Nuclear IκB-ζ, a target of TGF-β signaling in T cells, controls immune homeostasis and IFN-γ production.
Keywords: T lymphocyte, cytokine, transcriptional regulator
Abstract
The transcriptional regulator IκB-ζ is important for the control of apoptosis in keratinocytes. Thus, IκB-ζ-deficient mice develop autoimmune diseases, such as Sjögren’s syndrome. However, T cells also play a pivotal role in Sjögren’s syndrome. To study the role of IκB-ζ in T cells, we generated T cell-specific, IκB-ζ-deficient mice. We observed increased numbers of peripheral effector/memory CD4+ cells and IFN-γ-producing CD4+ cells in 3-week-old mice. We found that IκB-ζ can be up-regulated by TGF-β1 in naïve CD4+ T cells and that it negatively regulates IFN-γ expression. In addition, we generated Treg-specific, IκB-ζ deficient mice and found that IκB-ζ is dispensable for the plasticity and stability of Tregs. However, Tregs from T cell-specific, IκB-ζ-deficient mice have reduced immunoregulatory function. Thus, our data reveal a previously unappreciated role for IκB-ζ in IFN-γ production in T cells and the immunoregulatory function of Tregs.
Introduction
Inducible nuclear IκB-ζ is a member of the IκB family that was first identified in LPS-stimulated macrophages [1]. IκB-ζ is encoded by the primary response gene Nfkbiz, whose induction depends on NF-κB activation. IκB-ζ interacts with NF-κB through its 6 ankyrin repeats and regulates the expression of Il6 (a secondary response gene) in macrophages [2, 3]. We showed that IκB-ζ can also regulate the expression of NF-κB target genes, such as endothelial-leukocyte adhesion molecule 1 or neutrophil gelatinase-associated lipocalin [4]. Thus, IκB-ζ is a key transcriptional regulator of gene expression. Previous studies have shown that IκB-ζ is induced in T cells in response to TGF-β1 and IL-6 stimulation and that it cooperates with the transcriptional factor RORγt to induce IL-17 expression [5]. Therefore, IκB-ζ-deficient mice (Nfkbiz−/−) are resistant to Th17-dependent experimental autoimmune encephalomyelitis. However, IκB-ζ-deficient mice spontaneously develop a Sjögren’s syndrome-like autoimmune disease [6]. In addition, IκB-ζ-deficient mice have a higher proportion of effector/memory T cells in the periphery. Although this Sjögren’s syndrome-like autoimmune disease is triggered by apoptosis of lacrimal gland’s keratinocytes in IκB-ζ-deficient mice, T cells also play a pivotal role in this disease [6]. Therefore, the aim of this study was to determine whether IκB-ζ helps maintain T cell homeostasis. Here, we show that T cell-specific, IκB-ζ-deficient mice (Nfkbizflox/floxLck-Cre; cKO) have elevated levels of effector/memory CD4+ cells and IFN-γ-producing CD4+ cells. In addition, we found that IκB-ζ is up-regulated by TGF-β1 stimulation in a SMAD-dependent manner and that it negatively regulates Ifng gene expression. The plasticity and stability of Tregs play an important role in maintaining immune homeostasis and regulating IFN-γ production in T cells [7]. Thus, we generated Treg-specific, IκB-ζ-deficient mice and found that IκB-ζ in Tregs is dispensable for maintaining immune homeostasis, as well as plasticity and stability of Tregs. However, Tregs from cKO mice present reduced immunoregulatory function, indicating that IκB-ζ plays an important role in the generation of immunoregulatory function during the development of Tregs from naïve CD4+ T cells. Thus, IκB-ζ in T cells plays an important role for maintaining immune homeostasis and regulating IFN-γ production.
MATERIALS AND METHODS
Mice
C57BL/6 mice were purchased from Charles River Laboratories Japan (Yokohama, Japan). Nfkbizflox/flox [6], Lck-Cre [8], Foxp3YFP-Cre [9], and Smad2flox/flox [10] mice were established as described previously. All mice were raised under specific pathogen-free conditions in the animal facilities of Tohoku University. All animal protocols were approved by the Institutional Committee for the Use and Care of Laboratory Animals of Tohoku University (2013MA029, 2013MA031, and 2013MA032).
Cells
HEK293 cells were cultured in DMEM (Life Technologies, Carlsbad, CA, USA) containing 10% (v/v) heat-inactivated FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. CD4+CD25−CD62L+ T cells were prepared from mouse spleens by use of the CD4+CD62L+ Isolation Kit II (Miltenyi Biotec, Bergisch Gladbach, Germany), unless specified otherwise.
Plasmids, antibodies, and cytokines
Expression vectors for overexpressing FLAG-tagged versions of mIκB-ζ (L), IκB-ζ (S), and p65 were constructed with the pcDNA3 vector (Life Technologies, Madison, WI, USA) and were designated as pcDNA3-FLAG-mIκB-ζ (L), pcDNA3-FLAG-mIκB-ζ (S), and pcDNA3-FLAG-mp65, respectively. Plasmids for retroviral transduction [pMY-IRES-EGFP, pMY-FLAG-mIκB-ζ (L)-IRES-EGFP, and pMY-FLAG-mIκB-ζ (S)-IRES-EGFP] were constructed as described previously [6]. A reporter plasmid for the 600 bp mouse IFN-γ promoter was generated by cloning the mouse IFN-γ promoter (synthesized by GenScript, Piscataway, NJ, USA) into the KpnI and HindIII sites of the pGL4.12 vector (Promega, Madison, WI, USA), located upstream of the luciferase gene. The pcDNA3 and phRL-TK were obtained from Life Technologies (Madison, WI, USA) and Promega, respectively. The following anti-mouse antibodies were purchased from BioLegend (San Diego, CA, USA): FITC-conjugated anti-Ki67 (16A8); allophycocyanin-conjugated anti-CD25 (3C7), anti-CD62L (MEL-14), anti-GITR (DTA-1), and anti-IL-17A (TC11-18H10.1); FITC-conjugated anti-GITR (DTA-1); Alexa Fluor 488-conjugated anti-CD3 (17A2); PerCP/Cy5.5-conjugated anti-CD8α (53-6.7); Pacific Blue-conjugated anti-CD4 (GK1.5); PE-conjugated anti-Helios (22F6), anti-CD45RB (C363-16A), anti-CTLA-4 (UC10-4B9), anti-ICOS (7E.17G9), anti-F4/80 (BM8), anti-Gr-1 (RB6-8C5), and anti-B220 (RA3-6B2). PE-conjugated anti-CD44 (IM7), anti-IFN-γ (XMG1.7), anti-CD3 (145-2C11), anti-CD28 (37.51), and anti-IFN-γ (XMG1.2) antibodies were obtained from eBioscience (San Diego, CA, USA). allophycocyanin-conjugated anti-FOXP3 (FJK-16S) and anti-IL-2 (JES6-5H4) and PE-conjugated anti-Ki67 and anti-TNF-α (TN3-19.12) antibodies were from BD Biosciences (San Jose, CA, USA). The polyclonal anti-CD3 antibody used for immunohistochemistry was obtained from Sigma-Aldrich (St. Louis, MO, USA). Human rTGF-β1 was obtained from PeproTech (Rocky Hill, NJ, USA).
DSS-induced colitis model
Mice were supplied with 2% (w/v) DSS (MP Biomedicals, Santa Ana, CA, USA) in the drinking water for a period of 9 days to model DSS-induced colitis.
Flow cytometry analysis
Flow cytometry analysis was performed as described previously [11]. In brief, for intracellular cytokine staining, cell suspensions (1 × 106 cells/ml) were stimulated for 4 hours with 50 nM PMA (Sigma-Aldrich) and 250 nM ionomycin (BD Biosciences) in the presence of GolgiStop (BD Biosciences). Intranuclear staining of FOXP3 was performed with the FOXP3 staining buffer kit (eBioscience), according to the manufacturer’s protocol. The Annexin V apoptosis detection kit allophycocyanin and propidium iodide (eBioscience) were used for apoptosis/necrosis staining, according to the manufacturer’s protocol. Stained cells were subjected to flow cytometric analysis with a Gallios flow cytometer (Beckman Coulter, Brea, CA, USA). Data were analyzed with FlowJo software (Tree Star, Ashland, OR, USA).
Suppression assay
Purified CD4+CD25− T cells were labeled with CFSE (Wako Pure Chemical Industries, Osaka, Japan). CFSE-labeled CD4+CD25− cells (0.5 × 106 cells/ml) were cocultured with or without CD4+CD25+ T cells (0.125–0.5 × 106 cells/ml) in the presence of irradiated APCs (3 × 106 cells/ml) and anti-CD3 antibody (0.5 μg/ml) at 37°C in 5% CO2 for 3 days.
IBD model
To prepare pure naïve CD4+ T cells, CD4+CD25-CD62L+ T cells, purified from congenic C57BL/6 mice, were sorted into a CD45RBhigh population (>95%) by use of a BDAria II cell sorter (BD Biosciences). Then, the obtained naïve CD4+CD25−CD62L+CD45RBhigh T cells (3 × 105 cells) were mixed with CD4+CD25+ T cells (1 × 105 cells) from Nfkbizflox/flox or Nfkbizflox/floxLck-Cre mice and intraperitoneally injected into sex-matched, 8- to 12-week-old Rag2−/− mice.
Treg adoptive transfer
Purified CD4+CD25+ T cells (4 × 105 cells) from Nfkbizflox/flox (Control) or Nfkbizflox/flox Lck-Cre (cKO) mice were injected intravenously into sex-matched, 8- to 12-week-old Rag2−/− mice, which were observed daily and weighed weekly. Six weeks after cell transfer, the mice were euthanized for experiments.
Bisulfite sequencing
Genomic DNA was isolated from FOXP3-positive Tregs in 8-week-old Nfkbizflox/+Foxp3YFP-Cre and Nfkbizflox/floxFoxp3YFP-Cre males. Genomic DNA was then analyzed by modified bisulfite sequencing with the MethylEasy Xceed DNA modification kit (Human Genetic Signatures, Randwick, Australia), as described previously [7]. The Foxp3-CNS2 region was amplified by PCR with the primers 5′- TTTTGGGTTTTTTTGGTATTTAAGA -3′ and 5′- TTAACCAAATTTTTCTACCATTAAC -3′, using the EpiTaq HS polymerase (TaKaRa Bio, Shiga, Japan) and cloned into the pMD20-T-vector (TaKaRa Bio) by TA cloning.
Luciferase assays
HEK293 cells (1 × 105 cells) were transfected by the calcium phosphate-DNA coprecipitation method with an IFN-γ promoter reporter, pcDNA3, pcDNA3-FLAG-mp65, pcDNA3-FLAG-mIκB-ζ (L), and/or pcDNA3-FLAG-mIκB-ζ (S) and phRL-TK-Luc. Twenty-four hours after transfection, the medium was changed, and the cells were maintained for another 24 hours. Luciferase activities were measured by use of the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s instructions.
ChIP assays
Naïve CD4+ T cells were activated with plate-bound anti-CD3 and soluble anti-CD28 with TGF-β1 for 72 hours. ChIP was performed by use of an acetyl-histone H3 (Lys27) antibody or normal rabbit IgG, as described previously [12]. Immunoprecipitated and input DNA was then analyzed by quantitative PCR by use of SYBR Premix Ex Taq (TaKaRa Bio). The sequences of primers used are as follows: 5′- CCATGGTGGCGATTGATTCTGCAG -3′ and 5′- TTCCTGCAGATTGCCGTCTGGTCT -3′ for the Ifng enhancer; 5′- GCTCTGTGGATGAGAAAT -3′ and 5′- AAGATGGTGACAGATAGG -3′ for the Ifng promoter.
Real-time RT-PCR
Naïve CD4+ T cells (1 × 106 cells/ml) were cultured for 24 hours with soluble anti-CD28 (1 μg/ml) in plates coated with anti-CD3 (1 μg/ml), with or without TGF-β1 (2 ng/ml). Subsequently, total RNA was prepared by use of RNAiso Plus (TaKaRa Bio). mRNA levels of Tbx21 and Gapdh were quantified by real-time RT-PCR by use of the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), SYBR Premix Ex Taq (TaKaRa Bio), and a LightCycler 3302 (Roche Diagnostics, Mannheim, Germany). The sequences of the primers used were as follows: 5′- AGCAAGGACGGCGAATGTT -3′ and 5′- GGGTGGACATATAAGCGGTTC -3′ for Tbx21 and 5′-GAAGTCGCAGGAGACA-3′ and 5′-TCCCAGAGCTGAACGG-3′ for Gapdh.
Retroviral transduction
Recombinant retroviruses were prepared by transfecting the Plat-E packaging cells with indicated plasmids by use of the calcium phosphate-DNA coprecipitation method [13]. Naive CD4+ T cells (1 × 106 cells/ml) stimulated with plate-bound anti-CD3 and soluble anti-CD28 antibodies for 24 hours were transduced with fresh retroviral supernatant by centrifugation for 2 hours at 780 g in the presence of 10 μg/ml polybrene (Sigma-Aldrich). Cells were cultured further for 3 days with plate-bound anti-CD3 and soluble anti-CD28.
ELISAs
Mouse IFN γ ELISA Ready-SET-GO! (eBioscience) was used to quantify IFN-γ expression in culture supernatants, according to the manufacturer’s suggested protocol.
Lamina propria cell isolation
Mouse intestines were opened longitudinally and washed for 20 min at 37°C in RPMI 1640 containing 3% FBS, 20 mM HEPES, 5 mM EDTA, and 0.145 mg/ml DTT. Tissues were digested with scissors and washed with RPMI 1640 containing 20 mM HEPES and 2 mM EDTA. After removal of epithelial cells and fat tissue, the intestines were cut into small pieces and incubated for 30 min at 37°C in a shaking incubator with RPMI 1640 containing 20 mM HEPES, 0.2 mg/ml Liberase TL (Roche Applied Science, Basel, Switzerland), and 0.05% DNase (Sigma-Aldrich). The digested tissues were mashed on the 40 μm strainer and resuspended in 5 ml 30% Percoll (GE Healthcare Life Sciences, Pittsburgh, PA, USA). Percoll gradient separation was performed by centrifugation at 780 g for 10 min at 4°C. Percoll gradient excluded debris, allowing the collection of lamina propria lymphocytes, which were washed with RPMI 1640 and used immediately for experiments.
Histology
Tissues were fixed by immersion in 10% formalin in PBS and embedded in paraffin blocks. Sections (4 μm thick) were stained with H&E staining and examined by light microscopy.
Statistical analysis
The Student's t-test (2-tailed) was used to determine significant differences between 2 groups. For multiple comparisons, 1-way ANOVA, followed by Dunnett’s test, was used.
RESULTS
IκB-ζ in T cells control immune homeostasis in vivo
A previous study demonstrated that T cell-specific, IκB-ζ-deficient mice (Nfkbizflox/floxLck-cre) do not develop Sjögren’s syndrome diseases with age [6]. To investigate the role of IκB-ζ in T cells in vivo, we used T cell-specific, IκB-ζ-deficient mice (cKO). First, we examined the deficiency of IκB-ζ in immune cells in cKO mice and found that CD4+ and CD8α+ T cells expressed the Nfkbiz gene to a lower degree than other immune cells, including B cells, macrophages, and dendritic cells (Fig. 1A). These cKO mice appeared healthy and grew without any noticeable phenotypic abnormalities until ∼6 months of age. After 6 months, cKO mice developed splenomegaly, lymphadenopathy, and dense infiltration of leukocytes in multiple vital organs and tissues, including the liver (Fig. 1B–D). Although mice with a global IκB-ζ deficiency have previously been shown to exhibit high titers of ANA in the serum [6], we were not able to detect high titers of ANA with anti-dsDNA antibodies (a specific type of ANA) in cKO mice (Fig. 1E and F). Analysis of the serum in cKO mice showed that IFN-γ and IFN-γ-induced protein 10 were more highly expressed in cKO mice than in control mice (Supplemental Table 1). These results suggest that T cell-specific deletion of IκB-ζ disrupts immune homeostasis with age, a phenotype that differs from that observed in IκB-ζ globally deficient (Nfkbiz−/−) mice. At 3 weeks of age, cKO mice appeared healthy and exhibited normal-sized spleens and peripheral LNs compared with control mice (Fig. 2A). However, the absolute number of CD4+ cells and CD8α+ cells was lower in cKO mice than in control mice (Supplemental Fig. 1A–C). In addition, cKO mice showed a lower frequency of naïve CD4+ cells (CD44−CD62L+) and a higher frequency of effector/memory CD4+ cells (CD44+CD62L−) than control (Nfkbizflox/flox) mice (Fig. 2B and C). Furthermore, IFN-γ production in CD4+ T cells was significantly higher in cKO mice than in control mice, consistent with our finding in IκB-ζ-deficient mice (Fig. 2D) [6]. Additionally, we evaluated IL-2 and TNF-α production in CD4+ T cells and found that IL-2 production (but not TNF-α production) was slightly higher in cKO mice than in control mice (Supplemental Fig. 2A and B). These results suggest that T cell-specific, IκB-ζ-deficient mice exhibit immune dysregulation at a young age.
Figure 1. T Cell-specific, IκB-ζ-deficient mice exhibit disrupted immune homeostasis with age.
(A) Relative IκB-ζ expression in Nfkbizf/f (Control) and Nfkbizf/f Lck-Cre (cKO) mice. Data shown are the means ± sd of duplicate samples and are representative of 2 independent experiments. (B) Representative appearance (upper) and macroscopic view (lower) of 6- to 18-month-old Nfkbizf/f (Control) and Nfkbizf/f Lck-Cre (cKO) mice. (C) Absolute numbers of lymphocytes in the spleen and LNs of 6- to 18-month-old Nfkbizflox/flox (Control) and Nfkbizflox/floxLck-Cre (cKO) mice. (D) Histopathology of the liver. H&E staining or CD3 immunostaining of tissue sections from 6- to 18-month-old mice. (E and F) Serum concentrations of ANA (E) and anti-dsDNA (F) in Nfkbizflox/flox (Control) and Nfkbizflox/floxLck-Cre (cKO) mice. Original scale bars, 100 µm. Original magnification, 20× (D). *P < 0.05.
Figure 2. Characterization of T cell-specific, IκB-ζ-deficient mice.
(A) Lymphocyte cell counts in the spleen and LNs of 3-week-old Nfkbizflox/flox (Control) and Nfkbizflox/floxLck-Cre (cKO) mice (n = 9). (B and C) Activation status of CD4+ cells in the spleen and LNs of 3-week-old Control and cKO mice. Frequency of cells in each quadrant is shown as mean percentages of CD4+ cells that were (B) CD44−CD62L+ or (C) CD44+CD62L− ± sem (n = 4). (D) Flow cytometry analysis of IFN-γ-producing CD4+ cells isolated from the spleen and LNs of Nfkbizflox/flox (Control) and Nfkbizflox/floxLck-Cre (cKO) mice at 3 weeks of age (n = 6). (E) Frequencies of CD4+ FOXP3+ Tregs in the thymus, spleen, and LNs of Nfkbizflox/flox (Control) and Nfkbizflox/floxLck-Cre (cKO) mice at 3 weeks of age (means ± sem; n = 6). (F) Frequencies of Ki67+ cells in the CD4+ FOXP3+ Treg population harvested from the thymus, spleen, and LNs of 3-week-old Control and cKO mice are shown as means ± sem (n = 4). (G) Frequencies of Ki67+ cells in CD4+CD44-CD62L+ (naïve T cells) and CD4+CD44+CD62L− (effector T cells) cells harvested from the thymus, spleen, and LNs of 3-week-old control and cKO mice are shown as the means ± sem (n = 3). The horizontal bars represent the mean. *P < 0.05; **P < 0.01.
As Tregs play a prominent role in regulating immune homeostasis [14, 15], we decided to investigate whether cKO mice have defective Tregs. Our results showed that the frequencies of FOXP3+ Tregs in the thymus, spleen, and peripheral LNs were higher in cKO mice than in the control mice (Fig. 2E). This observation is corroborated with our finding that IκB-ζ-deficient Tregs had a substantially higher frequency of Ki67-expressing cells than Tregs from control mice (Fig. 2F). In addition. IκB-ζ-deficient effector/memory CD4+ T cells had a higher frequency of Ki67-expressing cells than effector/memory CD4+ T cells from control mice (Fig. 2G). Additionally, the number of necrotic cells (Annexin V+ propidium iodide+) among CD4+ or CD8α+ cells in the spleen was comparable between control and cKO mice (Supplemental Fig. 2C). Thus, these results suggest that the increase of Tregs in cKO mice was secondary to the increase in effector/memory T cells and that IκB-ζ expression in T cells is important for maintaining immune homeostasis.
Robust induction of IFN-γ in IκB-ζ-deficient T cells
TGF-β1 is a key cytokine that inhibits IFN-γ expression in CD4+ T cells [16]. Here, we found that T cell-specific, IκB-ζ-deficient mice had elevated levels of IFN-γ-producing CD4+ cells (Th1) in vivo (Fig. 2D). We performed in vitro culture assays to determine the function of TGF-β1 in naïve cKO CD4+ T cells. TGF-β prevented IFN-γ production from naïve CD4+ T cells in response to TCR stimulation (Fig. 3A–C). Our results demonstrated that naïve CD4+ T cells from cKO mice produced a large amount of IFN-γ in the presence of TGF-β1 stimulation (Fig. 3A–C). In addition, TGF-β-induced FOXP3 induction was comparable between control and IκB-ζ deficient T cells (Fig. 3D). Elevation of T-bet, the master regulator of IFN-γ production, was higher in IκB-ζ-deficient T cells when compared with the control T cells, even in the presence of TGF-β1 stimulation (Fig. 3E). Thus, IκB-ζ controls IFN-γ expression in T cells.
Figure 3. IκB-ζ-deficient T cells show high levels of IFN-γ expression in the presence of TGF-β.
(A) ELISA for IFN-γ in the supernatants of naïve CD4+ T cells from Nfkbizflox/flox (Control) and Nfkbizflox/floxLck-Cre (cKO) mice that were cultured for 72 hours. Data shown represent means ± sem (n = 3). (B) Flow cytometric analysis of IFN-γ expression in CD4+ T cells from control and cKO mice, cultured for 72 hours in the presence or absence of TGF-β1. Data are representative of 4 independent experiments. (C) Percent reduction of IFN-γ expression in CD4+ cells under the TGF-β1 condition compared with the non-TGF-β1 condition in B. Data shown represent the means ± sem (n = 4). (D) Flow cytometric analysis of FOXP3 expression in CD4+ T cells from control and cKO mice, cultured for 72 hours in the presence or absence of TGF-β1. Data are representative at least 3 independent experiments. (E) Expression of Tbx21 in cultured naive CD4+ T cells from control and cKO mice for 24 hours. Data shown represent means ± sem (n = 3). *P < 0.05.
TGF-β1 controls IκB-ζ expression in CD4+ T cells and negatively regulates IFN-γ gene expression
IκB-ζ expression in CD4+ T cells is reportedly hard to detect, although TGF-β1 + IL-6 stimulation can induce IκB-ζ expression, which positively regulates IL-17A gene expression [5]. Here, we found that only TGF-β1 stimulation in CD4+ T cells can induce IκB-ζ expression (Fig. 4A). In addition, we used CD4+ T cells deficient in Smad2, which is downstream of TGF-β signaling [17]. These cells did not show up-regulated IκB-ζ in response to TGF-β1 (Fig. 4B). As T cell-specific, Smad2-deficient mice have more IFN-γ-producing CD4+ T cells [10], we investigated whether induction of IκB-ζ via the TGF-β1-SMAD axis negatively regulates IFN-γ gene expression.
Figure 4. TGF-β-induced IκB-ζ represses Ifng gene expression.

(A) Expression of IκB-ζ in naïve CD4+ T cells cultured with/without TGF-β1 stimulation. (B) Expression of IκB-ζ in naive CD4+ T cells from Smad2flox/flox mice or Smad2flox/flox Lck-Cre mice that were cultured for 24 hours. (C and D) CD4+ T cells were retrovirally transduced to express IκB-ζ and GFP. (C) The numbers indicate the percentage of IFN-γ-producing GFP+CD4+ T cells. Data are representative of 3 independent experiments. (D) Relative IFN-γ-producing GFP+CD4+ T cells. The percentage of IFN-γ-producing GFP+CD4+ T cells from the IRES-GFP control is set as “1.” Data shown represent the means ± sd (n = 3). (E) Relative histone acetylation of the IFN-γ promoter region or CNS region in naïve CD4+ T cells from control or cKO mice. Purified naïve CD4+ T cells were cultured for 3 d in the presence of TGF-β1. Histone acetylation was analyzed by ChIP assays, performed by use of an acetyl-histone (AcH) H3 antibody and control IgG. Data represent means ± sd with triplicate samples. (F) IFN-γ reporter activity in HEK293 cells. Reporter activity in the absence of p65 and IκB-ζ expression vector was set to 1. Data shown reflect means ± sd (triplicate samples). Data are representative of 3 (A–E) or 2 (F) independent experiments. *P < 0.05; **P < 0.01.
IκB-ζ expresses 2 known splicing isoforms [18, 19]. IκB-ζ (L) is the major, long splicing isoform in macrophages and T cells [5]. IκB-ζ (S) is the short form and lacks the 99 N-terminal aa present in IκB-ζ (L). T cells that were retrovirally transduced with cDNA encoding the IκB-ζ (L) or IκB-ζ (S) isoforms produced decreased IFN-γ, relative to control T cells transduced with IRES-GFP (Fig. 4C and D). Thus, IκB-ζ plays an important role in controlling IFN-γ production.
We next analyzed the chromatin structure of the genomic Ifng locus in activated T cells. Two conserved, noncoding sequences in the genomic Ifng locus play a pivotal role in Ifng gene expression, namely, the CNS (∼5 kb upstream from the transcription start site) and promoter regions [20, 21]. We performed ChIP assays to study the acetylation of histone H3 (Lys27), a marker of active chromatin, and found that the Ifng promoter region was highly acetylated in IκB-ζ-deficient T cells under TGF-β1 stimulation compared with that in control T cells (Fig. 4E). However, the status of acetylated histone in the IFN-γ CNS region was comparable in control and IκB-ζ-deficient T cells (Fig. 4E).
To investigate further the role of IκB-ζ in Ifng gene expression, we developed an Ifng promoter luciferase reporter assay. It has been reported that the IFN-γ promoter can be activated by NF-κΒ [22]. In addition, IκB-ζ can regulate NF-κB target gene expression [23]. Thus, we investigated whether IκB-ζ controls NF-κB-induced IFN-γ gene expression. We confirmed that the IFN-γ promoter reporter can be activated in the presence of the NF-κB subunit p65 in HEK293 cells (Fig. 4F). Interestingly, induction of IFN-γ promoter reporter activity by p65 is less pronounced in the presence of IκB-ζ (L) or IκB-ζ (S) (Fig. 4F).
IκB-ζ-deficient mice are highly sensitive to DSS
The gut has a unique immune system and is known to produce large amounts of TGF-β; thus, it plays an important role in maintaining immune homeostasis [24, 25]. To investigate the role of IκB-ζ in T cells in the gut, we developed a murine model of DSS-induced colitis. After the cKO mice were given 2% DSS in their drinking water, they began to lose weight by 6–7 days, and severe injury to colonic villi was observed by 9 days of DSS treatment (Fig. 5A and B). We found that the percentage of Tregs in colonic lamina propria increased following DSS treatment, and expression levels were comparable between control and cKO mice (Fig. 5C and data not shown). We found that cKO mice have even more IFN-γ-producing CD4+ T cells in the colonic lamina propria after DSS treatment than control mice (Fig. 5D). Thus, IκB-ζ in T cells plays a pivotal role in maintaining immune homeostasis in the gut.
Figure 5. T Cell-specific, IκB-ζ-deficient mice have higher sensitivity to DSS-induced colitis.
(A) Relative weight loss of Nfkbizf/f (Control; n = 7) and Nfkbizf/f Lck-Cre (cKO; n = 5) mice after 2% DSS treatment. The horizontal bars represent the means ± sem. (B) Histopathology of the colon. H&E staining of tissue sections at 9 days after DSS treatment. Data are representative of 3 independent experiments. (C and D) Percentage of FOXP3+ Tregs (C) or IFN-γ production in CD4+ T cells (D) in colonic lamina propria from Nfkbizf/f (Control) and Nfkbizf/f Lck-Cre mice at 9 days after 2% DSS treatment (data for 2 mice pooled together for analysis). Original scale bars, 100 µm. Original magnification, 10× (B). **P < 0.01.
IκB-ζ-deficient Tregs show defective immunosuppression
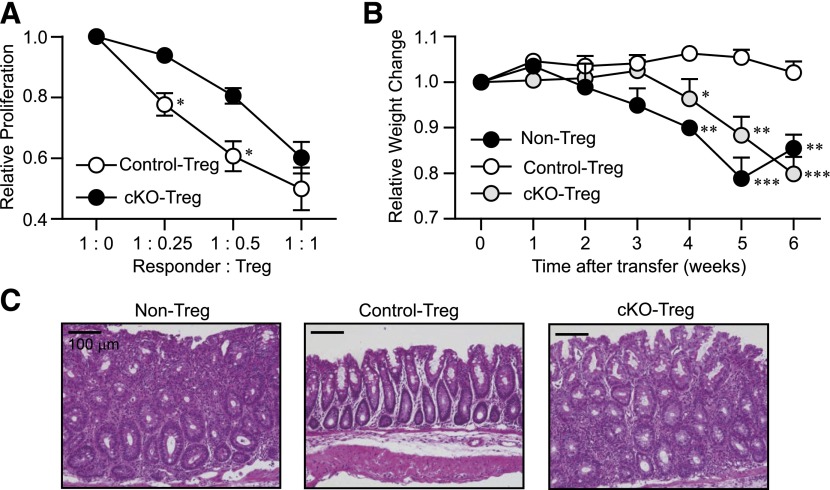
We next determined whether IκB-ζ deficiency affected Treg-mediated immunosuppressive activity. With the use of an in vitro suppression assay, we demonstrated that IκB-ζ-deficient Tregs suppressed the proliferation of responder cells (CD4+CD25− T cells) but to a far lesser extent than control Tregs (Fig. 6A). To determine whether this diminished ability to suppress T cell activity also occurred in vivo, we used a T cell transfer model of IBD [26, 27]. Our results showed that adoptive transfer of CD4+CD25−CD62L+CD45RBhigh T cells into Rag2−/− mice caused weight loss and inflammation of the colon (Fig. 6B and C). These side-effects were not observed when control Tregs were cotransferred into Rag2−/− mice with CD4+CD25−CD62L+CD45RBhigh T cells. By contrast, cotransfer with IκB-ζ-deficient Tregs failed to prevent weight loss and inflammation of the colon (Fig. 6B and C). In addition, when control or IκB-ζ-deficient Tregs were transferred into Rag2−/− mice on their own, no weight loss or inflammation was observed in the colon (Supplemental Fig. 3). These results show that IκB-ζ-deficient Tregs have a reduced immunoregulatory function.
Figure 6. IκB-ζ-deficient Tregs have defective immunosuppressive ability.
(A) In vitro suppression assay by use of CFSE-labeled CD4+CD25− T cells (Responder) and CD4+CD25+ Tregs from control or cKO mice. Responders in the absence of Tregs (1:0) were set as 1. Data represent the means ± sd of 3 experiments. (B and C) IBD model. Adoptive transfer of 3 × 105 CD4+CD45RBhigh T cells to Rag2−/− mice with/without 1 × 105 Tregs from control or cKO mice. Data are shown as the means + se (n = 6 from each sample set). Original scale bars, 100 µm. Original magnification ×20 (C). *P < 0.05; **P < 0.01; ***P < 0.001.
IκB-ζ does not affect Treg plasticity or stability
Next, we investigated whether IκB-ζ in Tregs plays an important role in maintaining immune homeostasis in cKO mice. To analyze the role of IκB-ζ in Treg stability and plasticity, we generated Treg-specific, IκB-ζ-deficient mice (Nfkbizflox/floxFoxp3YFP-cre). We examined Nfkbiz gene expression in immune cells (B cells, CD4+FOXP3− cells, CD4+FOXP3+ cells, CD8+ T cells, and CD11b+ cells) of Treg-specific, IκB-ζ-deficient mice and found that only CD4+FOXP3+ Tregs had reduced Nfkbiz gene expression (Fig. 7A). These Treg-specific, IκB-ζ-deficient mice (Nfkbizflox/floxFoxp3YFP-cre) appeared healthy and grew without any noticeable phenotypic abnormalities for >6 months. The percentage of Tregs did not differ between control (Nfkbizflox/+Foxp3YFP-cre) and Treg-specific, IκB-ζ-deficient mice (Nfkbizflox/floxFoxp3YFP-cre; Fig. 7B). In addition, the proportions of naïve (CD44−CD62L+) and effector/memory (CD44+CD62L− and CD44+CD62L+) CD4+ T cells did not differ between Treg-specific, IκB-ζ-deficient mice and control mice (Fig. 7C). Furthermore, the levels of costimulatory molecules (CTLA-4, GITR, and ICOS) in FOXP3+ Tregs and the levels of cytokine production in CD4+ cells were comparable between control and Treg-specific, IκB-ζ-deficient mice (Fig. 7D and E). In addition, the status of CNS2 demethylation, which is associated with stable expression of FOXP3 [28], was comparable in CD4+CD25+FOXP3+ Tregs from 8-week-old male Nfkbizflox/floxFoxp3YFP-cre and Nfkbizflox/+Foxp3YFP-cre mice (Fig. 7F).
Figure 7. IκB-ζ is dispensable for the stability of Foxp3 gene expression.
(A) Relative IκB-ζ expression in immune cells (B cells, CD4+FOXP3− cells, CD4+FOXP3+ cells, CD8+ T cells, and CD11b+ cells) from Nfkbizf/+ Foxp3YFP-Cre and Nfkbizf/f Foxp3YFP-Cre mice. Data shown are the means ± sd of duplicate samples and are representative of 2 independent experiments. (B and C) Helios staining (B) or the activation status of CD4+ cells (C) in the spleens of 6- to 9-months-old male Nfkbizflox/+Foxp3YFP-cre and Nfkbizflox/floxFoxp3YFP-cre mice. Data shown are from 1 experiment that was representative of 3 independent experiments. (D) Costimulatory molecule expression in Tregs isolated from the spleens of 6- to 9-month-old Nfkbizflox/+Foxp3YFP-cre and Nfkbizflox/floxFoxp3YFP-cre mice. Relative values were obtained by comparing the mean fluorescence intensities ± sem; n = 3. Red, Nfkbizflox/+Foxp3YFP-cre; blue line, Nfkbizflox/floxFoxp3YFP-cre. (E) Cytokine production from CD4+ T cells in spleens of 6- to 9-month-old male Nfkbizflox/+Foxp3YFP-cre (open circles) and Nfkbizflox/floxFoxp3YFP-cre (filled circles) mice. Horizontal bars represent the calculated means. (F) Bisulfite sequencing of genomic DNA from YFP-positive Tregs isolated from the spleens of 8-week-old male Nfkbizflox/+Foxp3YFP-cre or Nfkbizflox/floxFoxp3YFP-cre mice. CpG methylation status in the CNS2 region of the Foxp3 gene was analyzed and is shown in sector graphs; 21–25 cDNA clones were sequenced from each subset. (G and H) Flow cytometry analysis of 6- to 8-month-old female Nfkbizflox/+Foxp3YFP-cre/+ and Nfkbizflox/floxFoxp3YFP-cre/+ mice. The percentages of FOXP3+ Tregs in the spleens of the indicated mice are shown.
Next, we examined Treg mosaic model female mice (Nfkbizflox/+Foxp3YFP-cre/+ and Nfkbizflox/floxFoxp3YFP-cre/+) to analyze the stability of Foxp3 gene expression with increasing age [9]. Six to 8 months after birth, these mice appeared healthy, and the proportion of YFP-Cre-positive Tregs from Nfkbizflox/floxFoxp3YFP-cre/+ mice was even higher than that in Nfkbizflox/+Foxp3YFP-cre/+ mice (Fig. 7G and H). These results show that IκB-ζ is dispensable for the stability of Foxp3 gene expression. In addition, these results show that IκB-ζ plays an important role for the development of immunoregulatory function during Treg generation from naïve CD4+ T cells, but it is dispensable for immunoregulatory function after the generation of Foxp3+ Tregs from naïve CD4+ T cells.
DISCUSSION
In this study, we examined the role of IκB-ζ in T cell homeostasis. We found that IκB-ζ in T cells acts as a key molecule in maintaining homeostasis and is up-regulated by TGF-β1 stimulation. TGF-β1 is an immune-regulatory cytokine that is also important for the generation of FOXP3+ Tregs from naïve CD4+ T cells [29, 30]. In addition, activation of the classic TGF-β signaling molecule, SMAD2, in T cells is important for maintaining immune homeostasis through the repression of IFN-γ production and Treg induction [10]. We found that SMAD2 plays a pivotal role in IκB-ζ induction in T cells by TGF-β1, which is the key event in regulating IFN-γ production, but not for Treg production. In addition, we have shown previously that the IL-6 downstream molecule STAT-3 positively regulates IκB-ζ expression [6]. Thus, IκB-ζ would be regulated by the TGF-β-Smad axis and the IL-6-STAT-3 axis.
Previously, IκB-ζ overexpression in T cells was found not to affect Th17 production in response to CD3 + CD28, without TGF-β + IL-6 stimulation [5]. As TGF-β + IL-6-induced RORγt and IκB-ζ are essential transcriptional factors for inducing Th17 cell differentiation, IκB-ζ or RORγt expression alone was insufficient to induce Th17 cell differentiation. In addition, TGF-β-induced FOXP3 forms a complex with RORγt and prevents RORγt transcriptional activity related to IL-17 gene expression [31]. Thus, TGF-β-induced IκB-ζ expression would be insufficient to promote Th17 cell differentiation mediated by RORγt.
Many studies have reported that IκB-ζ forms a complex with NF-κB and controls NF-κB target gene expression [1, 3, 23]. The IFN-γ promoter has 2 putative NF-κB-binding elements, and its expression is dependent on NF-κB transcriptional activity through TCR stimulation [22, 32]. Although further studies are required to understand the mechanism of IFN-γ regulation by IκB-ζ in T cells, it is possible that IκB-ζ is recruited to the IFN-γ promoter region and controls NF-κB-dependent IFN-γ gene expression.
The stability and plasticity of Tregs are also important for controlling Th1 responses in the periphery [7, 33]. We confirmed that IFN-γ production from CD4+ cells in Treg-specific, IκB-ζ-deficient mice did not differ from that seen in control mice (Nfkbizf/+ Foxp3YFP-cre). Thus, high IFN-γ production from CD4+ T cells in T cell-specific, IκB-ζ-deficient mice is dispensable for the stability and plasticity of IκB-ζ-deficient Tregs. As FOXP3 physically associates with NF-κB and suppresses its transcriptional activity, FOXP3+ Tregs showed less IFN-γ expression than observed in non-Tregs [34]. We also confirmed that IFN-γ production from Tregs is comparable between control (Nfkbizf/+ Foxp3YFP-cre) and Treg-specific, IκB-ζ-deficient mice (Nfkbizf/f Foxp3YFP-cre; data not shown).
Taken together, our findings indicate that IκB-ζ is a target of TGF-β signaling, controls IFN-γ gene expression, and plays an important role in maintaining immune homeostasis.
AUTHORSHIP
Tak.Y. conceived of and directed this study, designed and performed experiments, analyzed data, and contributed to the writing of this the manuscript. S.K. performed critical experiments. K.O. and A.Y. provided critical materials, discussion, and contributions to the writing of this manuscript. W.C. and Tat.M. supervised experiments and contributed to the writing of this manuscript.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research on Innovative Areas 25118702 from the Ministry of Education (to Tak.Y.);a Grant-in-Aid for Young Scientists B (Grant 24790458); Grant-in-Aid for Challenging Exploratory Research 26670019 from the Japan Society for the Promotion of Science (to Tak.Y.); and grants from the Takeda Science Foundation, Uehara Memorial Foundation, Novartis Foundation, and Sumitomo Foundation (to Tak.Y.). This work was partly supported by the Cooperative Research Project Program of the Joint Usage/Research Center at the Institute of Development, Aging and Cancer, Tohoku University. The authors are grateful to Dr. Alexander Y. Rudensky (Memorial Sloan-Kettering Cancer Center) for providing the Foxp3YFP-cre mice. The authors also thank Drs. Ono Masao, Atsushi Okuma (Tohoku University), and Maekawa Yoichi (Gifu University) for discussion.
Glossary
- ANA
antinuclear antibodies
- CD62L
cluster of differentiation 62 ligand
- ChIP
chromatin immunoprecipitation
- cKO
conditioned knockout
- CNS
conserved noncoding sequence
- DSS
dextran sodium sulfate
- EGFP
enhanced GFP
- FOXP3
forkhead box P3
- GITR
glucocorticoid-induced TNFR
- HEK
human embryonic kidney
- IBD
inflammatory bowel disease
- IRES
internal ribosome entry site
- L
long form
- LN
lymph node
- mIκB
mouse IκB
- Nfkbiz
NF-κB inhibitor ζ
- Rag2−/−
recombination-activating gene 2-deficient
- RORγt
retinoic acid-related orphan receptor γt
- S
short form
- Tbx21
T-box transcription factor 21
- Treg
regulatory T cell
- YFP
yellow fluorescent protein
Footnotes
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
DISCLOSURES
The authors declare no commercial or financial conflict of interest.
REFERENCES
- 1.Yamazaki S., Muta T., Takeshige K. (2001) A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J. Biol. Chem. 276, 27657–27662. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., Takeda K., Akira S. (2004) Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature 430, 218–222. [DOI] [PubMed] [Google Scholar]
- 3.Yamazaki S., Muta T., Matsuo S., Takeshige K. (2005) Stimulus-specific induction of a novel nuclear factor-kappaB regulator, IkappaB-zeta, via Toll/interleukin-1 receptor is mediated by mRNA stabilization. J. Biol. Chem. 280, 1678–1687. [DOI] [PubMed] [Google Scholar]
- 4.Matsuo S., Yamazaki S., Takeshige K., Muta T. (2007) Crucial roles of binding sites for NF-kappaB and C/EBPs in IkappaB-zeta-mediated transcriptional activation. Biochem. J. 405, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamoto K., Iwai Y., Oh-Hora M., Yamamoto M., Morio T., Aoki K., Ohya K., Jetten A. M., Akira S., Muta T., Takayanagi H. (2010) IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature 464, 1381–1385. [DOI] [PubMed] [Google Scholar]
- 6.Okuma A., Hoshino K., Ohba T., Fukushi S., Aiba S., Akira S., Ono M., Kaisho T., Muta T. (2013) Enhanced apoptosis by disruption of the STAT3-IκB-ζ signaling pathway in epithelial cells induces Sjögren’s syndrome-like autoimmune disease. Immunity 38, 450–460. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi R., Nishimoto S., Muto G., Sekiya T., Tamiya T., Kimura A., Morita R., Asakawa M., Chinen T., Yoshimura A. (2011) SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-gamma and IL-17A production. J. Exp. Med. 208, 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahama Y., Ohishi K., Tokoro Y., Sugawara T., Yoshimura Y., Okabe M., Kinoshita T., Takeda J. (1998) Functional competence of T cells in the absence of glycosylphosphatidylinositol-anchored proteins caused by T cell-specific disruption of the Pig-a gene. Eur. J. Immunol. 28, 2159–2166. [DOI] [PubMed] [Google Scholar]
- 9.Rubtsov Y. P., Rasmussen J. P., Chi E. Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W. R. Jr., Muller W., Rudensky A. Y. (2008) Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558. [DOI] [PubMed] [Google Scholar]
- 10.Takimoto T., Wakabayashi Y., Sekiya T., Inoue N., Morita R., Ichiyama K., Takahashi R., Asakawa M., Muto G., Mori T., Hasegawa E., Saika S., Hara T., Nomura M., Yoshimura A. (2010) Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185, 842–855. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama T., Li J., Vaque J. P., Konkel J. E., Wang W., Zhang B., Zhang P., Zamarron B. F., Yu D., Wu Y., Zhuang Y., Gutkind J. S., Chen W. (2011) Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat. Immunol. 12, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agata Y., Katakai T., Ye S. K., Sugai M., Gonda H., Honjo T., Ikuta K., Shimizu A. (2001) Histone acetylation determines the developmentally regulated accessibility for T cell receptor gamma gene recombination. J. Exp. Med. 193, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C., Okayama H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hori S., Nomura T., Sakaguchi S. (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061. [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi S., Sakaguchi N., Asano M., Itoh M., Toda M. (1995) Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164. [PubMed] [Google Scholar]
- 16.Lin J. T., Martin S. L., Xia L., Gorham J. D. (2005) TGF-beta 1 uses distinct mechanisms to inhibit IFN-gamma expression in CD4+ T cells at priming and at recall: differential involvement of Stat4 and T-bet. J. Immunol. 174, 5950–5958. [DOI] [PubMed] [Google Scholar]
- 17.Schmierer B., Hill C. S. (2007) TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8, 970–982. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura H., Kanehira K., Okita K., Morimatsu M., Saito M. (2000) MAIL, a novel nuclear I kappa B protein that potentiates LPS-induced IL-6 production. FEBS Lett. 485, 53–56. [DOI] [PubMed] [Google Scholar]
- 19.Haruta H., Kato A., Todokoro K. (2001) Isolation of a novel interleukin-1-inducible nuclear protein bearing ankyrin-repeat motifs. J. Biol. Chem. 276, 12485–12488. [DOI] [PubMed] [Google Scholar]
- 20.Lee D. U., Avni O., Chen L., Rao A. (2004) A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J. Biol. Chem. 279, 4802–4810. [DOI] [PubMed] [Google Scholar]
- 21.Qiao Y., Giannopoulou E. G., Chan C. H., Park S. H., Gong S., Chen J., Hu X., Elemento O., Ivashkiv L. B. (2013) Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity 39, 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., Young H. A. (1997) Interaction of NF-kappaB and NFAT with the interferon-gamma promoter. J. Biol. Chem. 272, 30412–30420. [DOI] [PubMed] [Google Scholar]
- 23.Motoyama M., Yamazaki S., Eto-Kimura A., Takeshige K., Muta T. (2005) Positive and negative regulation of nuclear factor-kappaB-mediated transcription by IkappaB-zeta, an inducible nuclear protein. J. Biol. Chem. 280, 7444–7451. [DOI] [PubMed] [Google Scholar]
- 24.Coombes J. L., Siddiqui K. R., Arancibia-Cárcamo C. V., Hall J., Sun C. M., Belkaid Y., Powrie F. (2007) A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gad M., Brimnes J., Claesson M. H. (2003) CD4+ T regulatory cells from the colonic lamina propria of normal mice inhibit proliferation of enterobacteria-reactive, disease-inducing Th1-cells from scid mice with colitis. Clin. Exp. Immunol. 131, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mottet C., Uhlig H. H., Powrie F. (2003) Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J. Immunol. 170, 3939–3943. [DOI] [PubMed] [Google Scholar]
- 27.Singh B., Read S., Asseman C., Malmström V., Mottet C., Stephens L. A., Stepankova R., Tlaskalova H., Powrie F. (2001) Control of intestinal inflammation by regulatory T cells. Immunol. Rev. 182, 190–200. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y., Josefowicz S., Chaudhry A., Peng X. P., Forbush K., Rudensky A. Y. (2010) Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W., Jin W., Hardegen N., Lei K. J., Li L., Marinos N., McGrady G., Wahl S. M. (2003) Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni A. B., Huh C. G., Becker D., Geiser A., Lyght M., Flanders K. C., Roberts A. B., Sporn M. B., Ward J. M., Karlsson S. (1993) Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. USA 90, 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L., Lopes J. E., Chong M. M., Ivanov I. I., Min R., Victora G. D., Shen Y., Du J., Rubtsov Y. P., Rudensky A. Y., Ziegler S. F., Littman D. R. (2008) TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai W., Yu M., Huang M. N., Okoye F., Keegan A. D., Farber D. L. (2011) Transcriptional control of rapid recall by memory CD4 T cells. J. Immunol. 187, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu L. F., Boldin M. P., Chaudhry A., Lin L. L., Taganov K. D., Hanada T., Yoshimura A., Baltimore D., Rudensky A. Y. (2010) Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142, 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettelli E., Dastrange M., Oukka M. (2005) Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. USA 102, 5138–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]