 sec, rapidly decant or aspirate the medium, and flash freeze the cell pellet in liquid nitrogen. Store the cells at -80 °C until downstream analysis.
2. Small-scale Synchrony – Optimal for Microscopy
From a freezer stock or a plate, grow a 5 ml O/N culture shaking at 28 °C in M2G.
Dilute in 15 ml of M2G (Tables 1-2) and grow until mid-log (OD600 = 0.5-0.6).
Spin at 6.4 k x g for 10 min at 4 °C in a JA-20 rotor, and resuspend in 1 ml cold M2 (Tables 1-2) and transfer to a 2 ml microcentrifuge tube.
Spin at 15 k x g for 3 min in a microcentrifuge tube to pellet cells, aspirate off the supernatant, put the pellet on ice, and resuspend in 900 µl of cold M2.
Add 900 µl of cold PVP coated colloidal silica and spin for 20 min at 15 k x g at 4 °C in a microcentrifuge tube.
(Critical) Aspirate or pipet off the top stalked/predivisional cell band and collect the bottom swarmer band into a new microcentrifuge tube.
Wash the swarmer cells two times in 1 ml of cold M2 while centrifuging at 15 k x g for 3 min.
Before the final spin, move the cells into a pre-chilled 1 ml glass test tube and measure the OD600 of the cells compared to a blank of M2.
Resuspend the final cell pellet into 28 °C M2G at an OD between 0.3 – 0.4 and shake/roll cells at 28 °C. NOTE: Typical yields are between 2 and 4 ml of swarmer cell culture.
For microscopy experiments, at the desired time points place 1 µl of cells onto an M2G agarose pad for imaging.
Representative Results
Synchronization typically yields two bands of cells (Figure 1B): the swarmer band, which has a higher density, and a stalked/predivisional cell band of lower density. To ensure efficient synchronization common controls include monitoring the OD600 and measuring the levels of CtrA protein by western blot at distinct cell cycle time points. The OD600 should increase by approximately 2 fold during the course of the cell cycle (Figure 2). The western blot for the cell cycle master regulator CtrA is a useful control to verify a good synchrony (Figure 2). CtrA is utilized in the swarmer cell to block DNA replication and is degraded upon the onset of DNA replication 10. CtrA is then synthesized later in the cell cycle and activates transcription of a host of developmental genes including many components of the flagellum 11,12. A successful synchrony will have this oscillating pattern of CtrA protein levels.
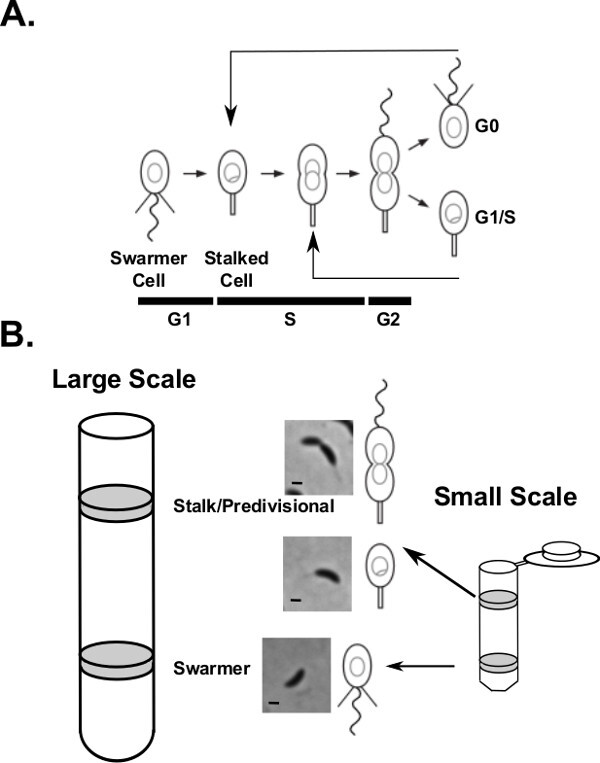 Figure 1. The cell cycle of Caulobacter crescentus.(A) A cartoon schematic of the swarmer cell cycle. The swarmer cells differentiate into replication competent stalked cells by retracting pili, ejecting flagella, and initiating DNA replication. The circles and theta structures shown inside the cell outlines represent quiescent and replicating chromosomes. Cells then progress through the cell cycle building a single flagellum at the pole opposite the stalk. Upon division, two unique cell types are generated, a replication blocked motile swarmer cell and a replication competent stationary stalked cell. (B) Representative results for density centrifugation. Lower density stalked and predivisional cells float near the top of the gradient while dense swarmer cells end up toward the bottom of the tube. Scale bars are 1 µm in phase microscopy images. Please click here to view a larger version of this figure.
Figure 1. The cell cycle of Caulobacter crescentus.(A) A cartoon schematic of the swarmer cell cycle. The swarmer cells differentiate into replication competent stalked cells by retracting pili, ejecting flagella, and initiating DNA replication. The circles and theta structures shown inside the cell outlines represent quiescent and replicating chromosomes. Cells then progress through the cell cycle building a single flagellum at the pole opposite the stalk. Upon division, two unique cell types are generated, a replication blocked motile swarmer cell and a replication competent stationary stalked cell. (B) Representative results for density centrifugation. Lower density stalked and predivisional cells float near the top of the gradient while dense swarmer cells end up toward the bottom of the tube. Scale bars are 1 µm in phase microscopy images. Please click here to view a larger version of this figure.
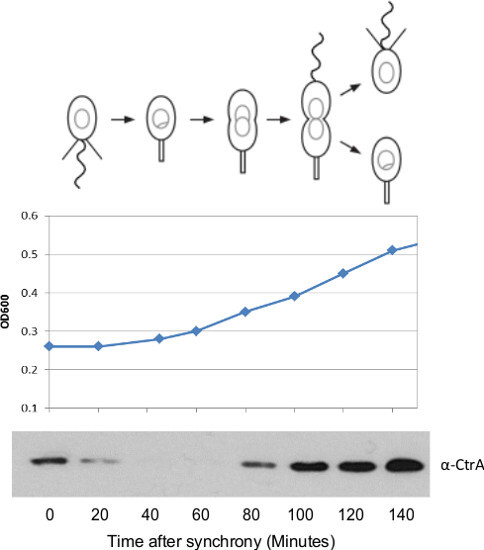 Figure 2. Representative results for a successful swarmer cell synchrony. The cell mass measured by OD600 should slowly increase and ultimately double throughout the course of the assay. 1 ml aliquots were pipetted into plastic cuvettes from a large cell synchrony and the OD600 measured using a spectrophotometer at the indicated time points. Additionally, the CtrA cell cycle master regulatory protein should be present in the swarmer cell to block DNA replication, followed by a rapid degradation coincident with the initiation of DNA replication. CtrA is then regenerated later in the cell cycle to activate expression of many developmental genes including critical flagellar and pili components. Western blots for cell cycle master regulatory protein CtrA were performed by taking 1 ml aliquots of a large-scale synchrony. The cells were resuspended in 250 µl Laemmlli sample buffer per OD600, separated on a 10% TRIS-GLY PAGE, transferred to PVDF, and blotted with anti-CtrA antibody. Failed synchrony procedures lead to CtrA western blots with no change in protein levels. α-CtrA antibody 12 was incubated at a 1:10,000 dilution for 1.5 hr in 3% milk TBST and washed 3 times in TBST. Goat-α-rabbit secondary was then added at 1:10,000 dilution in 3% milk TBST for 1 hr, washed 3 times with TBST, and imaged on film using a chemiluminescent detection kit. Please click here to view a larger version of this figure.
Figure 2. Representative results for a successful swarmer cell synchrony. The cell mass measured by OD600 should slowly increase and ultimately double throughout the course of the assay. 1 ml aliquots were pipetted into plastic cuvettes from a large cell synchrony and the OD600 measured using a spectrophotometer at the indicated time points. Additionally, the CtrA cell cycle master regulatory protein should be present in the swarmer cell to block DNA replication, followed by a rapid degradation coincident with the initiation of DNA replication. CtrA is then regenerated later in the cell cycle to activate expression of many developmental genes including critical flagellar and pili components. Western blots for cell cycle master regulatory protein CtrA were performed by taking 1 ml aliquots of a large-scale synchrony. The cells were resuspended in 250 µl Laemmlli sample buffer per OD600, separated on a 10% TRIS-GLY PAGE, transferred to PVDF, and blotted with anti-CtrA antibody. Failed synchrony procedures lead to CtrA western blots with no change in protein levels. α-CtrA antibody 12 was incubated at a 1:10,000 dilution for 1.5 hr in 3% milk TBST and washed 3 times in TBST. Goat-α-rabbit secondary was then added at 1:10,000 dilution in 3% milk TBST for 1 hr, washed 3 times with TBST, and imaged on film using a chemiluminescent detection kit. Please click here to view a larger version of this figure.
| Na2HPO4 | 17.4 g |
| KH2PO4 | 10.6 g |
| NH4Cl | 10 g |
| H2O | Resuspend in 1 L & autoclave |
Table 1. 20X M2 Salts Recipe.
| 20X M2 Salts | 50 ml | |
| 0.5 M MgSO4 | 1 ml | |
| 20% Glucose | 10 ml | Substitute for H2O in M2 |
| Ferrous Sulfate Chelate Solution | 1 ml | |
| 0.1 M CaCl2 | 5 ml | Add last to avoid precipitation |
| H2O | Fill up to 1 L | |
| Sterile filter with 0.22 µM filter |
Table 2. M2 and M2G recipe.
Discussion
The bacterial cell cycle is a fundamental process in life and is important for the study of growth and as a target for next generation antibiotics. Here, we detailed the rapid synchronization procedures for C. crescentus NA1000, a model organism for the study of the bacterial cell cycle and asymmetric cell division. This method is amendable to western blot, gene expression profiling, and fluorescence microscopy assays to investigate the spatiotemporal regulation of the bacterial cell cycle.
The protocol is quick and yields healthy synchronized cells. Other synchronization methods require long starvation of cells and rely on the stringent response to arrest the cell cycle in the same state, leading to cells that are dissimilar to cultures in logarithmic growth 13. While “baby machines” and other surface attachment methods have been used successfully14,15, they typically have low yields of synchronized cells. The density centrifugation protocols presented here allow higher purity and yields of synchronized C. crescentus NA1000 swarmer cells.
Additional complications can lead to potential problems throughout the synchrony procedure. Have a liquid mount phase microscope handy to optically check the purity of cells before synchronization and to check the purity of swarmers. A good synchrony will yield >95% swarmer cells. Check the OD600 on a spectrophotometer to ensure the cells are doubling during the time course of the synchrony.
Cells don’t synchronize as well in the presence of antibiotics or some mutations. If the strain does not carry a replicating plasmid, try to remove the antibiotics from the growth medium to improve swarmer cell yield. Cells grown in the presence of antibiotics or mutant strains will also have a slower cell cycle, so it is important to check the division time by microscopy for comparison to healthy strains.
Avoid vortexing the cells, as this will shear off the flagella and lower the quality of the synchrony. In preparation for the synchrony protocol don’t grow the cells above OD600 0.5. This ensures healthy growth that isn’t altered by high cell density. Be careful when aspirating/pipetting off supernatants containing the stalked and predivisional cell populations. In particular, do not to touch the swarmer band which may lead to contamination and/or a loss in yield of the swarmer cells. The cell pellet will be very loose in the first wash of cells after removing them from the Colloidal silica. Be careful when removing the supernatant not to disrupt the pellet.
Occasionally NA1000 cells can lose synchronization capacity due to a loss of DNA in the prophage region of the genome 7. In this case, no swarmer band is observed or the swarmer band intensity is dramatically reduced. If possible, collect cells if present in the swarmer band and streak onto a PYE plate to re-isolate the synchronizable cells. If working with a mutant strain, it is often useful to reintroduce the mutation into a fresh batch of NA1000. Conversely, mutation of critical cell cycle regulatory proteins can, in some cases, disrupt the ability of cells to synchronize16.
M2G medium was initially described with a higher 20 mM phosphate concentration 8,17; however, current studies use a lower 10 mM concentration as presented in Table 1 18,19. Alternative to the standard M2G medium, it is possible to use richer PYE medium. Here the unsynchronized growth and growth after synchronization can be substituted for PYE.
C. crescentus NA1000 provides a valuable experimental tool to study the bacterial cell cycle. Additionally, due to the single copy of the chromosome in the swarmer cell, this bacterium has also become a powerful model to study the structure of the bacterial chromosome.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank members of the Shapiro lab and Erin Schrader for comments on the manuscript. The authors acknowledge financial support from: NIH postdoctoral fellowship F32 GM100732 to JMS and NIH grants R01 GM51426 and R01 GM32506 to LS.
References
- McAdams HH, Shapiro L. System-level design of bacterial cell cycle control. FEBS Lett. 2009;583:3984–3991. doi: 10.1016/j.febslet.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Shapiro L. The architecture and conservation pattern of whole-cell control circuitry. J. Mol. Biol. 2011;409:28–35. doi: 10.1016/j.jmb.2011.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Shapiro L. Initiating bacterial mitosis: understanding the mechanism of ParA-mediated chromosome segregation. Cell Cycle. 2010;9:4033–4034. doi: 10.4161/cc.9.20.13521. [DOI] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks ME, et al. The genetic basis of laboratory adaptation in Caulobacter crescentus. J. Bacteriol. 2010;192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol. Microbiol. 2014;93:853–866. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Ferullo DJ, Cooper DL, Moore HR, Lovett ST. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen ST, Newton A. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Bates D, et al. The Escherichia coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle. Mol. Microbiol. 2005;57:380–391. doi: 10.1111/j.1365-2958.2005.04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, et al. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the caulobacter cell cycle. PLoS Genet. 2013;9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britos L, et al. Regulatory response to carbon starvation in Caulobacter crescentus. PLoS One. 2011;6:e18179. doi: 10.1371/journal.pone.0018179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol. Microbiol. 2011;80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


