Abstract
Peripheral Artery Disease (PAD) is a common manifestation of atherosclerosis, characterized by lower leg ischemia and myopathy in association with leg dysfunction. In the present study, Spontaneous and coherent anti-Stokes Raman scattering (CARS) spectroscopic techniques in CH-stretching spectral region were evaluated for discriminating healthy and diseased tissues of human gastrocnemius biopsies of control and PAD patients. Since Raman signatures of the tissues in the fingerprint region are highly complex and CH containing moieties are dense, CH-stretching limited spectral range was used to classify the diseased tissues. A total of 181 Raman spectra from 9 patients and 122 CARS spectra from 12 patients were acquired. Due to the high dimensionality of the data in Raman and CARS measurements, principal component analysis (PCA) was first performed to reduce the dimensionality of the data (6 and 9 principal scores for Raman and CARS, respectively) in the CH-stretching region, followed by a discriminant function analysis (DFA) to classify the samples into different categories based on disease severity. The CH2 and CH3 vibrational signatures were observed in the Raman and CARS spectroscopy. Raman and CARS data in conjunction with PCA-DFA analysis were capable of differentiating healthy and PAD gastrocnemius with an accuracy of 85.6% and 78.7%, respectively.
OCIS codes: (300.0300) Spectroscopy; (300.6230) Spectroscopy, coherent anti-Stokes Raman scattering; (300.6450) Spectroscopy, Raman; (170.0170) Medical optics and biotechnology
1. Introduction
Spontaneous Raman spectroscopy is a common technique used to characterize materials using their vibrational signatures. Each peak in the spectrum corresponds to a specific molecular vibration mode, such that the spectrum could be a direct function of the molecular composition of the material. In this non-destructive technique, external labels are not required for identifying biochemical changes in various tissues [1–4]. Many efforts have been made to classify the Raman spectra of normal and abnormal tissues, demonstrating the correlation between diseased situation of the tissues and their chemical compositions [5–7].
The main drawback of spontaneous Raman spectroscopy is its weak Raman scattering efficiency (lower than 1 in 107) that requires high laser power, long acquisition time, or large accumulation number to obtain high-quality Raman spectra or mapping [8]. Although the sensitivity can be improved by applying a gold-coated glass substrate [9], which has better fluorescence background suppression and surface plasmonic enhancement, this drawback still has severely limited the applications of Raman microscopy in biomedical studies. On the other hand, coherent anti-Stokes Raman scattering (CARS), a nonlinear optical process, provides the same Raman vibrational signatures but stronger signals, which is suited for applications that require rapid analysis. In CARS, it involves the interaction of four waves designated as pump (p), Stokes (s), probe (p′), and anti-Stokes, where pump and probe waves are usually fixed to the same frequency (ωp = ωp′). When the frequency difference between the pump and the Stokes beams matches with the resonant vibrational frequency of a Raman active molecular vibration, the molecular vibration is coherently driven by the excitation fields, thereby generating a strong anti-Stokes signal at ωas = 2ωpump – ωStokes. CARS benefits, unlike spontaneous Raman, from the constructive interference of light scattered by spectrally overlapping vibrational modes within the focal volume, thus enabling rapid acquisition and analysis [10]. When compared to spontaneous Raman spectroscopy, CARS also has many other advantages such as no fluorescence interference and high spatial resolution.
The CH region of organic biomolecular compounds is much more congested and is much harder to differentiate than the fingerprint region. However, the signal-to-noise ratio for the CH region is better than that of the fingerprint region and important information can be collected from relative changes in the CH-stretching region of CH containing moieties in biomolecular compounds. Relative to collecting broad spectra, limiting the spectral range increases acquisition rates, which is essential for mapping tissues over larger areas. Together, the chemical information in the fingerprint (500-1600 cm−1) and CH-stretching (2840-3000 cm−1) regions of Raman spectra have been used to characterize and stratify cancerous tissues with high sensitivity and selectivity [11,12].
Since CARS is particularly sensitive to CH-rich molecules [13,14], this technique could yield sufficient spectral features in the CH-stretching region to identify tissue components and to discriminate certain diseases such as peripheral artery disease (PAD). PAD is a common manifestation of atherosclerosis, characterized by lower leg ischemia and myopathy in association with leg dysfunction [15]. It is present in at least 25% of individuals over the age of 70 years [16,17]. Intermittent claudication, i.e., limping in response to walking-induced pain relieved by rest, is the major clinical manifestation of PAD [17,18]. The pathophysiology of PAD is complex and is often observed with comorbid conditions such as hypertension, diabetes, dyslipidemia, cigarette smoking, and/or physical inactivity. Patients with PAD are at high risk for myocardial infarction, stroke, and all-cause mortality [19]. Those patients have up to six times the 10-year mortality of age-matched controls [20,21]. PAD has long been under-diagnosed, and disease in up to half of those affected by PAD is yet to be detected [17]. Reasons for this include the high prevalence of asymptomatic disease, inappropriate use of screening and diagnostic tools, and poor awareness of PAD [22]. The methods used for diagnosis of PAD include ankle-brachial index (ABI), doppler and ultrasound (Duplex) imaging, computed tomographic angiography (CT), and magnetic resonance angiography (MRA). Each method has its own advantages and disadvantages.
It was reported that oxidative damage to gastrocnemius myofibers in PAD patients increased with advancing disease and was associated with reduced myofiber cross-sectional area [23]. A discriminant model based on myofiber morphometrics separated PAD from control patients and provided a grading of muscle degeneration within clinical stages of PAD [24]. Human enterovirus (HEV) has been implicated in the pathogenesis of a number of muscle diseases and has been detected in the skeletal muscle of patients with PAD and is associated with severity of the disease [25]. In addition to the efforts for a better physiological understanding of the pathogenesis and treatment options for PAD patients, an effective and reliable method is needed to diagnose the disease in its early stage. Since tissue samples have many chemical constituents and their biomolecular vibrational signatures are quite complex, Raman analysis of only one or two pathology-related peaks is not expected to classify reliably the disease status [5]. The solution is to apply multivariate analysis in a specific spectral region, to extract useful information and identify variables important for classification. Several studies have used Raman spectroscopy with multivariate statistical analysis for biological samples to detect diseases such as cancer [26–29], Huntington’s disease [30], and asthma [31]. Multivariate analysis such as principal component analysis (PCA) is one of the most commonly used methods for transforming spectral data [32]. This leads to the classification method known as principal component-discriminant function analysis (PC-DFA) [33]. The objectives of the present study are to 1) analyze healthy and PAD diseased human gastrocnemius muscle biopsies using both Raman and CARS techniques in the CH-stretching region and 2) use PC-DFA multivariate analysis to evaluate the ability of Spontaneous Raman and CARS to discriminate healthy from PAD gastrocnemius.
2. Materials and method
2.1. Tissue sample preparation
The experimental protocol was approved by the Institutional Review Board of the VA Nebraska and Western Iowa Medical Center, and all subjects gave informed consent. The currently used clinical PAD staging [34] is based on the presentation of the patient. Specifically, patients at: 1) Stage I are asymptomatic, 2) Stage II present with intermittent claudication (walking-induced pain relieved by rest), 3) Stage III experience ischemic leg pain at rest, and 4) Stage IV develop necrosis and gangrene in the affected lower limb(s). In this study, we analyzed tissue from control patients (healthy) and from patients with Stage II disease (functional limb ischemia; moderate) and Stage III/IV disease (critical limb ischemia; severe).
The full description of the tissue sample collection and preparation can be found in previous study [24,35]. Biopsies from human gastrocnemius (calf muscle) were acquired with a Bergstrom needle, fixed in methacarn, and embedded in paraffin. The 4 µm-thick tissue specimens were mounted on 100 nm nanostructured gold-coated slides (Platypus Technologies, Madison, WI) for Raman measurements and on regular glass slides for CARS measurements, respectively. Prior to spectral acquisitions, the slide specimens were deparaffinized with xylene, rehydrated through a series of ethanol washes and allowed to air dry. Since Spontaneous Raman and CARS techniques are based on intrinsic molecular vibrations, they do not require labeling.
2.2. Raman spectroscopy measurements
Raman spectra were recorded using a Renishaw micro-Raman microscope (InVia Reflection) with 633-nm diode laser excitation. The laser power was 10 mW. The laser beam was focused onto the center of the myofiber surface by a 50 × microscope objective with a numerical aperture of 0.75 (Leica n PLAN EPI 50 × /0.75) to a spot size of 1 μm in diameter. In the experiment, myofibers were randomly chosen for recording the Raman spectra. The center of the myofiber excited by the laser, which represented the entire myofiber, was photo-bleached for 30 s and then the spectrum was recorded with an exposure time of 1 s and accumulated by 50 times. Thus it is about 2 min for one Raman spectrum acquisition. The tissue specimens were mounted on gold-coated glass slides for better suppression of fluorescence background and surface plasmonic enhancement. The Raman data sets used in the study are shown in Table 1. A total of 9 tissues specimens from different patients were tested and about 20 spectra per slide were recorded.
Table 1. Raman data sets tested in this study.
| Patient Code | Healthy / disease severity | Number of spectra |
|---|---|---|
| H333 | Healthy | 20 |
| H372 | Healthy | 21 |
| H438 | Healthy | 20 |
| H368 | Moderate | 20 |
| H369 | Moderate | 20 |
| H378 | Moderate | 20 |
| H373 | Severe | 20 |
| H409 | Severe | 20 |
| H415 | Severe | 20 |
2.3. CARS spectroscopy measurements
The femtosecond (fs) broadband CARS microscopy based on a photonic crystal fiber light source was used to measure the muscle tissue specimens. The details about this setup have been described elsewhere; note, however, that the CARS signal was detected with a spectrometer instead of a photomultiplier tube [36]. The powers for the pump and the broadband Stokes beams before entering the microscope were 40 and 60 mW, respectively. Although the instantaneous power is quite high, the ultrashort pulse duration significantly minimizes the heat effect, which occurs and results sample damage when continuum wave lasers are used. As for Raman spectroscopy, myofibers were randomly chosen for recording the CARS spectra and the spectrum from the center of the myofiber can represent the entire myofiber. Because CARS collects anti-Stokes Raman signals, interference from fluorescence is minimal and therefore no photo-bleaching was performed prior to the data collection unlike in Raman spectroscopy. Each CARS spectrum was recorded with an exposure time of 1 s and accumulated by 10, so the total acquisition time for a single CARS spectrum is ~10 s. The tissues measured by CARS were mounted on regular glass slides to keep the best optical transmission. The CARS data sets are shown in Table 2. In CARS experiments, 12 tissue specimens from 12 different patients were analyzed and about 10 spectra from each slide were recorded.
Table 2. CARS data sets tested in this study.
| Patient Code | Healthy / disease severity | Number of spectra |
|---|---|---|
| H333 | Healthy | 10 |
| H418 | Healthy | 9 |
| H433 | Healthy | 8 |
| H438 | Healthy | 9 |
| H345 | Moderate | 10 |
| H368 | Moderate | 12 |
| H369 | Moderate | 10 |
| H378 | Moderate | 11 |
| H350 | Severe | 10 |
| H373 | Severe | 12 |
| H409 | Severe | 11 |
| H415 | Severe | 10 |
2.4. Data preprocessing and PC-DFA analysis
Data preprocessing of the raw Raman spectra included baseline correction and normalization techniques. Baseline corrections were performed by Vancouver Raman algorithm with five-point boxcar smoothing and a 5th order polynomial fit. Vancouver Raman algorithm is a robust iterative modified multi-polynomial fitting algorithm that removes intrinsic autofluorescence, background signals and improves signal to noise ratios [37]. After baseline correction, the spectra were normalized using the standard normal variate technique, which is designed to remove multiplicative error and preserve the same contribution to the results for each spectrum. Raw CARS spectra were preprocessed only by the standard normal variate technique to minimize multiplicative error. Since CARS is free of the autofluorescence, no baseline correction was applied.
The preprocessed spectra were then analyzed by the PC-DFA method utilizing “mixOmics” package in R software. The PCA reduced the dimensionality of the Raman (499 independent variables from 2500 to 3200 cm−1) and CARS spectra (551 independent variables from 2500 to 3100 cm−1) to 6 and 9 principal component scores (PCs), respectively. DFA was performed on these PCs which served as input independent variables. Classification efficacy was determined by cross-validation method (“leave-one-out” method). This method removes one spectrum at a time from the data set and predicts the classification of this spectrum by finding the discriminant model built with the other data.
3. Results and discussion
The CH-containing chemical groups, which are rich in lipid and protein molecules, produce overlapping vibrational bands from CH, CH2 and CH3 groups which complicate a detailed assignment of the band structures of most biomolecular compounds. Although CH regions give less information about most components of the biological samples, possible differences between healthy and diseased tissues can be determined by evaluation of spectral signatures in CH region using PC-DFA multivariate analysis. The details of mode assignments for CH-region can be seen in Table 3.
Table 3. Peak assignments of the Raman spectra of the CH region.
| Peak position (cm−1) | Major assignments |
|---|---|
| 2900 and 2915 | CH symmetric stretching |
| 2845–2850 | CH2 symmetric stretching |
| 2880–2910 | CH2 asymmetric stretching |
| 2865–2885 | CH3 symmetric stretching |
| 2960 and 2970 | CH3 asymmetric stretching |
| 2904–2906, 2918 and 2954 | CH2-fermi (multiple) |
| 2925–2940 | CH3-fermi resonance |
*Assignments are mainly based on reference [38].
3.1. PC-DFA results for Raman measurements in the CH region
To have a complete view of the Raman spectra of three levels of PAD, all preprocessed spectra (2200 – 3550 cm−1) from the same PAD levels were simply averaged and then normalized at 3300 cm−1, as shown in Fig. 1(a). As can be seen, the tissue spectra consisted of CH2 and CH3 stretching bands of lipids or proteins. The peak at 2845 cm−1 is from CH2 symmetric stretching. The peak located at 2875 cm−1 is attributed to CH3 (asymmetric stretching) and the larger band around 2930 cm−1 is assigned to CH3 (fermi resonance) groups (Table 3). The relative intensity of the peaks at 2845 (CH2) and 2875 cm−1 (CH3) increased as disease level increased, indicating the possibility of structural changes in the alkyl sides of lipids and/or proteins because of PAD.
Fig. 1.
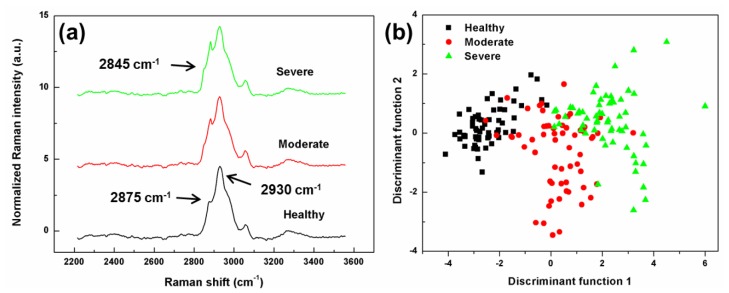
(a) Averaged preprocessed Raman spectra of healthy tissue and two PAD levels: moderate and severe. The spectra have been normalized at 3300 cm−1 and offset vertically for clarity. (b) PC-DFA plot showing the two discriminant function scores of a total 181 Raman spectra from healthy tissue and two levels of PAD.
In the PC-DFA analysis, the CH region 2500-3200 cm−1 of the preprocessed Raman was used. Figure 1(b) shows the PC-DFA plot for healthy tissue and PAD stages. Each colored point represents a spectrum which is plotted as its discriminant function DF1 and DF2 scores. It is obvious that the three groups are recovered in clusters separated well from each other; however, there were a few overlaps mostly between severe and moderate levels, which could be thought as reasonable due to the biochemical similarity of the diseased tissues.
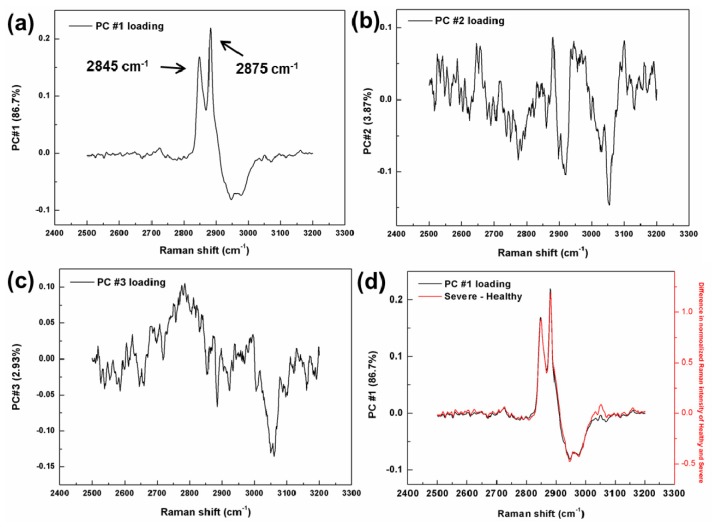
In PCA, a highly complex chemical system (499 independent variables from 2500 to 3200 cm−1) was reduced to a small number of PCs (6) that effectively carry all the important information from the spectral data. Figure 2 shows the first three PC loadings which correspond to the variation of PCs as a function of wavenumber. PC #1 carries the most important data with 86.7% of the total variance. The two bands at 2845 and 2875 cm−1 in the loading plot of PC #1 clearly show CH2 and CH3 stretching vibrations, respectively. As can be seen, both bands at 2845 and 2875 cm−1 become more obvious after the processing. The PC #2 and PC #3 carried only 3.87% and 2.93% of the total variance, respectively.
Fig. 2.
Raman spectra of first three principal component loadings of the PCA, where (a) is PC #1, (b) PC #2, and (c) PC #3; (d) PC #1 loading of the PCA plotted with the spectrum obtained by subtracting the averaged healthy spectrum from that of severe.
To determine the differences between healthy and diseased tissues and check if the first PC loading acquired from PCA was reliable, the averaged healthy spectrum was simply subtracted from the averaged severe spectrum. This derived spectrum and PC #1 loading spectrum were plotted in the Fig. 2(d), which revealed high similarity between two spectra. A strong correlation between these spectra showed that CH2 and CH3 vibration modes could be a significant indicator for PAD detection.
The classification accuracy of PC-DFA, as shown in Table 4, was determined by the cross-validation method as described earlier. As can be seen in Table 4, total 2 spectra of the healthy group in the first data set was misclassified as moderate and none of the healthy group was classified as severe; for diseased cases, some of the spectra were correctly identified as moderate or severe while others were misclassified to the healthy group. Most misclassifications were in the neighboring class rather than extreme class. For three categories: healthy, moderate, and severe, the overall accuracy of Raman spectral data found was to be 85.6% (Table 4). It should be noted that each Raman spectrum was acquired from the center of each myofiber in the tissues. Actually, the myofibers in the muscle tissues are extremely heterogeneous. In order to analyze the spectral variations between myofiber to myofiber than within the myofibers, we assumed that the center of the myofiber would represent the entire myofiber and the spectrum obtained from the center would reflect the condition of the whole fiber. In the experiment, we took spectra from similar anatomical locations (center) in each myofiber for demonstration of the effectiveness of the approach. The sample selection here was random and the center of the myofibers was identified by the operator based on his experience with the architecture of the tissue. Our approach may, therefore, introduce subjectivity in the sample selection process which may affect the final classification results. Moreover, although the tissues evaluated clearly belonged either to healthy control subjects or patients with PAD, not every myofiber was healthy in the samples from healthy subjects nor every myofiber was damaged in the samples from PAD patients. This effect may be another important reason for the misclassification in the model. Considering these biochemical variations, the overall accuracy of 85.6% is acceptable for PAD classification.
Table 4. Classification accuracy of PC-DFA based on Raman data (cross-validation method).
| Patient Code | Healthy / disease severity | PC-DFA as Healthy | PC-DFA as Moderate | PC-DFA as Severe |
|---|---|---|---|---|
| H333 | Healthy (20) | 19 (95.0%) | 1 (5.0%) | 0 |
| H372 | Healthy (21) | 21 (100.0%) | 0 | 0 |
| H438 | Healthy (20) | 19 (95.0%) | 1 (5.0%) | 0 |
| H368 | Moderate (20) | 3 (15.0%) | 16 (80.0%) | 1 (5.0%) |
| H369 | Moderate (20) | 4 (20.0%) | 8 (40.0%) | 8 (40.0%) |
| H378 | Moderate (20) | 0 | 20 (100.0%) | 0 |
| H373 | Severe (20) | 0 | 1 (5.0%) | 19 (95.0%) |
| H409 | Severe (20) | 0 | 6 (30.0%) | 14 (70.0%) |
| H415 | Severe (20) | 0 | 1 (5.0%) | 19 (95.0%) |
|
| ||||
| Overall accuracy | 85.6% | |||
3.2. PC-DFA results of CARS spectra in the CH region
CARS is sensitive to the same CH vibrational signatures as Raman spectroscopy. As discussed earlier, CARS has many advantages compared to spontaneous Raman spectroscopy, such as no fluorescence interference, strong vibrational signals, rapid analysis, and high spatial resolution. Although the fs pulse applied and the nonresonant background influence in CARS imply the lower spectral resolution that reduces spectral quality in the measurement, the information extracted from CARS spectra in the CH region by PC-DFA could be still useful for detection of PAD disease. To have a view of the CARS spectra of healthy tissue and two levels of PAD, all preprocessed spectra from the same levels were averaged and then normalized at 3200 cm−1, as shown in Fig. 3(a). Because of low spectral resolution, the CH2 and CH3 stretching bands in CARS spectrum were not separated properly. Different excitation laser wavelengths (633 nm for Raman, and 800 nm for CARS) caused slight changes in peak locations. Figure 3(b) shows the PC-DFA plot of the CARS data for healthy tissue and PAD tissues. The three groups were mostly recovered in clusters separated from each other, as observed by Raman spectroscopy. However, there was still some overlap among healthy and two levels of diseased samples.
Fig. 3.
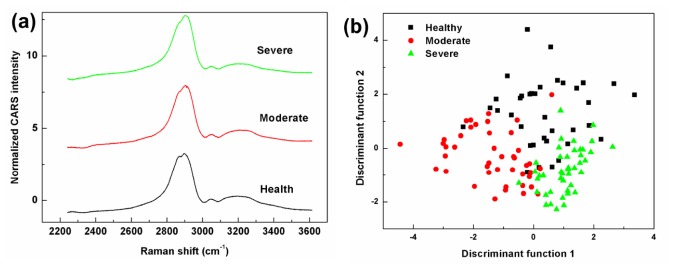
(a) Averaged preprocessed CARS spectra of healthy tissue and two PAD levels: moderate and severe. The spectra have been offset vertically for clarity. (b) A PC-DFA plot showing the first two discriminant function scores of a total of 122 CARS spectra from healthy tissue and two levels of PAD.
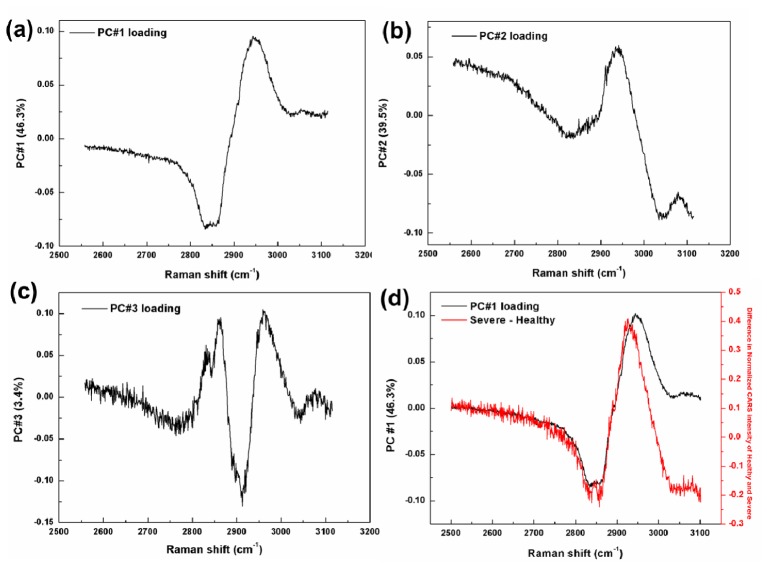
Similar to the Raman analyses, PCA was performed first in the CH region (2500-3100 cm−1) for the CARS data set to reduce the data dimensions (551 independent variables) to a smaller number of PCs (9) that effectively carry most the important information from the spectra. Figure 4 shows the first three PC loadings which correspond to the variation of PCs as a function of wavenumber for CARS analysis.
Fig. 4.
CARS spectra of first three principal components loadings of the PCA, where (a) is PC #1, (b) PC #2, and (c) PC #3; (d) PC #1 loading of the PCA plotted with the spectrum obtained by subtracting the averaged healthy spectrum from that of the severe.
PC #1 carried the most important data with 46.3% of the total variance. The wide band around 2920 cm−1 in the loading plot of PC #1 clearly shows the CH3 symmetric stretching mode. However, the symmetric stretching vibration peak of CH2 was not detected because of low resolution of CARS technique. The next two highest loading plots of PC #2 and PC #3 represented as 39.5% and 3.4% of the total variance, respectively. The spectrum derived by subtraction of the averaged healthy spectrum from the averaged severe spectrum, and PC #1 loading spectrum were plotted in the Fig. 4(d). As can be seen, there is a similarity between the PC #1 loading plot and the difference plot as observed with the Raman data.
The classification accuracy of the PC-DFA of the CARS data determined by the cross-validation method is presented in Table 6. Despite correct identification, there were also misclassifications of some healthy, moderate and severe spectra as observed with Raman data. Most misclassifications were in the neighboring class. The overall accuracy based on the CARS data was 78.7% (Table 5). In our experiment, the average time for one Raman spectrum acquirement is about ~2 min including 30 s photon-bleaching time, 1 s exposure time, and 50 accumulation times. While, the average time for acquiring one CARS spectrum is about 10 s. Although the overall accuracy was less than the one found for Raman data (85.6%), it is still competitive since CARS microscopy enables high-speed collection compared to spontaneous Raman spectroscopy, considering the minute-scale operation is quite long for in clinical application.
Table 5. Classification accuracy of PC-DFA based on CARS data (Cross-validation method).
| Patient Code | Healthy / disease severity | PC-DFA as Healthy | PC-DFA as Moderate | PC-DFA as Severe | |
|---|---|---|---|---|---|
| H333 | Healthy (10) | 7 (70.0%) | 0 | 3 (30.0%) | |
| H418 | Healthy (9) | 5 (55.6%) | 2 (22.2%) | 2 (22.2%) | |
| H433 | Healthy (8) | 5 (62.5%) | 1 (12.5%) | 2 (12.5%) | |
| H438 | Healthy (9) | 6 (66.7%) | 1 (11.1%) | 2 (22.2%) | |
| H345 | Moderate (10) | 1(10.0%) | 9 (90.0%) | 0 | |
| H368 | Moderate (12) | 1 (8.3%) | 10 (83.3%) | 1 (8.3%) | |
| H369 | Moderate (10) | 0 | 4 (40.0%) | 6 (60.0%) | |
| H378 | Moderate (11) | 0 | 10 (90.1%) | 1 (9%) | |
| H350 | Severe (10) | 0 | 1(10%) | 9 (90%) | |
| H373 | Severe (12) | 0 | 0 | 12 (100%) | |
| H409 | Severe (11) | 0 | 0 | 11 (100%) | |
| H415 | Severe (10) | 2 (20%) | 0 | 8 (80%) | |
|
| |||||
| Overall accuracy | 78.7% | ||||
3.3. Classification performance and discussion towards clinical applications
Classification accuracy of tissues from healthy individuals and patients with moderate or severe PAD, based on the spectra from each level is presented in Table 6. Identification of moderate and severe disease was similar for both methods, however; Raman was significantly more sensitive on identification of healthy tissues compared to CARS. Only two healthy spectra were misclassified among 61 healthy spectra (Table 6). In order to evaluate the sensitivity and specificity of Raman and CARS performance, the spectral data were divided into two categories (healthy and diseased). The sensitivity of 94.2% and specificity of 96.7% were calculated in Raman test, while the sensitivity of 95.4% and specificity of 63.9% were calculated in CARS test. Although CARS suffers a low specificity rate, it seems to be sensitive on identification of PAD severity, which indicates that CARS may be a competitive tool compared to Raman for differentiating tissues from patients with moderate and severe PAD, by PC-DFA. Actually the setup, spectral analysis method, and classification performance of CARS in our study still have room for improvement. For example, the spectral resolution of the fs broadband CARS used in this study is estimated to be ~30 cm−1, which however can be improved using a picosecond (ps) laser and a tunable optical parametric oscillator (OPO) ultrafast laser [39]. It was reported that the spectral resolution can be improved to ~10 cm−1 and the resonant CARS spectra can be extracted [40], which in principle has potential to have better classification performance. Moreover, many research efforts have been made to design and develop CARS probes [41], even the CARS endoscopy [42]; therefore if these maturing CARS probe technique involved, CARS has potential towards clinical applications.
Table 6. Classification accuracy of Raman and CARS.
| PC-DFA as Healthy |
PC-DFA as Moderate |
PC-DFA as Severe |
|
|---|---|---|---|
| Healthy Raman (61) | 59 (96.7%) | 2 | 0 |
| Healthy CARS (36) | 23 (63.9%) | 4 | 9 |
| Moderate Raman (60) | 7 | 45 (75.0%) | 9 |
| Moderate CARS (43) | 2 | 33 (76.7%) | 8 |
| Severe Raman (60) | 0 | 8 | 52 (86.7%) |
| Severe CARS (43) | 2 | 1 | 40 (93.0%) |
In a previous study, we focused on the fingerprint region (400-1800 cm−1) [35]. We averaged all spectra from different myofiber for one patient and used these averaged data in a partial least squares regression model. Thus the data set included a total of 15 averaged spectra (5 for healthy, 5 for each disease levels). However, the current work represents larger numbers of spectra. We used a total of 181 spectra from 9 patients for Raman spectroscopy and a total 122 of spectra from 12 patients for CARS. We showed that CH region could be a good choice for PAD application. The overall accuracy is 85.6% for Raman and 78.7% for CARS, respectively. Because not every fiber was damaged by disease or not every myofiber was healthy in healthy tissues, some misclassifications in both measurements were unavoidable. Biochemical variations among patients and myofiber lowered the classification accuracy compared to our previous Raman study.
Other studies such as Doppler, ultrasound, computed tomographic angiography etc. measure changes in the blood supply to the legs to help with the diagnosis and staging of PAD. However, the pathophysiology of PAD and its effects on the ischemic lower limbs cannot be explained simply by changes in the blood flow of the affected legs [43]. Our work responds to the need for the development of new methods that can directly evaluate the skeletal muscle as the end-organ affected in the ischemic legs. Our methods and findings represent an initial attempt to quantify the damage produced by PAD in the affected leg muscle. Measurements of skeletal muscle degeneration such as those obtained using Raman and CARS spectroscopy can complement the measures obtained from tests of blood flow, effectively expanding the information available to vascular specialists; thereby improving their ability to appropriately stage and treat patients with PAD.
4. Conclusions and future work
We performed Raman measurements to identify biochemical changes in CH containing moieties in the tissues and therefore, to determine damage to the muscle. Both the spontaneous Raman and CARS spectroscopy, in conjunction with PC-DFA, were applied to the problem of diagnosis of PAD. The overall accuracies for Raman and CARS were 85.6% and 78.7%, respectively. Considering the biochemical variation of the tissue samples, both overall accuracies are high. Because of the lower spectral resolution in CARS, accuracy for this technique was expected to be lower, however; CARS is still competitive compared to Raman because of its nature advantages, especially allowing fast data collection. Future research work could involve maturing CARS probe techniques for fast and better diagnostic performance and also make it towards clinical applications.
References and Links
- 1.Bonifacio A., Beleites C., Vittur F., Marsich E., Semeraro S., Paoletti S., Sergo V., “Chemical imaging of articular cartilage sections with Raman mapping, employing uni- and multi-variate methods for data analysis,” Analyst (Lond.) 135(12), 3193–3204 (2010). 10.1039/c0an00459f [DOI] [PubMed] [Google Scholar]
- 2.Movasaghi Z., Rehman S., Rehman I. U., “Raman spectroscopy of biological tissues,” Appl. Spectrosc. Rev. 42(5), 493–541 (2007). 10.1080/05704920701551530 [DOI] [Google Scholar]
- 3.Koljenović S., Bakker Schut T. C., van Meerbeeck J. P., Maat A. P., Burgers S. A., Zondervan P. E., Kros J. M., Puppels G. J., “Raman microspectroscopic mapping studies of human bronchial tissue,” J. Biomed. Opt. 9(6), 1187–1197 (2004). 10.1117/1.1805555 [DOI] [PubMed] [Google Scholar]
- 4.Nijssen A., Bakker Schut T. C., Heule F., Caspers P. J., Hayes D. P., Neumann M. H. A., Puppels G. J., “Discriminating basal cell carcinoma from its surrounding tissue by Raman spectroscopy,” J. Invest. Dermatol. 119(1), 64–69 (2002). 10.1046/j.1523-1747.2002.01807.x [DOI] [PubMed] [Google Scholar]
- 5.Chen P., Shen A., Zhou X. D., Hu J. M., “Bio-Raman spectroscopy: a potential clinical analytical method assisting in disease diagnosis,” Anal. Methods 3(6), 1257–1269 (2011). 10.1039/c1ay05039g [DOI] [Google Scholar]
- 6.Puppels G. J., de Mul F. F., Otto C., Greve J., Robert-Nicoud M., Arndt-Jovin D. J., Jovin T. M., “Studying single living cells and chromosomes by confocal Raman microspectroscopy,” Nature 347(6290), 301–303 (1990). 10.1038/347301a0 [DOI] [PubMed] [Google Scholar]
- 7.Buschman H. P., Deinum G., Motz J. T., Fitzmaurice M., Kramer J. R., van der Laarse A., Bruschke A. V., Feld M. S., “Raman microspectroscopy of human coronary atherosclerosis: Biochemical assessment of cellular and extracellular morphologic structures in situ,” Cardiovasc. Pathol. 10(2), 69–82 (2001). 10.1016/S1054-8807(01)00064-3 [DOI] [PubMed] [Google Scholar]
- 8.van Manen H. J., Kraan Y. M., Roos D., Otto C., “Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes,” Proc. Natl. Acad. Sci. U.S.A. 102(29), 10159–10164 (2005). 10.1073/pnas.0502746102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X. N., Allen J., Black P. N., Baldacchini T., Huang X., Huang H., Jiang L., Lu Y. F., “Coherent anti-Stokes Raman scattering and spontaneous Raman spectroscopy and microscopy of microalgae with nitrogen depletion,” Biomed. Opt. Express 3(11), 2896–2906 (2012). 10.1364/BOE.3.002896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Napoli C., Pope I., Masia F., Watson P., Langbein W., Borri P., “Hyperspectral and differential CARS microscopy for quantitative chemical imaging in human adipocytes,” Biomed. Opt. Express 5(5), 1378–1390 (2014). 10.1364/BOE.5.001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gniadecka M., Philipsen P. A., Sigurdsson S., Wessel S., Nielsen O. F., Christensen D. H., Hercogova J., Rossen K., Thomsen H. K., Gniadecki R., Hansen L. K., Wulf H. C., “Melanoma diagnosis by Raman spectroscopy and neural networks: structure alterations in proteins and lipids in intact cancer tissue,” J. Invest. Dermatol. 122(2), 443–449 (2004). 10.1046/j.0022-202X.2004.22208.x [DOI] [PubMed] [Google Scholar]
- 12.Kast R. E., Serhatkulu G. K., Cao A., Pandya A. K., Dai H., Thakur J. S., Naik V. M., Naik R., Klein M. D., Auner G. W., Rabah R., “Raman spectroscopy can differentiate malignant tumors from normal breast tissue and detect early neoplastic changes in a mouse model,” Biopolymers 89(3), 235–241 (2008). 10.1002/bip.20899 [DOI] [PubMed] [Google Scholar]
- 13.Le T. T., Duren H. M., Slipchenko M. N., Hu C. D., Cheng J. X., “Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans,” J. Lipid Res. 51(3), 672–677 (2010). 10.1194/jlr.D000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan X., Cheng J. X., Xie X. S., “Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy,” J. Lipid Res. 44(11), 2202–2208 (2003). 10.1194/jlr.D300022-JLR200 [DOI] [PubMed] [Google Scholar]
- 15.Belch J. J. F., Topol E. J., Agnelli G., Bertrand M., Califf R. M., Clement D. L., Creager M. A., Easton J. D., Gavin J. R., 3rd, Greenland P., Hankey G., Hanrath P., Hirsch A. T., Meyer J., Smith S. C., Sullivan F., Weber M. A., Prevention of Atherothrombotic Disease Network , “Critical issues in peripheral arterial disease detection and management: a call to action,” Arch. Intern. Med. 163(8), 884–892 (2003). 10.1001/archinte.163.8.884 [DOI] [PubMed] [Google Scholar]
- 16.Criqui M. H., Fronek A., Barrett-Connor E., Klauber M. R., Gabriel S., Goodman D., “The prevalence of peripheral arterial disease in a defined population,” Circulation 71(3), 510–515 (1985). 10.1161/01.CIR.71.3.510 [DOI] [PubMed] [Google Scholar]
- 17.Hirsch A. T., Criqui M. H., Treat-Jacobson D., Regensteiner J. G., Creager M. A., Olin J. W., Krook S. H., Hunninghake D. B., Comerota A. J., Walsh M. E., McDermott M. M., Hiatt W. R., “Peripheral arterial disease detection, awareness, and treatment in primary care,” JAMA 286(11), 1317–1324 (2001). 10.1001/jama.286.11.1317 [DOI] [PubMed] [Google Scholar]
- 18.Wood A. J. J., Hiatt W. R., “Medical treatment of peripheral arterial disease and claudication,” N. Engl. J. Med. 344(21), 1608–1621 (2001). 10.1056/NEJM200105243442108 [DOI] [PubMed] [Google Scholar]
- 19.Muller M. D., Reed A. B., Leuenberger U. A., Sinoway L. I., “Physiology in medicine: peripheral arterial disease,” J. Appl. Physiol. 115(9), 1219–1226 (2013). 10.1152/japplphysiol.00885.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Criqui M. H., Langer R. D., Fronek A., Feigelson H. S., Klauber M. R., McCann T. J., Browner D., “Mortality over a period of 10 years in patients with peripheral arterial disease,” N. Engl. J. Med. 326(6), 381–386 (1992). 10.1056/NEJM199202063260605 [DOI] [PubMed] [Google Scholar]
- 21.Leng G. C., Fowkes F. G. R., Lee A. J., Dunbar J., Housley E., Ruckley C. V., “Use of ankle brachial pressure index to predict cardiovascular events and death: a cohort study,” BMJ 313(7070), 1440–1443 (1996). 10.1136/bmj.313.7070.1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimminiello C., Kownator S., Wautrecht J. C., Carvounis C. P., Kranendonk S. E., Kindler B., Mangrella M., Borghi C., PANDORA Study Investigators , “The PANDORA study: peripheral arterial disease in patients with non-high cardiovascular risk,” Intern. Emerg. Med. 6(6), 509–519 (2011). 10.1007/s11739-011-0511-0 [DOI] [PubMed] [Google Scholar]
- 23.Weiss D. J., Casale G. P., Koutakis P., Nella A. A., Swanson S. A., Zhu Z., Miserlis D., Johanning J. M., Pipinos I. I., “Oxidative damage and myofiber degeneration in the gastrocnemius of patients with peripheral arterial disease,” J. Transl. Med. 11(1), 230 (2013). 10.1186/1479-5876-11-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cluff K., Miserlis D., Naganathan G. K., Pipinos I. I., Koutakis P., Samal A., McComb R. D., Subbiah J., Casale G. P., “Morphometric analysis of gastrocnemius muscle biopsies from patients with peripheral arterial disease: objective grading of muscle degeneration,” Am. J. Physiol. Regul. Integr. Comp. Physiol. 305(3), R291–R299 (2013). 10.1152/ajpregu.00525.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J. K. S., Zhu Z., Casale G., Koutakis P., McComb R. D., Swanson S., Thompson J., Miserlis D., Johanning J. M., Haynatzki G., Pipinos I. I., “Human enterovirus in the gastrocnemius of patients with peripheral arterial disease,” J Am Heart Assoc 2(4), e000082 (2013). 10.1161/JAHA.113.000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendall C., Stone N., Shepherd N., Geboes K., Warren B., Bennett R., Barr H., “Raman spectroscopy, a potential tool for the objective identification and classification of neoplasia in Barrett’s oesophagus,” J. Pathol. 200(5), 602–609 (2003). 10.1002/path.1376 [DOI] [PubMed] [Google Scholar]
- 27.Krafft C., Sobottka S. B., Schackert G., Salzer R., “Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors,” Analyst (Lond.) 130(7), 1070–1077 (2005). 10.1039/b419232j [DOI] [PubMed] [Google Scholar]
- 28.Krishna C. M., Sockalingum G. D., Kegelaer G., Rubin S., Kartha V. B., Manfait M., “Micro-Raman spectroscopy of mixed cancer cell populations,” Vib. Spectrosc. 38(1-2), 95–100 (2005). 10.1016/j.vibspec.2005.02.018 [DOI] [Google Scholar]
- 29.Moreno M., Raniero L., Arisawa E. A. L., do Espirito Santo A. M., dos Santos E. A. P., Bitar R. A., Martin A. A., “Raman spectroscopy study of breast disease,” Theor. Chem. Acc. 125(3-6), 329–334 (2010). 10.1007/s00214-009-0698-6 [DOI] [Google Scholar]
- 30.Muratore M., “Raman spectroscopy and partial least squares analysis in discrimination of peripheral cells affected by Huntington’s disease,” Anal. Chim. Acta 793, 1–10 (2013). 10.1016/j.aca.2013.06.012 [DOI] [PubMed] [Google Scholar]
- 31.Sahu A., Dalal K., Naglot S., Aggarwal P., Murali Krishna C., “Serum based diagnosis of asthma using Raman spectroscopy: an early phase pilot study,” PLoS ONE 8(11), e78921 (2013). 10.1371/journal.pone.0078921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong Y. H., Lim M., Liu Q., “Comparison of principal component analysis and biochemical component analysis in Raman spectroscopy for the discrimination of apoptosis and necrosis in K562 leukemia cells,” Opt. Express 20(20), 22158–22171 (2012). 10.1364/OE.20.022158 [DOI] [PubMed] [Google Scholar]
- 33.Hamasha K., Mohaidat Q. I., Putnam R. A., Woodman R. C., Palchaudhuri S., Rehse S. J., “Sensitive and specific discrimination of pathogenic and nonpathogenic Escherichia coli using Raman spectroscopy-a comparison of two multivariate analysis techniques,” Biomed. Opt. Express 4(4), 481–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner A. W., Afaq A., “Management of lower extremity peripheral arterial disease,” J. Cardiopulm. Rehabil. Prev. 28(6), 349–357 (2008). 10.1097/HCR.0b013e31818c3b96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cluff K., Kelly A. M., Koutakis P., He X. N., Huang X., Lu Y. F., Pipinos I. I., Casale G. P., Subbiah J., “Surface-enhanced Raman spectral biomarkers correlate with Ankle Brachial Index and characterize leg muscle biochemical composition of patients with peripheral arterial disease,” Physiol. Rep. 2(9), e12148 (2014). 10.14814/phy2.12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X., He X. N., Xiong W., Gao Y., Jiang L. J., Liu L., Zhou Y. S., Jiang L., Silvain J. F., Lu Y. F., “Contrast enhancement using silica microspheres in coherent anti-Stokes Raman spectroscopic imaging,” Opt. Express 22(3), 2889–2896 (2014). 10.1364/OE.22.002889 [DOI] [PubMed] [Google Scholar]
- 37.Zhao J., Lui H., McLean D. I., Zeng H., “Automated Autofluorescence Background Subtraction Algorithm for Biomedical Raman Spectroscopy,” Appl. Spectrosc. 61(11), 1225–1232 (2007). 10.1366/000370207782597003 [DOI] [PubMed] [Google Scholar]
- 38.Lu R., Gan W., Wu B. H., Zhang Z., Guo Y., Wang H. F., “C-H stretching vibrations of methyl, methylene and methine groups at the vapor/alcohol (n = 1-8) interfaces,” J. Phys. Chem. B 109(29), 14118–14129 (2005). 10.1021/jp051565q [DOI] [PubMed] [Google Scholar]
- 39.Evans C. L., Xie X. S., “Coherent anti-Stokes Raman scattering microscopy: chemical imaging for biology and medicine,” Annu Rev Anal Chem (Palo Alto Calif) 1(1), 883–909 (2008). 10.1146/annurev.anchem.1.031207.112754 [DOI] [PubMed] [Google Scholar]
- 40.Di Napoli C., Pope I., Masia F., Watson P., Langbein W., Borri P., “Hyperspectral and differential CARS microscopy for quantitative chemical imaging in human adipocytes,” Biomed. Opt. Express 5(5), 1378–1390 (2014). 10.1364/BOE.5.001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balu M., Liu G., Chen Z., Tromberg B. J., Potma E. O., “Fiber delivered probe for efficient CARS imaging of tissues,” Opt. Express 18(3), 2380–2388 (2010). 10.1364/OE.18.002380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Légaré F., Evans C. L., Ganikhanov F., Xie X. S., “Towards CARS Endoscopy,” Opt. Express 14(10), 4427–4432 (2006). 10.1364/OE.14.004427 [DOI] [PubMed] [Google Scholar]
- 43.Hirsch A. T., Haskal Z. J., Hertzer N. R., Bakal C. W., Creager M. A., Halperin J. L., Hiratzka L. F., Murphy W. R. C., Olin J. W., Puschett J. B., Rosenfield K. A., Sacks D., Stanley J. C., Taylor L. M., White C. J., White J., White R. A., “ACC/AHA 2005 Practice Guidelines for the Management of Patients With Peripheral Arterial Disease (Lower Extremity, Renal, Mesenteric, and Abdominal Aortic),” Circulation 113(11), e463–e465 (2006). [DOI] [PubMed] [Google Scholar]