Abstract
Experimentation demonstrates long-range surface plasmon polariton waveguides as a useful biosensor to selectively detect gram negative or gram positive bacteria in human urine having a low concentration of constituents. The biosensor can detect bacteria at concentrations of 105 CFU/ml, the internationally recommended threshold for diagnostic of urinary tract infection. Using a negative control urine solution of bacterial concentration 1000☓ higher than the targeted bacteria, we obtain a ratio of 5.4 for the positive to negative signals.
OCIS codes: (170.0170) Medical optics and biotechnology; (170.7230) Urology; (280.1415) Biological sensing and sensors; (240.6680) Surface plasmons; (130.3120) Integrated optics devices; (230.7390) Waveguides, planar; (250.5403) Plasmonics
1. Introduction
The gold standard for the diagnosis of Urinary Tract Infection (UTI) is the detection of pathogen along with the presence of clinical symptoms. The best detection and identification of the pathogen remains the culturing of patient urine sample [1]. This technique provides very good selectivity of the pathogen and superior sensitivity but requires 24 to 48 hours in a microbiology laboratory environment. Nitrite dipsticks provide results in less than one hour but cannot detect organisms which are unable to reduce nitrate to nitrite [2]. In addition, dipsticks have poor sensitivity especially with diluted urine or low colony count samples. In the labeled biosensing category, flow cytometry is a family of automated systems providing results in less than one minute for detection levels below 105 CFU/ml [3-5]. Selectivity in flow cytometry can be obtained by double staining with fluorescent dyes [6]. However, fluorescence detection requires labeling by well-trained personnel, which can add several hours to perform the detection. Here we investigate the ability of a label-free long-range surface plasmon polariton (LRSPP) waveguide biosensor to detect and identify bacteria as gram positive or gram negative species in human urine samples. Experimental results demonstrate the performance of the sensor, from which we define a protocol supporting the diagnostic of UTI.
Label-free biosensors such as surface plasmon resonance (SPR) [7], LRSPP resonance [8,9] and LRSPP waveguides [10] can provide detection of bacteria in clean fluids [8,11,12]. They are capable of detection selectivity similar to enzyme-linked immunosorbent assay (ELISA). Thus, they have the potential, in principle, to improve clinical diagnosis [13,14] of many diseases if conclusive detection in complex human fluids such as blood serum and urine can be achieved. We demonstrate here that this is possible using a simple non-specific binding mitigation strategy and by careful comparison with negative controls.
Here we use a surface functionalized with antibody to selectively capture target bacteria on the LRSPP waveguide. Test devices are manufactured in a wafer-based fabrication process [15,16] and are interrogated by a laser-based test system [17]. Injecting the content of syringes in the fluidic channel of the test device, the biosensing area is exposed to attachment chemistries then antibodies to functionalize the surface in preparation for a test. The test fluid containing the target or control bacteria is then injected. If present at a high enough concentration, the target bacteria is selectively adsorbed by the antibody, disrupting the LRSPP mode propagation. By measuring and analyzing the optical response of the waveguide with a power detector and an infrared camera, we establish the presence of the target bacteria.
2. Materials and methods
2.1 Chemical and biochemical reagents
Our labeling convention to identify solutions prepared is to use an acronym with a date in the format: AAAAmmdd, where AAAA is the biochemical acronym; mm is the month and dd is the day of creation. A complete list of the labeled solutions and preparation protocols used in experimentation can be found in Table 3 of Appendix A. Unless stated differently, all chemicals were acquired from Sigma-Aldrich (S-A). Chemicals used includes: distilled deionized water (DIH2O) from a Barnstead Nanopure system D11931. Phosphate buffered saline (PBS, 0.01 M, pH 7.4), sodium dodecyl sulfate (SDS)(71725-50G), 2-isopropanol semiconductor grade (IPA), acetone HPLC grade ≥ 99.9%, heptane, glycerol (electrophoresis grade), LB broth (Lennox), PBS Glycerol (PBSG) solutions were filtered through Millex-GP filters (PES membrane 0.22 μm). Labeled fluids Gprot (P4689-1MG): protein G (50 μg/ml) dissolved in PBSG0715 and used to functionalize the bare gold surface of a waveguide. Labeled fluids GNeg (AB41202): gram negative antibody (50 μg/ml) in PBSG0715 buffer solutions. Labeled fluids GPos (AB20344): gram positive antibodies (50 μg/ml) diluted in PBS. Labeled fluids BSA (A0281-250): bovine serum albumin (100 μg/ml) dissolved in PBSG. Labeled fluids Urine: A human urine sample was collected from the donor on the day of experimentation [18]. All urine samples were centrifuged at 3000 rpm for 7 min and the supernatant filtered through Millex-GP filters. Labeled fluids EColi: Escherichia coli XL1 Blue were donated by the Canadian Blood Services (CBS). Labeled fluid SEPI: Staphylococcus epidermidis ATCC 12228 bacteria were donated by the CBS. Inoculation of bacteria into LB Broth was done aseptically (CAREG laboratory of uOttawa). Weekly cultures of the bacteria were grown in 10 ml vials and incubated at 37° C. The cultured bacteria were centrifuged at 3000 RPM for 7 min and the supernatant removed with a syringe and replaced with a PBS, PBSG or urine test solution as required by the specific experiment.
2.2 Biosensor fabrication
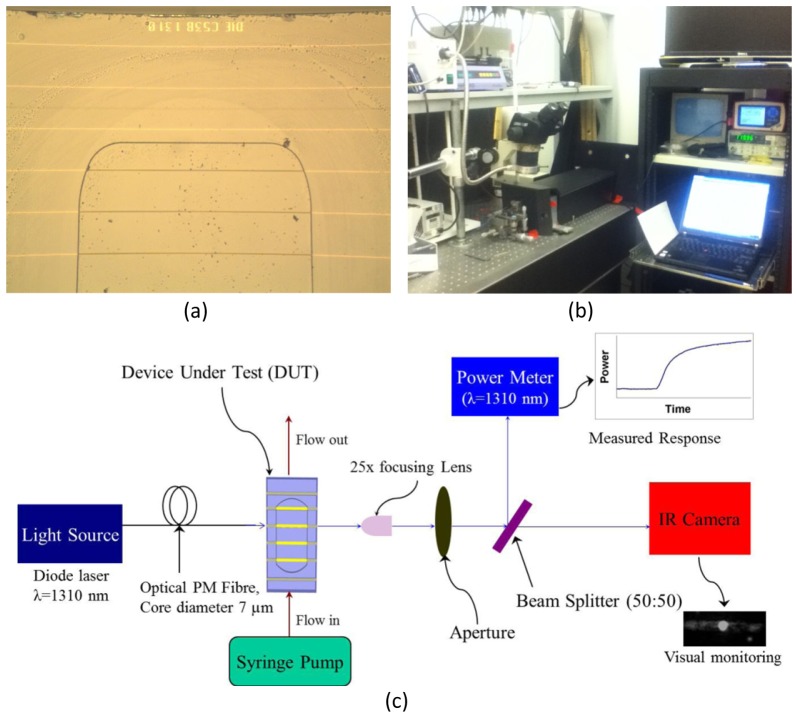
The biosensors were fabricated by cladding a Au stripe, 5 μm wide and 35 nm thick, with top and bottom CYTOP layers, each 8 μm thick, on a 4-inch Si wafer. Au evaporation and lift-off were used to define the Au features using an optical lithography technique. The top CYTOP cladding was etched down to the Au stripe surface to form a large fluidic channel of sensing length L = 1.65 mm. The Au stripe is on a CYTOP pedestal of height ~400 nm created by over-etching the fluidic channels. A thick layer of photoresist was deposited onto the wafer before it was sent for dicing into ∼300 dies per wafer. The detailed fabrication process of the sensors was described in previous work [15,16]. A microscope image under 50☓ magnification of a portion of a die after experimentation is shown in Fig. 1(a). The sensor dies utilized originated from wafer 4D1-A reticule C53B1310, and were 3.8 mm long by 6.4 mm wide. Each die had several straight uniform Au waveguides, 5 μm wide and 35 nm thick, of which stripes 9 and 13 were mostly used for sensing experiments as they are located in the centre of the fluidic channels. Stripe 10 was also used for sensing experiments, as a failed (unresolved) step-in-width grating which is mostly uniform and centrally located within the fluidic channel. Stripes 1, 3 and 21 are fully cladded and were used for alignment trials and waveguide quality checks.
Fig. 1.
(a) Microscope image at 50☓ magnification of the top portion of die C53B1310 after use. (b) Photograph of the interrogation system. (c) Block diagram of the LRSPP waveguide biosensor interrogation system [©2013 Optical Society of America; adapted from [17].
2.3 Die cleaning process
To ensure the cleanliness of the waveguide facets for efficient optical input and output coupling, a fresh sensor die was cleaned by ultra-sonication (FB-11201, Fisher Scientific) in heptane for 5 min to remove any possible debris formed during dicing of the wafer. The sensor die was then left immersed in two sequential acetone baths for 5 and 30 min, respectively, to completely remove the dicing photoresist. After a thorough washing in IPA and drying with nitrogen gas (N2), the sensor die was placed in a digital UV ozone system (PSD-UV-4, Novascan) to remove any possible organic matter from the Au surface. The die was then washed intensely with IPA and distilled/de-ionized water (DDI H2O), followed by Nitrogen (N2) drying. Cleaned sensor die assembled in the test jig were all primed with DDI H2O before assembly onto the test system.
2.4 Surface functionalization process
Once inserted into the test system, the surface of a Au stripe was functionalized by injecting GProt solution for 20 min at a rate of 20 μL/min. Following this step, and for a gram negative selective surface, Gneg was injected for 10 min at a rate of 20 μL/min followed by stop flow for 80 min which results in the formation of a monolayer of gram negative antibody. For a gram positive selective surface, GPos was injected for 10 min at a rate of 20 μL/min followed by stop flow for 80 min which results in the formation of a monolayer of gram positive antibody. In both cases, we complete the functionalization by injecting BSA for 5 min at a rate of 20 μL/min. BSA injection was selected as a simple non-specific binding mitigation strategy, as BSA would block open adsorption sites without interfering with the monolayer of antibody. Generally however, a flat sensorgram response was observed during BSA injection, indicating low vulnerability to non-specific binding and valid surface functionalization.
Upon completion of experimental runs, which could last up to 36 hrs (2160 min), the Au surface of the sensor die was discarded or fully regenerated. The regeneration process starts by flowing 2 ml of SDS then 2 ml of DIH20. The die is then removed from the jig and deposited in a vial of SDS for periods of 24 to 96 hours to dissolve the lipopolysaccharide membrane of the bacteria. The die is further cleaned by rinsing and depositing in vials with Acetone, IPA and DIH2O to remove debris. Nitrogen gas (N2) is used to dry the surface and a microscope inspection provides necessary quality control. The regenerated surface of a sensor die is then placed in a UV/ozone chamber before starting a new experimental run. Measurement of the optical insertion loss with a RI-controlled fluid after cleaning was used as a quality measure before re-using a die in a new experimental run.
2.5 Mechanical test system
A block diagram of the test system is shown in Fig. 1(b). It was integrated from commercial components. A bacteriological control area is delimited by all components of the fluidic circuit. A closed fluid circuit starts from an input syringe, followed by a 50 cm long segment of Pico tubing (IDEX 550 µm outer dia., 250 µm inner dia.), inserted into the syringe end. The other end of the input tubing was glued to a hole in a Plexiglas cover bearing an o-ring to seal the fluidic channel of the test device. A similar segment of output tubing was glued to another hole in the cover and returns from the cover to the stainless steel needle at the end of the output syringe. A hermetically-sealed connection is obtained by gluing the tubing inside the needle and wrapping it with tape. The output syringe completes the fluidic circuit. The piston of the output syringe is fixed into a syringe pump to precisely control the rate of flow. The syringe pump (PicoPlus, Harvard Apparatus) and associated syringes where located on a shelf 30 cm above the optical setup. Injection of fluid was normally done by pulling during experimental runs, and pushing to prime the line or clear an air bubble from the fluidic circuit.
Two multi-axis positioning stages were used to align the laser beam, one out of a polarized fiber and one out of the test device, providing precise alignment accuracy and stability. For the fiber-to-waveguide alignment, a 6-axis stage was used to manipulate the fiber holder and fiber. For test device alignment, a 3-axis stage to manipulate the fluidic jig assembly was used. Alignment was performed just before time zero and all of the positioner actuators were fixed for the duration of the experiment.
2.6 Optical test system
All Optical components were fixed onto an anti-vibration table. The light source was a PM-fiber pigtailed laser diode (NLK1B5GAAA, NEL) in a laser mount (LDM-4980, ILX) controlled by current and temperature controller (LDX3220, ILX) delivering 14.5 dBm of output power at a free-space optical wavelength of 1310 nm with a set current of 120 mA and a set temperature of 30° C. A set current of 50 mA (delivering 11.1 dBm) is sometime used. The optical PM fiber (PMJ-3S3A-1300-7/125-1-1-1, OZ Optics Ltd.) used to excite a sensor was 0.5 m long, cleaved and fixed into a fiber holder. A 25☓ optical lens (25/0.50, Melles Griot) was used to focus the output light onto an optical sensor (S144C, Thorlabs) connected to a power meter (PM100, Thorlabs). An optical aperture was used during alignment of the laser beam and to reduce the power detected at the sensor due to background light. An optical beam splitter (BSW29, Thorlabs) was used to split the output beam in order to provide an image of the mode on an IR camera (Micronviewer 7290A, electrophysics). Without a sensor in the set-up, the detected power was typically 7.9 dBm. Thus, we estimate the loss through the cleaved optical fiber, lens, aperture and beam splitter to be 6.6 dB, of which 5 dB comes from the beam splitter at the set angle. The typical insertion loss of a 3.8 mm long cladded waveguide was measured to be 27 dB. Thus the maximum output power that we can obtain with a LRSPP waveguide biosensor in the set-up is about −19.1 dBm, providing more than 11 dB of dynamic range on the power detector (rated for −30 dBm at 1310 nm).
In our experimental arrangement, and when operating above LRSPP cutoff, the biosensing region is defined by the area of the Au stripe exposed in the fluidic channel, shown in Fig. 1(a). On die C53B1310 the sensing area is 5 μm wide by 1.6 mm long. The probing depth of LRSPPs is about 2 μm [17], which is about 5☓ larger than conventional SPR.
3. Bulk sensing of urine with an LRSPP waveguide
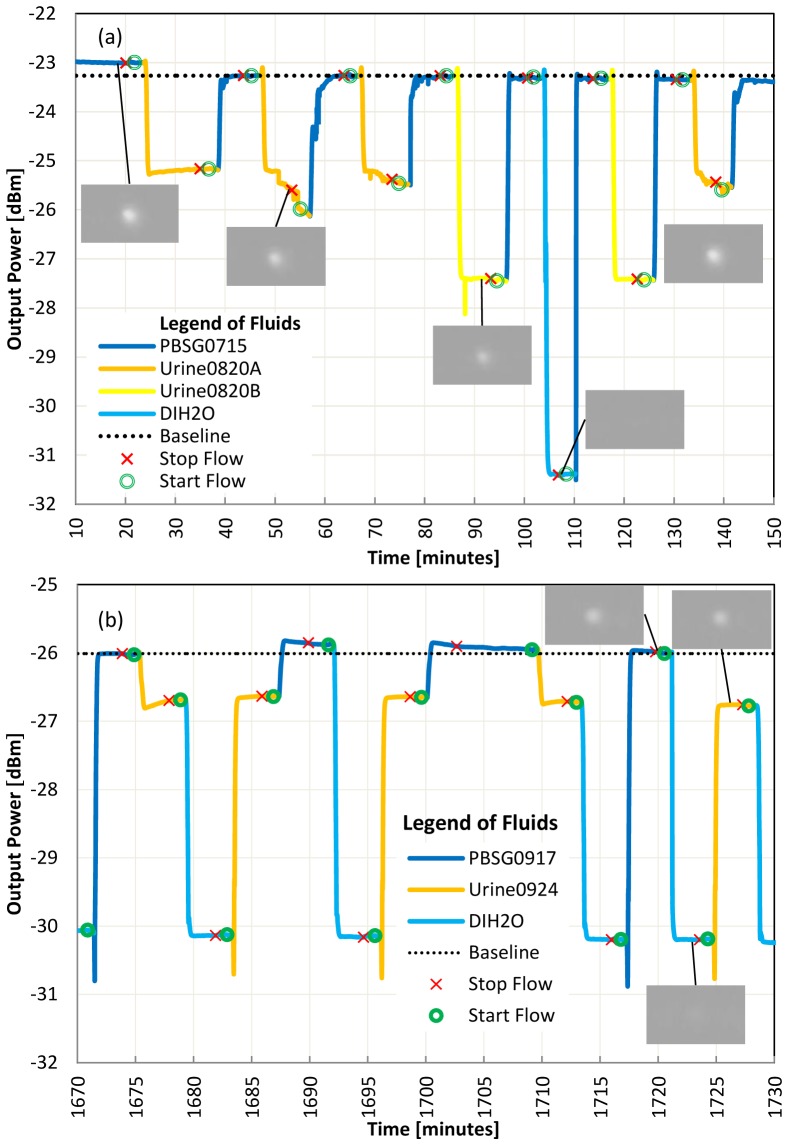
The response of LRSPP waveguide biosensors due to directly injecting human urine is shown in Fig. 2. A bare Au surface was used in Fig. 2(a) whereas a gram negative antibody on protein G surface was used in Fig. 2(b). These two surface functionalizations could be used for different urinalysis experimentation (as will be shown later). Four water-based solutions, namely PBSG0715, Urine0820A, Urine0820B and water, of varying refractive index (RI), were injected in sequence. Considering LRSPP mode cutoff and radiation [19], we expect a reduction in output power as the difference between the RI of CYTOP and that of the solution increases, because the RI asymmetry between the top and bottom claddings of the waveguide increases. The images of the output mode, shown as insets to Fig. 2, confirm excitation of the LRSPP mode for all solutions except pure water (DIH2O) which has a RI 15.1 mRIU below that of CYTOP. With a large RI asymmetry in the top (test fluid) and bottom (CYTOP) claddings, the LRSPP mode becomes cut-off, and propagation occurs in the form of radiative modes that leak into the higher RI cladding, but that may still exhibit localization near the metal stripe [19]. The output observed for the case of DIH2O consists essentially of background optical energy with the LRSPP not being evident. Note that on the IR camera, the output is magnified by 25☓ and is limited in diameter by the aperture.
Fig. 2.
(a) Bulk sensing of high concentration and low concentration urine, flow rate = 20 μl/min, laser power = 14.5 dBm, Die C53B1310 first cleaned on 07 Aug 2014. No functionalization (bare Au surface). (b) Use of DIH2O to separate urine from PBSG, flow = 80 μl/min, laser power = 11.1 dBm, Die C53B1310 first cleaned on 23 sept 2015. Surface functionalized with gram negative antibody on protein G.
The RI of each test solution used was measured at 1310 nm using a refractometer (Model 2010, Metricon, Prism 200-P1). The RI of urine varies depending on the concentration of its constituents. The major constituents of urine are water (H2O), Urea (H2NCONH2), chloride (Cl-), sodium (Na+) and potassium (K+), and it has been shown that the RI of urine varies linearly with the solute weight fraction [20]. Figure 2(a) confirms that urine of lower constituent concentration (Urine0820B, RI = 1.32798) generates a lower output power than urine of higher constituent concentration (Urine0820A, RI = 1.32991). Both urines have a RI that is lower than that of CYTOP (1.3346), so the higher-index solution brings the waveguide closer to symmetry thus reducing its insertion loss and producing a higher output power [21].
We sometimes note in Fig. 2(b) a slope or binding curve at the transition between 2 fluids. With more careful observation, we note that PBSG and urine may interact if we don`t separate their injection with DIH2O or urine in low concentration. It is unclear at this time which urine constituent causes the interaction but always separating the injection of both solutions with DIH2O seems to reduce the effect and provides repeatable absolute power levels and faster settling to a stable response. Even if all fluids were filtered with a 0.2 μm syringe filters, we suspect that Urine0820A still includes large particles causing sudden variations of power. Unlike other experimentation, the test urines Urine0820A and Urine0820B were not centrifuged before filtering.
In Fig. 2(a) we note a 0.2 dB drop in output power for PBSG0715 at 45 min compared to the level at 20 min. We think that proteins contained in urine may have adsorbed on the bare Au surface. This is not seen in Fig. 2(b) probably because the gold surface was already functionalized with protein G and a gram negative antibody.
The urine RI at 1310 nm is close enough to the RI of CYTOP for the waveguide to support a bound LRSPP mode of propagation, thus enabling biosensing applications in urine. Care must be taken in analyzing results when the RI of the sample becomes close to that of water because the detected power is no longer dominated by the LRSPP mode. In such cases, the sensitivity is reduced but the output remains related to biological material on the sensing area due to the propagation of radiative modes. Bulk sensing could be used to monitor the aggregate concentration of constituents in urine after proper calibration.
4. Selective bacteria detection
4.1 Gram negative bacteria detection
In order to demonstrate the selective detection of gram negative bacteria in urine, we prepared test solutions with freshly grown gram positive and gram negative bacteria. The first experiment was conducted in a clean PBSG solution and the second experiment was conducted in a urine solution. A PBS solution with heat killed bacteria was also used.
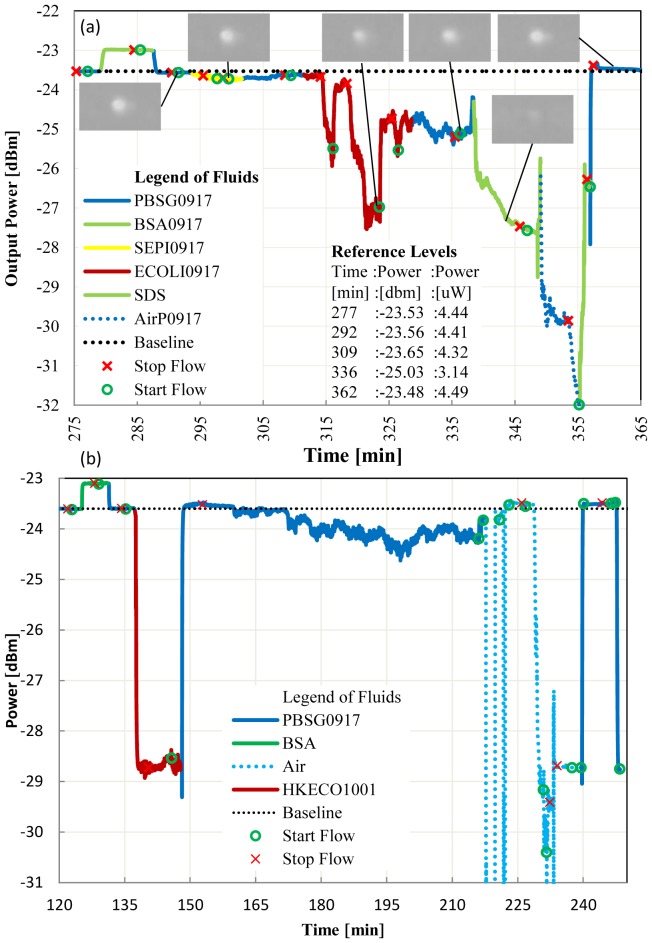
In Fig. 3(a) we demonstrate the selective detection of gram negative bacteria in PBSG. A sensor die was first functionalized with protein G and gram negative antibody as previously described (not shown in the response). A BSA solution was first injected for 5 min to confirm the quality of the Au surface functionalization (no adsorption is observed). The solution containing gram positive bacteria was then injected as a negative control. To ensure contact between bacteria and the functionalized surface, we stop the flow to allow the bacteria to sink onto the biosensing area. Re-starting the flow returns the signal to within 0.09 μW of the baseline signal (i.e., Pt(292) - Pt(309) = 0.09 μW, where Pt stands for power at the observed time t in parentheses). We then inject the solution with gram negative bacteria and stop the flow again. This time the power drops by 1.18 μW (Pt(309) - Pt(336) = 1.18 μW) indicating adsorption of the bacteria by the antibody on the waveguide surface. Defining a positive-to-negative ratio (P/N) as the ratio of the difference of these powers, we obtain P/N = 13.1. A threshold of P/N ≥ 2 could be used as a decision threshold for the selective detection of the bacteria. In addition, we show that by flowing 0.5% SDS solution for 15 min, the antibody-antigen link is broken [22, 23], the bacteria are washed away from the biosensing area, and the baseline is recovered (in this case to within 0.05 μW).
Fig. 3.
(a) Live E.coli detection in PBSG, flow = 20 μl/min, laser power = 14.5 dBm, gram negative antibody surface, bacteria growth time of 4.3 hours in LB Broth. (b) Dead E.coli detection in PBSG, flow = 20 μl/min, laser power = 14.5 dBm, gram negative antibody surface.
In Fig. 3(b) we injected heat killed gram negative bacteria in a PBS solution (HKECO1001). The large signal change between the PBSG0917 (RI = 1.3285) and the HKECO1001 (RI = 1.3235) signal is mainly caused by a bulk change in RI between the solutions. At time 145 min, we note a small perturbation (0.2 dB) in signal as the heat killed bacteria fall onto the biosensing area. A fluidic limitation is observed after 160 min in Fig. 3(b): After approximately 10 min without flow, our stable signal becomes noisy; we speculate that, even if we had been injecting a bacteria-free solution for 5 min, some bacteria from the tubing have back-flowed onto the biosensing area. Regardless of the cause, we avoid stopping the flow for more than 10 min in subsequent experimentation. We observe recovery of the signal to essentially the baseline level at 240 min after removing the air bubbles.
UTI diagnostic by culture provides a count of living colony. With LRSPP waveguide biosensors, it is interesting to note differences in signals when using live or dead bacteria. In Fig. 3(b) with heat killed bacteria little adsorption was observed as indicated by recovery of the signal power after exposing the waveguide to heat killed bacteria. In the following experimental sequence (not shown), heat killed bacteria in urine were injected and adsorbtion was observed by a drop of power of 0.44 μW. We suspect that duration of the stop flow, which was 5 min with PBS and 6 min with urine, is a control parameter that needs to be adjusted based on the concentration of constituents in the fluid because it affects the fluid density. Changes in the density of the fluid will affect the buoyancy of a given bacteria directly affecting the time required for a bacteria to contact the surface. Consideration of the kinetics of the antibody antigen adsorption may also be necessary in selecting the duration of the stop flow. Finally, we also note that with heat killed bacteria the signal is stable once a clean fluid replaces the solution with bacteria, as observed in Fig. 3(b) at 149 min. Incontrast, to live bacteria in Fig. 3(a), at 330 min a noisy signal is observed. Further study of this difference may prove useful to identify live vs dead bacteria.
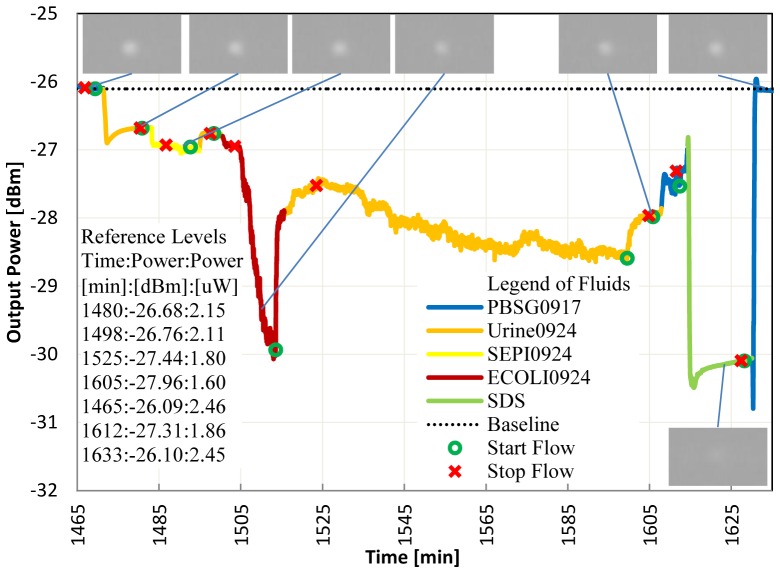
In Fig. 4, the sensor die was also functionalized with protein G and gram negative antibody. We repeat the same experimentation protocol as in Fig. 3(a) but now in urine solutions. We obtain a P/N ratio of [Pt(1480)-Pt(1525)]/[Pt(1480)-Pt(1498)] = 7.5 confirming the selective detection of gram negative bacteria in urine. We also observe in Fig. 4 from 1535 to 1595 min a very noisy signal, which may be caused in part by back-flow of material onto the waveguide surface; but it is not impossible that live bacteria also contribute to the noise. Subsequent cleaning with SDS recovers the signal to the baseline level.
Fig. 4.
Live E.coli detection in urine, flow = 20 μl/min, laser power = 11.1 dBm, gram negative antibody surface, bacteria growth time of 4.3 hrs in LB broth.
4.2 Gram positive bacteria detection
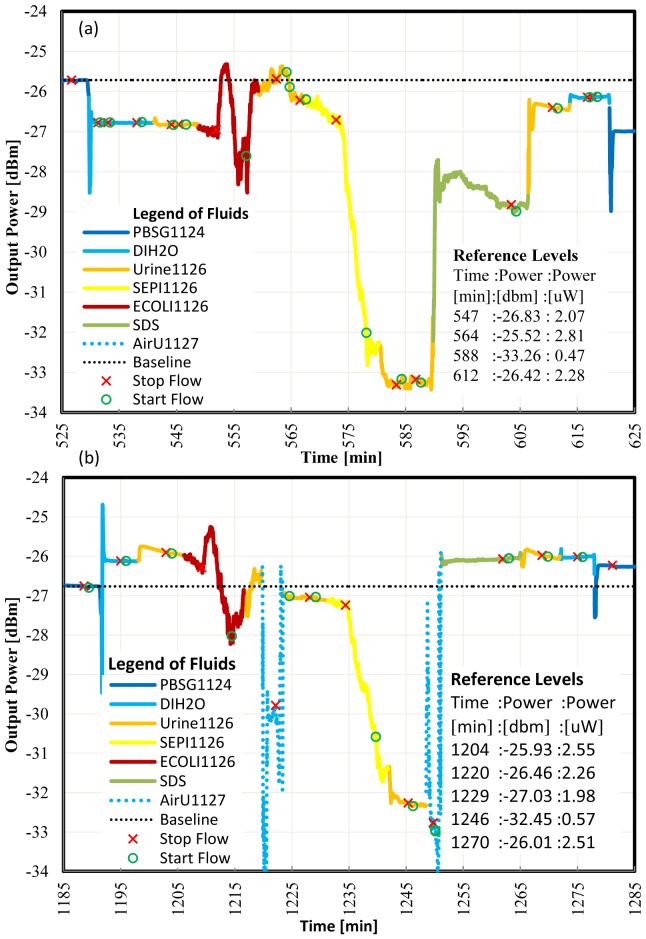
In order to demonstrate the detection of gram positive bacteria in urine, a sensor die was functionalized with protein G then gram positive antibody. We injected solutions with freshly grown gram negative (negative control) then gram positive bacteria in urine. Figure 5(a) reports the results of the first experiment and Fig. 5(b) of 1 repeat of the same experiment on the following day. The high concentration of bacteria in the negative control solution causes large signal fluctuations in Fig. 5(a) during the stop flow. The signal then stabilizes at 558 min after re-starting the flow. We note that most of the signal power is recovered. The small change in power observed before and after the negative control is attributed to nonspecific binding. The gram positive bacteria solution causes a large signal change with no recovery when we re-start the flow at 578 min. Similar observations can be made relative to the repeated experiment in Fig. 5(b), where additionally the inadvertent injection of air bubbles had to be managed during the experiment. Calculating the P/N ratio for both experiments, we obtain P/N = [Pt(564)-Pt(588)]/[Pt(564)-Pt(547)] = 3.1 for Fig. 5(a) and P/N = [Pt(1229)-Pt(1246)]/[Pt(1204)- Pt(1220)] = 4.8 for Fig. 5(b). This confirms the selectivity of gram positive bacteria in urine.
Fig. 5.
(a) Live S.epi detection in urine, flow = 20 μl/min, laser power = 14.5 dBm, gram positive antibody surface. (b) Repeat of live S.epi detection in urine, flow = 20 μl/min, laser power = 14.5 dBm, gram positive antibody surface.
In Fig. 5(b) at 1270 and 1275 min, the same output power is measured for Urine1126 and DIH2O. From the images of the outputs on the IR camera (not shown), we noted a visible LRSPP mode for Urine1126 which was not present for DIH2O. In both images significant background radiation was present.
4.3 Gram negative bacteria detection: low bacterial concentration with contamination
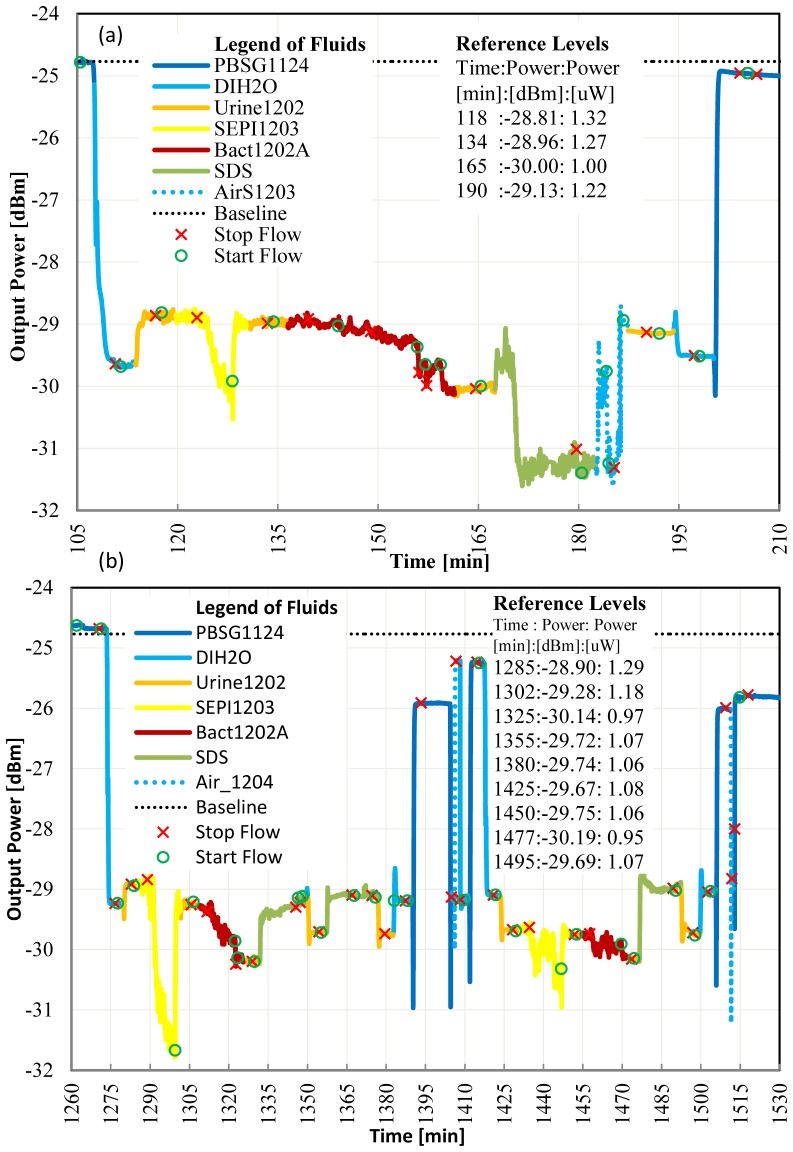
In the experiments reported in Fig. 6, the device was functionalized with a gram negative antibody, and the concentration of gram positive bacteria in our negative control solution was 1000☓ larger than the concentration of the target gram negative bacteria in our positive solution. Despite this, and despite the low RI of the urine used (RI = 1.32276) which is below the RI of PBSG (RI = 1.32351), we still obtain P/N = [Pt(134)-Pt(165)]/[Pt(118)-Pt(134)] = 5.4 for the first experiment (Fig. 6(a)), and P/N = [Pt(1305)-Pt(1325)]/[Pt(1305)-Pt(1285)] = 1.9 and P/N = [Pt(1450)-Pt(1477)]/[Pt(1425)-Pt(1450)] = 5.5 for the first and second repeats, respectively (Fig. 6(b)). After the experiments, a plate culture of the target gram negative bacteria (Bact1202A) revealed the presence of contaminating bacteria with lower concentration then the target.
Fig. 6.
a) Live gram negative bacteria detection with contamination in urine, flow = 20 μl/min, laser power = 14.5 dBm, gram negative antibody surface. b) Repeat of live gram negative bacteria detection in the presence of contamination in urine, flow = 20 μl/min, laser power = 14.5 dBm, gram negative antibody surface.
We note that in the first repeat (Fig. 6(b)), recovery after SDS flow was good but not excellent, as a drop of 0.1μW at 190 min relative to 118 min, and a drop of 0.27μW at 1355 min relative to 1285 min, is observed. This suggests that more nonspecific binding occurred during the second repeat. In our protocol, the die surface was immersed in PBSG1124 overnight unlike the test repeat reported in Fig. 5 where the die surface was immersed in SDS overnight. A repeat of the SDS clean at min 1360 of Fig. 6(b) didn’t improve the recovery of the signal power. To better clean the surface, we intentionally pushed air into the system for a few seconds improving the signal amplitude slightly (Fig. 6(b), Pt(1405) and Pt(1514)). We speculate that the surface tension of a fluid following air is effective in removing some non-specifically bound material from the biosensing area. In the second repeat, the P/N ratio improved compared to the first repeat mainly due to very little non-specific binding (Pt(1425) - Pt(1450) = 0.02 μW).
4.4: Selective bacteria detection: summary of results
A summary of the P/N ratios demonstrating selective detection of bacteria is collected in Table 1. All of our experiments, except for the one listed in the penultimate row, produced a P/N ratio greater than 2.
Table 1. Summary of P/N ratio demonstrating selective detection of bacteria.
| Experimental Figure; Sequence | Test Fluid | Negative Control | Detected Bacteria | P/N | ||
|---|---|---|---|---|---|---|
| Label | Label | Growth time [hrs] | Label | Growth time [hrs] | ||
| Figure 3(a);1 | PBSG0917 | SEPI0917 | 4.3 | ECOLI0917 | 4.3 | 13.1 |
| Figure 4;1 | Urine0924 | SEPI0924 | 4.3 | ECOLI0924 | 4.3 | 7.5 |
| Figure 5(a);1 | Urine1126 | ECOLI1126 | 7.0 | SEPI1126 | 18 | 3.1 |
| Figure 5(b);2 | Urine1126 | ECOLI1126 | 7.0 | SEPI1126 | 18 | 4.8 |
| Figure 6(a);1 | Urine1202 | SEPI1203 | 17 | Bact1202A | 5.0 | 5.4 |
| Figure 6(b);2 | Urine1202 | SEPI1203 | 17 | Bact1202A | 5.0 | 1.9 |
| Figure 6(b);3 | Urine1202 | SEPI1203 | 17 | Bact1202A | 5.0 | 5.5 |
After completing the detection experiments, samples of test fluids with bacteria were plated using Fischer Scientific Trypticase Soy Agar plates (B21185). Using a dilution and plate count technique, the concentration of bacteria was estimated and is reported in Table 2. From these bacterial concentration measurements, we can approximate the sensitivity of the LRSPP waveguide biosensor. In our experimentation, the SEPI0917 test solution had the lowest bacterial concentration. This solution was grown for 4.3 hrs and caused a signal power change of 0.7μW when injected and flow stopped from 295 to 298 min of Fig. 3(a). From the SEPI1117 and SEPI1126 concentration measurements of Table 2, we estimate that 24 bacterial generations are produced in 12 hrs (or 30 min per generation). We can therefore extrapolate the concentration at 4.3 hrs of growth to 3☓104 CFU/ml. Thus, this biosensor is capable of detecting bacterial concentrations below 1☓105 CFU/ml, which is the internationally recognized threshold for the diagnostic of UTI. Further optimization of the test protocol and sensor design is required to estimate selectivity and sensitivity of this diagnostic approach using infected patient urine.
Table 2. Count of bacteria concentration.
| Label | Bacteria | Colony count | CFU / ml | ||
|---|---|---|---|---|---|
| Gram | Growth time [hrs] | Dilution | Plate count | ||
| SEPI1117 | positive | 6 | 1:1000 | 232 | 2☓105 |
| SEPI1126 | positive | 18 | 1:10000000000 | 300 | 3☓1012 |
| Bact1202A | negative | 5 | 1:10000 | 210 | 2☓106 |
| EColi1117 | negative | 6 | 1:100000000 | 171 | 2☓1010 |
5. Conclusion
Our experiments demonstrate that LRSPP waveguides can selectively detect gram negative or gram positive bacteria in human urine. A first approximation of the sensitivity of the biosensor indicates that it is relevant to the diagnostic of UTI with a detection threshold less than 105 CFU/ml. In addition, the direct use of human urine in the detector greatly simplifies its future use at the point-of-care. We also expect that the test time after sample urine collection from a patient would be minutes considering that ready-to-test dies and their functionalization can be prepared ahead of time.
A clinical urine collection protocol to optimize specificity and sensitivity of a UTI diagnostic test using this technology seems feasible. Currently, the collection of urine one hour after drinking one liter of water helped to demonstrate the robustness of the biosensor with a low concentration of urine constituents, but this could also reduce the quantity of bacteria in the sample of an infected patient. Mid-stream urine is normally collected for UTI testing to reduce the risk of contamination. In Fig. 6 trials, a P/N ratio greater than 2 was observed for all, except in 1 case (P/N = 1.9). The experiments reported in Fig. 6 are particularly promising in that a P/N ratio of 5 is obtained even when 2 types of bacteria are present in the test solution and a negative control at 1000☓ greater concentration was used. It is unknown whether large urine constituents such as leukocytes, fungi or blood cells will interfere with the detection, but considering that they typically flow away from the biosensing zone, and that control over the test protocol can be exercised, we are hopeful that this will not be a serious issue.
Acknowledgments
We are grateful to the Ontario Centres of Excellence (OCE) for funding this work under project number 21107. We are also grateful to Canadian Blood Services (Sandra Ramirez, sandra.ramirez@blood.ca) for donating two bacteria strains: Escherichia coli (E.coli) XL1 Blue and Staphylococcus epidermidis (S.epi) ATCC 12228.
Appendix A.
Table 3. List of Fluids; labeling: AAAAmmdd where ¨mm¨ is the month and ¨dd¨ is the day of creation.
| Fluid label | Part Number | Origin | Description | Refr. Index |
|---|---|---|---|---|
| Bact1202A | XL1 blue with contamination | CBS | E.coli bacteria grown for 5 hrs and transferred to Urine1202. Note that the culture was contaminated before incubation with an unknown bacteria measured to have a lower concentration then E.coli and probably a gram positive species. | 1.32276 |
| BSA0917 | A0281-250 mg | S-A | Albumin from bovine serum (100 μg/ml). 2 mg of bovine serum albumin was dissolved in 2 ml of PBS and stored in one vial of 2 ml (1 mg/ml) at 4° C. On the day of use, 100 μl from the BSA vial is diluted in 900 μl of PBSG0917 resulting in a 100 μg/ml solution for use in the experiments. | N/A |
| BSA1202 | A0281-250 mg | S-A | Albumin from Bovine Serum (100 μg/ml). This batch mixed from 100 μl of PBS with 900 μl of PBSG1124. Same protocol as BSA0917. | N/A |
| DIH2O | D11931 | Barnstead | Ultra pure water, 18.2 MΩ-cm, < 1CFU/ml, < 1μM/ml (theoretical RI = 1.3206) | 1.31977 |
| ECOLI0917 | XL1 blue | CBS | E.coli bacteria grown for 4.3 hrs then transferred to PBSG0917. | 1.32854 |
| ECOLI0924 | XL1 blue | CBS | E.coli bacteria grown for 4.3 hrs then transferred to Urine0924. | N/A |
| ECOLI1117 | XL1 blue | CBS | E.coli bacteria grown for 6 hrs, then transferred to Urine1120 and concentration measured with plate culture. | N/A |
| ECOLI1126 | XL1 blue | CBS | E.coli bacteria grown for 7 hrs then transferred to Urine1126. | N/A |
| GNeg | AB41202 | ABCAM | 50 μg/ml of gram negative antibody in solution of PBSG0715. 100 μl of gram negative antibody was diluted in 3900 μl of PBSG0715 and stored in 10 vials of 0.4 ml (50 μg/ml) at 4° C. | N/A |
| GPos | AB20344 | ABCAM | 50 μg/ml of gram positive antibody in solution of PBSG0917. 0.5 ml of gram positive antibodies was diluted in 0.5 ml of PBS and stored in 2 vial of 0.5 ml (50 μg/ml) at 4° C. | N/A |
| GProt0715 | P4689-1MG | S-A | 1 mg of protein G (immunoglobulin-binding protein expressed in group C and G Streptococcal bacteria) was dissolved in 4 ml of PBSG0715 solution and stored in 4 vials of 1 ml (0.25 mg/ml) by freezing at −20° C. On the day of use, a protein G vial was thawed and 200 μL diluted in 800 μL of filtered PBSG solution resulting in a 50 μg/ml solution used to functionalize the bare gold surface of a waveguide with a monolayer of protein G. | N/A |
| HKECOL1001 | XL1 blue | CBS | E.coli bacteria grown for 7 hrs in LB broth, transferred to PBS solution, then heat killed in oven at 80° C for 60 min. | N/A |
| HKECOL1008 | XL1 blue | CBS | HKECOL1001 bacteria transferred to Urine1008 by spinning at 3000 RPM for 7 min. | N/A |
| LB Broth | L3022 | S-A | Mix one pouch in 500 ml of DIH2O. | N/A |
| PBS | P-5368 | S-A | Mix one pouch in 1L DIH2O ml (theoretical RI = 1.3329). | 1.32351 |
| PBSG0715 | 49767-250 ml | S-A | 66.66 g of glycerol in 500 ml of PBS. | 1.33425 |
| PBSG0917 | 49767-250 ml | S-A | 33.33 g of glycerol in 500 ml of PBS. | 1.32854 |
| PBSG1124 | 49767-250 ml | S-A | PBSG0917 sterilized in a microwave oven for 20 min. Final concentration: approximately 10 g/100 ml. | 1.33152 |
| SDS | 71725-50G | S-A | 5 g of sodium dodecyl sulfate in 1L of DIH2O. | N/A |
| SEPI0917 | ATCC 12228 | CBS | S.epi bacteria grown for 4.3 hrs then transferred to PBSG0917. | 1.32854 |
| SEPI0924 | ATCC 12228 | CBS | S.epi bacteria grown for 4.3 hrs then transferred to Urine0924. | N/A |
| SEPI1117 | ATCC 12228 | CBS | S.epi bacteria grown for 6 hrs then concentration measured with plate culture. | N/A |
| SEPI1126 | ATCC 12228 | CBS | S.epi bacteria grown for 18 hrs then transferred to Urine1126. | N/A |
| SEPI1203 | ATCC 12228 | CBS | S.epi bacteria grown for 17 hrs then transferred to Urine1202. | 1.32276 |
| Urine0820A | Human | anon. | First urine of the day collected on 20 Aug. 2014. Donor collected urine in the morning after fasting for 12 hrs. | 1.32991 |
| Urine0820B | Human | anon. | 2nd urine of the day collected on 20 Aug. 2014. Urine collected after drinking 1L of water and waiting one hour. | 1.32798 |
| Urine0924 | Human | anon. | 2nd urine of the day collected on 24 Sep. 2014 (as above). | N/A |
| Urine1126 | Human | anon. | 2nd urine of the day collected on 26 Nov. 2014 (as above). | N/A |
| Urine1202 | Human | anon. | 2nd urine of the day collected on 2 Dec. 2014 (as above). | 1.32276 |
References and links
- 1.Schmiemann G., Kniehl E., Gebhardt K., Matejczyk M. M., Hummers-Pradier E., “The Diagnosis of Urinary Tract Infection: A Systematic Review,” Dtsch. Arztebl. Int. 107(21), 361–367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simerville J. A., Maxted W. C., Pahira J. J., “Urinalysis: a comprehensive review,” Am. Fam. Physician 71(6), 1153–1162 (2005). [PubMed] [Google Scholar]
- 3.Broeren M. A., Bahçeci S., Vader H. L., Arents N. L., “Screening for urinary tract infection with the Sysmex UF-1000i urine flow cytometer,” J. Clin. Microbiol. 49(3), 1025–1029 (2011). 10.1128/JCM.01669-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marschal M., Wienke M., Hoering S., Autenrieth I. B., Frick J.-S., “Evaluation of 3 different rapid automated systems for diagnosis of urinary tract infections,” Diagn. Microbiol. Infect. Dis. 72(2), 125–130 (2012). 10.1016/j.diagmicrobio.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Zhang Y., Xu D., Shao W., Lu Y., “Evaluation of the Sysmex UF-1000i for the Diagnosis of Urinary Tract Infection,” Am. J. Clin. Pathol. 133(4), 577–582 (2010). 10.1309/AJCP1GT2JXOCQBCZ [DOI] [PubMed] [Google Scholar]
- 6.Van Dilla M. A., Langlois R. G., Pinkel D., Yajko D., Hadley W. K., “Bacterial characterization by flow cytometry,” Science 220(4597), 620–622 (1983). 10.1126/science.6188215 [DOI] [PubMed] [Google Scholar]
- 7.Liedberg B., Nylander C., Lundstrom I., “Surface plasmon resonance for gas detection and biosensing,” Sens. Act. 4, 299–304 (1983). 10.1016/0250-6874(83)85036-7 [DOI] [Google Scholar]
- 8.Vala M., Etheridge S., Roach J., Homola J., “Long-range surface plasmons for sensitive detection of bacterial analytes,” Sens. Act. B 139(1), 59–63 (2009). 10.1016/j.snb.2008.08.029 [DOI] [Google Scholar]
- 9.Chabot V., Miron Y., Grandbois M., Charette P. G., “Long range surface plasmon resonance for increased sensitivity in living cell biosensing through greater probing depth,” Sens. Act. B 174, 94–101 (2012). 10.1016/j.snb.2012.08.028 [DOI] [Google Scholar]
- 10.Berini P., “Long-range surface plasmon polaritons,” Adv. Opt. Photonics 1, 484–588 (2009). [Google Scholar]
- 11.Torun O., Hakkı Boyacı I., Temür E., Tamer U., “Comparison of sensing strategies in SPR biosensor for rapid and sensitive enumeration of bacteria,” Biosens. Bioelectron. 37(1), 53–60 (2012). 10.1016/j.bios.2012.04.034 [DOI] [PubMed] [Google Scholar]
- 12.Fratamico P. M., Strobaugh T. R., Medina M. B., Gehring A. G., “Detection of Escherichia coli O157:H7 using a surface plasmon resonance biosensor,” Biotechnol. Tech. 12(7), 571–576 (1998). 10.1023/A:1008872002336 [DOI] [Google Scholar]
- 13.Treviño J., Calle A., Rodríguez-Frade J. M., Mellado M., Lechuga L. M., “Single- and multi-analyte determination of gonadotropic hormones in urine by Surface Plasmon Resonance immunoassay,” Anal. Chim. Acta 647(2), 202–209 (2009). 10.1016/j.aca.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 14.Wong W. R., Krupin O., Sekaran S. D., Mahamd Adikan F. R., Berini P., “Serological Diagnosis of Dengue Infection in Blood Plasma Using Long-Range Surface Plasmon Waveguides,” Anal. Chem. 86(3), 1735–1743 (2014). 10.1021/ac403539k [DOI] [PubMed] [Google Scholar]
- 15.Chiu C., Lisicka-Skrzek E., Tait R. N., Berini P., “Fabrication of Surface Plasmon Waveguides and Devices in Cytop with Integrated Microfluidic Channels,” J. Vac. Sci. Technol. B 28(4), 729–735 (2010). 10.1116/1.3449187 [DOI] [Google Scholar]
- 16.Hassan S., Khodami M., Tait R. N., Berini P., “Fabrication of long-range surface plasmon-polariton Bragg gratings with microfluidic channels in Cytop claddings,” Microelectron. Eng. 135, 38–44 (2015). 10.1016/j.mee.2015.03.001 [DOI] [Google Scholar]
- 17.Krupin O., Asiri H., Wang C., Tait R. N., Berini P., “Biosensing using straight long-range surface plasmon waveguides,” Opt. Express 21(1), 698–709 (2013). 10.1364/OE.21.000698 [DOI] [PubMed] [Google Scholar]
- 18.ethics@uottawa.ca, Bureau d'éthique et d'intégrité à la recherche, 75 Laurier Avenue East, Université d`Ottawa, K1N 6N5, numéro dossier H06–14–01, 23 juin 2014 (personal communication, 2014).
- 19.Breukelaar I., Charbonneau R., Berini P., “Long-range surface plasmon-polariton mode cutoff and radiation in embedded strip waveguides,” J. Appl. Phys. 100(4), 043104 (2006). 10.1063/1.2244479 [DOI] [PubMed] [Google Scholar]
- 20.D. F. Putnam “Composition and concentrative properties of human urine,” Prepared by McDonnell Douglas Astronautics company – western division, Huntington Beach, Calif. 92647, Contractor Report NASA CR-1802, (1971).
- 21.Wong W. R., Krupin O., Adikan F. R. M., Berini P., “Optimization of long-range surface plasmon waveguides for attenuation-based biosensing,” J. Lightwave Technol. in press.
- 22.Su X., Zhang J., “Comparison of surface plasmon resonance spectroscopy and quartz crystal microbalance for human IgE quantification,” Sens. Act. B 100(3), 309–314 (2004). 10.1016/j.snb.2004.01.020 [DOI] [Google Scholar]
- 23.Dutra R. F., Mendes R. K., Lins da Silva V., Kubota L. T., “Surface plasmon resonance immunosensor for human cardiac troponin T based on self-assembled monolayer,” J. Pharm. Biomed. Anal. 43(5), 1744–1750 (2007). 10.1016/j.jpba.2006.12.013 [DOI] [PubMed] [Google Scholar]