Abstract
Specialized immune cells that infiltrate the tumor microenvironment regulate the growth and survival of neoplasia. Malignant cells must elude or subvert anti-tumor immune responses in order to survive and flourish. Tumors take advantage of a number of different mechanisms of immune “escape,” including the recruitment of tolerogenic DC, immunosuppressive regulatory T cells (Tregs), and myeloid-derived suppressor cells (MDSC) that inhibit cytotoxic anti-tumor responses. Conversely, anti-tumor effector immune cells can slow the growth and expansion of malignancies: immunostimulatory dendritic cells, natural killer cells which harbor innate anti-tumor immunity, and cytotoxic T cells all can participate in tumor suppression. The balance between pro- and anti-tumor leukocytes ultimately determines the behavior and fate of transformed cells; a multitude of human clinical studies have borne this out. Thus, detailed analysis of leukocyte subsets within the tumor microenvironment has become increasingly important. Here, we describe a method for analyzing infiltrating leukocyte subsets present in the tumor microenvironment in a mouse tumor model. Mouse B16 melanoma tumor cells were inoculated subcutaneously in C57BL/6 mice. At a specified time, tumors and surrounding skin were resected en bloc and processed into single cell suspensions, which were then stained for multi-color flow cytometry. Using a variety of leukocyte subset markers, we were able to compare the relative percentages of infiltrating leukocyte subsets between control and chemerin-expressing tumors. Investigators may use such a tool to study the immune presence in the tumor microenvironment and when combined with traditional caliper size measurements of tumor growth, will potentially allow them to elucidate the impact of changes in immune composition on tumor growth. Such a technique can be applied to any tumor model in which the tumor and its microenvironment can be resected and processed.
Keywords: Medicine, Issue 98, Chemerin, tumor microenvironment, leukocyte subsets, NK cells, chemoattractant, melanoma, leukocyte, migration, immunophenotype
Introduction
The balance between tumor growth and promotion and regression is, in part, dependent on the balance of pro- and anti- tumor infiltrating leukocytes present in the microenvironment1,2. In order to study the tumor microenvironment (TME) and specifically identify the infiltrating leukocyte subpopulations, we developed a method for evaluation of subcutaneous tumors in a murine tumor model. The importance of studying the tumor microenvironment is well known and supported in the literature. Numerous studies have shown that the balance of pro- and anti-tumor infiltrating immune cells can impact the outcomes of tumor growth, not only in mouse but also human studies (reviewed in3,4). For example, Curiel et al. showed that worsened clinical outcomes in ovarian cancer patients were correlated with the presence of increasing percentages of tumor-infiltrating regulatory CD4+ T cells (Tregs)5. Our own work also showed the effect of a novel leukocyte chemoattractant on the ratio of leukocyte subsets in a mouse melanoma model6, which also correlated with decreased tumor growth. Thus, the detailed analyses of the leukocyte subsets within a tumor is now more broadly recognized and increasingly important.
There are many ways to evaluate the tumor microenvironment for infiltrating leukocytes; for example groups have engineered transgenic mice to express various fluorescent proteins in order to image the TME7, classical immunohistochemistry and immunofluorescence of preserved sections8, including various imaging modalities such as MRI, PET, confocal microscopy9-11 - some with the ability to monitor intravitally10,12. These can be used with various molecular imaging agents, such as nanoparticles13 or novel contrast agents14 that label immune cells. Our method is a flow cytometry-based approach and has several advantages. First, the entire tumor microenvironment can be sampled; at the time of analysis, the entire subcutaneous tumor and surrounding periphery is surgically resected for processing. This eliminates any potentially sampling bias within a single tumor and gives a more “global” analysis of the tumor as a whole. Secondly, using multicolor flow cytometry to analyze the leukocyte subsets allows us to more specifically gauge the phenotype of infiltrating leukocytes. Depending on the number of colors used, very specific subsets can be identified. This is important as there are several leukocyte subsets within a particular cell type - or even in a general subtype classification – that have disparate functions that are potentially significant in determining the fate of the tumor. For example, plasmacytoid dendritic cells (pDC) have been implicated in anti-tumor immunity15. However, the CCR9+ subset of pDCs has been shown to be tolerogenic16, and shifting the balance of such a subset may have an impact on tumor growth.
Our method is appropriate for subcutaneous or other tumors that can be resected en bloc. In our hands, tumors were uniformly resected at the time of euthanasia. However, it is conceivable, as has been done in some studies, that a subcutaneous tumor could be fully resected with closure of the surrounding skin in a survival surgery17, thus allowing additional evaluation of the animal. The analysis is then performed on the resected tumor. Thus, the results represent a single timepoint in the tumor’s development. While this allows a detailed look into the microenvironment, it is also a static picture of what is no doubt a dynamic process. However, isolated leukocytes (e.g. via magnetic separation or density gradient) can then be analyzed separately from the tumor epithelia and stroma, or used in other, functional assays to further define their phenotype, as has been previously described18. This method, then, would be useful for any investigators interested in understanding the composition of the leukocytes within the tumor microenvironment at a given time point, whether in the setting of the natural disease course, or after a specific therapeutic perturbation. Although not done by us, variations of this procedure could also potentially be used to analyze specific portions of a tumor in isolation. For example, given the size of the tumor, the peripheral zone(s) could be dissected away from the central, possibly necrotic core of the tumor to give the researcher a more spatially segregated view of the tumor microenvironment.
In the burgeoning field of tumor immunology, there will no doubt be an exponentially growing number of studies evaluating novel immunomodulatory agents in murine tumor models. Several reports have highlighted the differences in specific leukocyte function within the tumor versus the peripheral environment. For example, Shafer-Weaver et al. showed in a mouse model that antigen-specific T effector CD8+ cells, while active in the periphery, were transformed into CD8+ T suppressor cells once they trafficked into the tumor microenvironment19. This was in part due to TGFβ, but other factors are likely involved as well. Thus, evaluating leukocyte subsets – numbers and ratios, as well as functional status – within the tumor itself will give a more accurate representation of the effect of a particular immunomodulation on tumor fate.
Our technique allows detailed analysis of the tumor and provides the researcher an opportunity to more closely identify changes in leukocyte populations than previous approaches.
Protocol
NOTE: All animal experiments were conducted in accordance with approved Stanford, Palo Alto VA HCS, and National Institutes of Health Institutional AnimalCare and Use Committee guidelines.
1. Preparing for Sample Collection and Processing (Time Required: ~10-15 min)
- Inoculate murine B16F0 melanoma cells (0.5-1 x 106) subcutaneously at or near the midline of the abdomen in female C57BL/6 mice as described previously6. Alternatively, inoculate tumor cells at additional anatomical sites (e.g. flank, mammary fat pad, etc). NOTE: Any number of tumors (implanted or spontaneous) can be used and evaluated using this protocol. For example, other commonly used syngeneic tumors in C57BL/6 include the colon line MC38, Lewis Lung Carcinoma (LLC), and the prostate TRAMP-C2 line. While human xenografts inoculated into immunodeficient mice can also be evaluated in this way, it should be noted that the profiles of the tumor-infiltrating leukocytes will necessarily be altered in accordance with the immunodeficient state of the host mouse.
- Use complete media (e.g. RPMI containing 10% FBS and additives) or another appropriate protein-containing buffer (e.g. HBSS or PBS +1% FBS or 1% BCS). Chill media or buffer to 4 °C prior to euthanization.
- Set up collection tubes and filters for resected tumors; place these on ice.
- Use one 50 ml conical tube per tumor to be resected. Insert a 40-70 µm cell suspension filter in each one. Alternatively, use 15 ml conical tubes with metal filter mesh. Use a 5 ml syringe plunger or thick pen to create a cup-shaped filter at the top of the 15 ml tube.
Prepare instruments for resection and tumor processing; use surgical forceps and fine scissors to resect and process tumors. Use 70% ethanol to clean instruments between resections/tumor samples. For larger number of samples (e.g. >10), we recommend simultaneous processing of samples by multiple researchers to avoid significant differences in processing time between tumor samples.
2. Tumor Resection (Time Required: ~5 min/tumor)
Euthanize B16 tumor-bearing mice individually using carbon dioxide or other approved method.
Secure the mouse on a surgical stage. Spray the tumor area with 70% ethanol and wipe excess off; this helps prevent spreading of any hair present. For non-nude mice, shave the skin area around the tumor prior to resection as needed.
- Using forceps and scissors, resect the subcutaneous tumor en bloc, including overlying and surrounding skin. The amount of “margin” skin taken can be varied, but typically resect 2-3 mm of skin around the tumor.
- Weigh resected tumors (+/- surrounding skin) at this point. Place the resected skin/tumor section in the filter secured within the plastic conical tube.
3. Preparing a Single Cell Tumor Suspension (Time Required: ~5-7 min/tumor)
Use forceps and scissors to mince tumor and overlying/surrounding skin. Mince the sample using scissors until all large sections of tissue are processed into 1-2 mm sections. NOTE: Typically, whole tumors are processed; alternatively, for larger tumors, representative sections of tumors could be minced and processed.
Using a plunger from a disposable 5 or 10 ml syringe, mechanically disaggregate the minced tumor tissue against the secured filter.
Using a pipettor, wash the processed tumor tissue with 5-10 mls of cold media or buffer. This will flush the single cells through the filter.
- Repeat the above two steps several times, ensuring that the total number and volumes of washings are approximately the same for similarly sized tumors.
- After washing the processed tumor tissue, observe small fragments of skin and/or other connective tissue should be left behind in the filter.
- Alternatively, for tumors where mechanical disaggregation alone may not be sufficient, digestion with collagenase and/or other enzymes can be performed prior to the mincing procedure. This will allow for a more thorough single cell suspension. However, it should be noted that such digestion protocols can affect surface antigen expression20, so caution should be taken in interpreting these results.
Wash and pellet the cells by adding media/buffer and centrifuging at 4 °C 500 x g for 5 min.
4. FACS Staining of Single Cell Suspension (Time Required: ~30-60 min)
Resuspend cells in ice cold FACS buffer (PBS + 1% FBS).
Count viable cells using trypan blue and a hemacytometer, or automated cell counter.
Determine total viable cell yield per volume. This data can be used to extrapolate absolute numbers of tumor infiltrating leukocytes in combination with FACS data.
- Determine the number of cells to be stained and analyzed by FACS. This will be influenced by several factors (e.g. tumor histology type, tumor age and size, average percentage of infiltrating leukocytes, etc.). NOTE: In B16 melanoma, we found a low percentage of tumor infiltrating leukocytes (TIL; typically <5%) compared to tumor cells. Thus, at least 1 x 106 total tumor (tumor + infiltrating leukocytes) cells were typically collected via FACS for analysis.
- Using the trypan-stained counting data, calculated volume of cells to be stained for FACS. For example, if TIL typically constitute 5% of total tumor cells (many of the tumor cells will be dead on trypan stain), factor this into the total number of tumor single cell suspension to be stained.
- For less common leukocyte subsets e.g. CCR9+ pDC) use a larger number of total tumor cells (e.g. 2-3 x 106) for FACS staining, given the relative low frequency of these cells in the single cell suspension.
- Stain the tumor single cell suspension for FACS.
- For live/dead discrimination, use a fixable stain or non-fixable stain (e.g. propidium iodide) can be used. If cell are stained without live/dead discrimination, caution must be advised given the potential for non-specific binding of antibodies.
- Stain the cell suspension for specific leukocyte subsets using a standard FACS staining protocol. Block cells to reduce non-specific antibody binding with PBS/FBS containing 1% rat serum and Fc block (anti-CD16/32) for 10 min prior to staining with antigen-specific antibodies. Include the correct isotype control antibodies to ensure staining is specific.
- At this point, after the FACS staining is complete, fix samples with 4% formaldehyde or other similar fixative agent. Store samples at 4 °C in the dark (as to avoid quenching of fluorophores) until time of collection and analysis. NOTE: Storing fixed samples for long periods of time may result in alteration in signal and intensity of the fluorophores21.
- Collect the samples on a flow cytometer. Optimal flow cytometer settings will vary depending on type of instrument and laser/detector setup. NOTE: Additionally, there can be variability between experiments even if using the same machine. Thus, perform appropriate machine setup (e.g. using calibration beads) to ensure appropriate compensation setting prior to FACS collection and analysis of each experiment.
- Analysis of leukoctye subsets
- Using FACS data analysis software, to analyze the data. For a live/dead stain, start by gating out dead cells. Use antibody against a leukocyte specific marker (e.g. anti-CD45) to identify leukocytes within the live cells.
- After this, use various gating schemes to identify specific subsets, depending on the staining protocol. For example, to identify CD4+ T cells, select the SSClo region from the CD45+ population and select the CD19-CD3+ cells; from this CD4+ T cells can be identified22.
- Analyze data and present it in a number of different ways to ensure consistency and identify patterns or trends (i.e. use total numbers, percentages, or ratios to quantify subsets). NOTE: For example, we looked at total live CD45+ cells as a percentage of total tumor cells collected (Figure 1). Additionally, we then calculated the percentage of each leukocyte subset (eg CD4+ T cells) within the tumor and compared these as ratios between the control and test groups (Figure 3).
Representative Results
Our results showed that forced over-expression of chemerin in murine B16 tumors augmented the percentage of tumor infiltrating leukocytes (TILs). Additionally, changes to the relative ratio of leukocyte subsets represented within the tumor microenvironment associated with chemerin expression were identified. Re-print with permission from Pachynski et al.6.
Figure 1 shows that there was a significant increase in total CD45+ cells (TILs) within in the tumors that expressed chemerin compared to controls, as determined by FACS analysis of resected day 17 tumors. Using stained, cryosectioned B16 tumors, the increase in infiltrating CD45+ cells by immunofluorescence of tumors was confirmed (Figure 2).
Further analysis of FACS data shows a relative increase, on average, of NK cells, T cells, and conventional DC (Lin-B220- CD11c+) in the tumors where chemerin was over-expressed compared to the control tumors (Figure 3). There was a relative decrease in the percentages of leukocyte subsets that have a role in suppressing or may suppress immune responses, specifically myeloid-derived suppressor cells (Lin-CD11b+GR1+) and pDC (Lin- B220+CD11cintPDCA1+)16,23.
An increasing number of studies have shown the ratio of leukocyte subsets within the tumor microenvironment to be an important factor, both in terms of tumor growth and clinical outcomes4,24,25. Our FACS data was further analyzed to reveal the number of T cells (total CD3+) and NK cells per tumor cell; these cell types were increasingly represented in the chemerin-expressing tumors compared to controls (Figure 4).
A direct comparison of effector (i.e. NK and T cells) and suppressor cell (i.e. MDSC and pDC) populations within the tumor microenvironment for both tumor groups was then performed. Figure 5 shows significant increases in the ratios of effector to suppressor cell populations within the chemerin-expressing tumors compared to controls.
Overall, the representative results show the effects of chemerin expression within the tumor microenvironment on leukocyte subset populations, and the favorable skewing of these populations and ratios in a manner consistent with the clinical benefit seen.
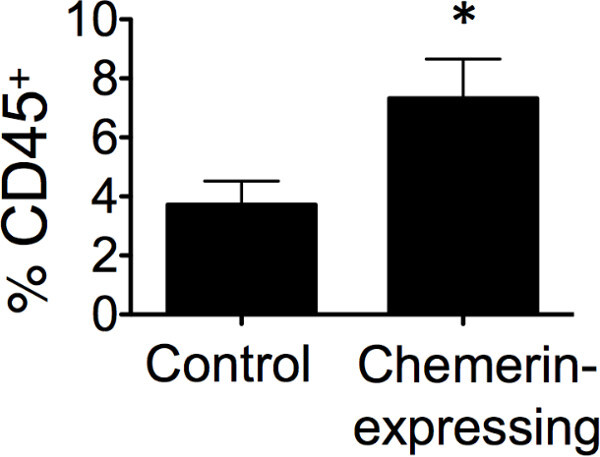 Figure 1: Increased tumor infiltrating leukocytes in chemerin-expressing tumors. Tumors excised from chemerin-expressing vs control on day 17 were analyzed for CD45+ leukocyte infiltration (% of viable cells) by flow cytometry; *p <0.05 by two-tailed Student’s t-test showing mean ± SEM of with 4 or more mice per group.
Figure 1: Increased tumor infiltrating leukocytes in chemerin-expressing tumors. Tumors excised from chemerin-expressing vs control on day 17 were analyzed for CD45+ leukocyte infiltration (% of viable cells) by flow cytometry; *p <0.05 by two-tailed Student’s t-test showing mean ± SEM of with 4 or more mice per group.
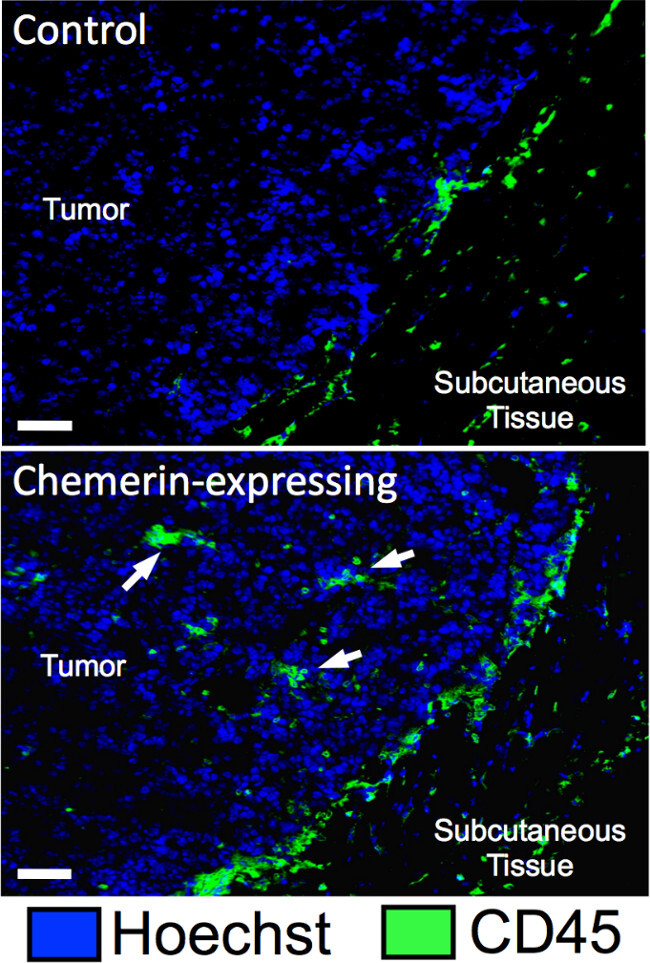 Figure 2: Imaging of B16 melanoma TIL. Immunofluorescence images illustrate CD45+ cell infiltrates (white arrows) in control and chemerin-expressing melanomas excised at day 9; bar represents 25 μm.
Figure 2: Imaging of B16 melanoma TIL. Immunofluorescence images illustrate CD45+ cell infiltrates (white arrows) in control and chemerin-expressing melanomas excised at day 9; bar represents 25 μm.
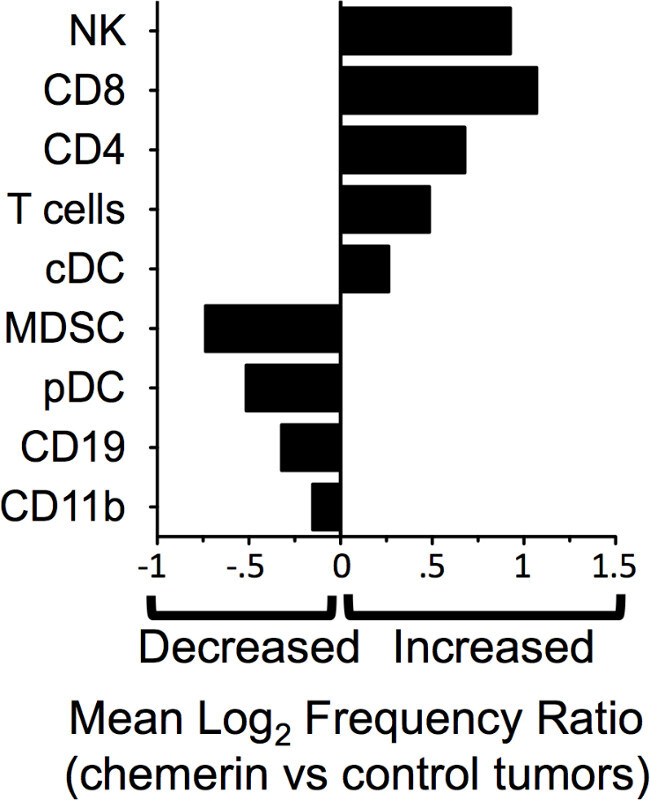 Figure 3: Altered representation of leukocyte subsets in chemerin-expressing tumors. FACS analyses data was converted via log2 ratio of TIL subset frequency in chemerin- expressing vs control tumors for pDC (plasmacytoid DC; Lin- CD11cintB220+PDCA1+), cDC (conventional DC; Lin-CD11chiB220-), CD4 (CD3+CD4+)T cells, CD8 (CD3+CD8+) T cells, total T cells (CD3+CD4+ and CD3+CD8+), NK cells (CD3-NK1.1+), CD11b (Lin-CD11b+) monocyte/macrophages, MDSC (myeloid derived suppressor cells; Lin-CD11b+GR1+), and CD19+ B cells (CD3-CD19+).
Figure 3: Altered representation of leukocyte subsets in chemerin-expressing tumors. FACS analyses data was converted via log2 ratio of TIL subset frequency in chemerin- expressing vs control tumors for pDC (plasmacytoid DC; Lin- CD11cintB220+PDCA1+), cDC (conventional DC; Lin-CD11chiB220-), CD4 (CD3+CD4+)T cells, CD8 (CD3+CD8+) T cells, total T cells (CD3+CD4+ and CD3+CD8+), NK cells (CD3-NK1.1+), CD11b (Lin-CD11b+) monocyte/macrophages, MDSC (myeloid derived suppressor cells; Lin-CD11b+GR1+), and CD19+ B cells (CD3-CD19+).
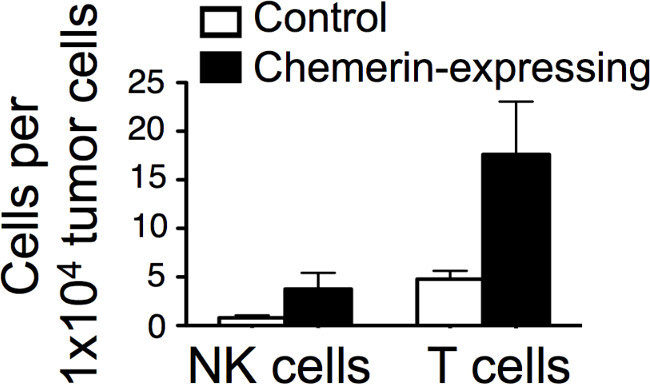 Figure 4: Altered ratios of effector to suppressor leukocytes in B16 tumors. Day 17 B16 melanoma tumors were analyzed by FACS as described. Individual leukocyte subsets were identified by gating. The ratio of NK or total T cells to MDSC or pDC in each tumor type was then calculated.
Figure 4: Altered ratios of effector to suppressor leukocytes in B16 tumors. Day 17 B16 melanoma tumors were analyzed by FACS as described. Individual leukocyte subsets were identified by gating. The ratio of NK or total T cells to MDSC or pDC in each tumor type was then calculated.
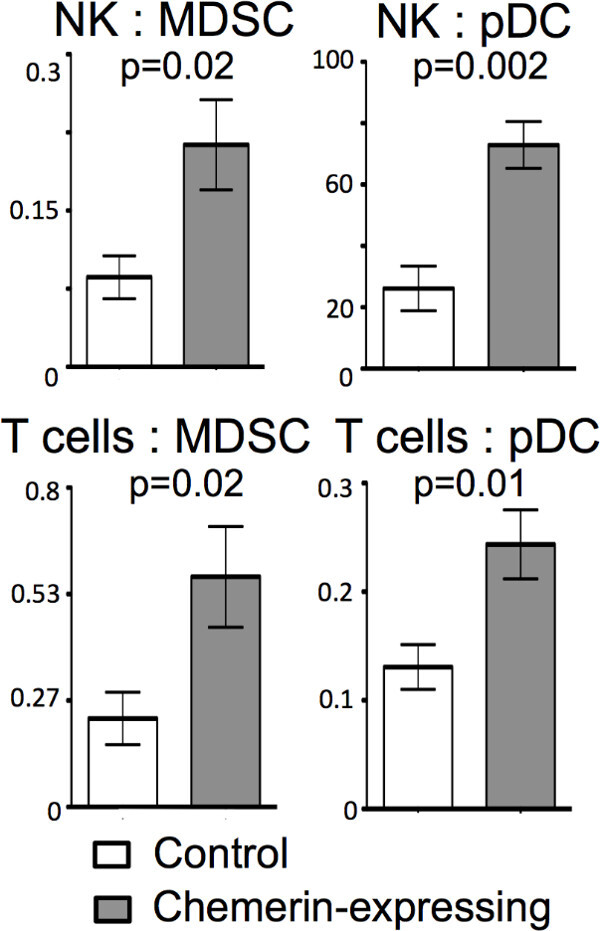 Figure 5: Increased effector cells in chemerin-expressing B16 tumors. Control or chemerin-expressing tumors were analyzed by FACS. Absolute numbers of T and NK cells per 10,000 total tumor cells were calculated using the FACS data. Results are from a representative experiment.
Figure 5: Increased effector cells in chemerin-expressing B16 tumors. Control or chemerin-expressing tumors were analyzed by FACS. Absolute numbers of T and NK cells per 10,000 total tumor cells were calculated using the FACS data. Results are from a representative experiment.
Discussion
Performing detailed analysis of the tumor microenvironment is critical in determining the mechanisms and effects of immunomodulation. With the increasing presence of immunotherapeutics in the human clinical realm, understanding the impact of these agents on the tumor infiltrating leukocytes is becoming requisite in defining their mechanism of action. In humans, there often exist clinical and/or logistic difficulties in obtaining and analyzing tumor tissue for leukocyte subset analysis, and thus analysis of the peripheral immune response is often performed. Performing preclinical studies in animal models allows the researcher to more fully explore the impact of immunomodulatory agents on the immune system within the tumor itself, thus giving additional insight into mechanisms that might affect clinical outcomes.
Our technique allows detailed analysis of tumor-infiltrating leukocytes. Because the entire tumor microenvironment can be sampled, a more global view of the immune presence within the tumor can be assessed. And, ultimately, the balance of pro-tumor suppressor cells and anti-tumor effector cells can have a significant impact on tumor growth and clinical outcomes 4. Thus, this type of analysis is valuable in providing the researcher with detailed information about the specific leukocyte subsets present. This detailed data may be used in combination with other functional data to further elucidate immune mechanisms within the tumor.
The relative simplicity of the protocol allows one to evaluate larger cohorts of tumors in order to minimize the variability within groups. Compared with the more typical and prevalent approach of imaging (either by IHC or immunofluorescence), our technique allows the identification of very specific leukocyte subsets by utilizing multi-color flow cytometry. The ability to sample either the entire tumor (including periphery) or a section of the tumor gives the researcher additional flexibility in terms of sampling and analysis. This also allows the compilation of significantly more data regarding leukocytes than could be obtained by analysis of multiple tissue sections by imaging, thus giving a more accurate picture of the tumor microenvironment.
We utilized this technique on subcutaneous tumors, which are widely used in mouse models given the relative ease of tumor inoculation and measurements. Thus, this protocol would be applicable for likely a majority of tumor models used. However, the technique could also be applied to tumors resected from orthotopic or spontaneous tumor models, albeit with additional steps for tumor resection. And while B16 melanoma did not require enzyme digestion to obtain a single cell suspension, there may be other tumor types where this step is required to obtain a uniform, representative single cell suspension. There are also advantages that imaging of tumor sections affords that cannot be assessed with our technique. For example, more specific data on leukocyte localization within the tumor microenvironment can be obtained through IHC/IF, and this may often be a valuable adjunct to the data provided by FACS analysis.
In conclusion, our technique for evaluation of leukocyte subsets within the tumor microenvironment allows the researcher to perform detailed analyses of the immune presence within a tumor using a relatively simple protocol. It can be applied to a majority of mouse tumor models and can help elucidate valuable information about the leukocyte type, number, and ratios present within the tumor.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by grants R01-CA169354 and R01-047822 from the National Institutes of Health and a Merit Award from the Department of Veterans Affairs (ECB). RKP was supported by NIH T32 CA009287-30A1, an ASCO Young Investigator Award, California Breast Cancer Research Project Fellowship, and an American Cancer Society Mentored Research Scholar Grant; BAZ was supported by NIH grant AI079320. JM was supported by fellowships from NIH T-32 training grant T32-AI07290- 25, T32-AI07290-24 and American Cancer Society post-doctoral fellowship PF-12-052-01-CSM.
References
- Mantovani A, et al. Chemokines in the recruitment and shaping of the leukocyte infiltrate of tumors. Semin Cancer Biol. 2004;14(3):155–160. doi: 10.1016/j.semcancer.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6(10):715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Pachynski RK, et al. The chemoattractant chemerin suppresses melanoma by recruiting natural killer cell antitumor defenses. J Exp Med. 2012;209(8):1427–1435. doi: 10.1084/jem.20112124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RM. Transgenic nude mice ubiquitously expressing fluorescent proteins for color-coded imaging of the tumor microenvironment. Methods Mol Biol. 2014;1194:353–365. doi: 10.1007/978-1-4939-1215-5_20. [DOI] [PubMed] [Google Scholar]
- Mansfield JR. Imaging in cancer immunology:phenotyping immune cell subsets in situ in FFPE tissue sections. MLO Med Lab Obs. 2014;46(6):12–13. [PubMed] [Google Scholar]
- Serres S, O'Brien ER, Sibson NR. Imaging angiogenesis, inflammation, and metastasis in the tumor microenvironment with magnetic resonance imaging. Adv Exp Med Biol. 2014;772:263–283. doi: 10.1007/978-1-4614-5915-6_12. [DOI] [PubMed] [Google Scholar]
- Schietinger A, et al. Longitudinal confocal microscopy imaging of solid tumor destruction following adoptive T cell transfer. Oncoimmunology. 2013;2(11):e26677. doi: 10.4161/onci.26677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AS, Radu CG, Ribas APET. imaging of the immune system: immune monitoring at the whole body level. Q J Nucl Med Mol Imaging. 2010;54(3):281–290. [PubMed] [Google Scholar]
- Kilarski WW, et al. Intravital immunofluorescence for visualizing the microcirculatory and immune microenvironments in the mouse ear dermis. PLoS One. 2013;8(2):e57135. doi: 10.1371/journal.pone.0057135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibollahi P, et al. Fluorescent nanoparticle imaging allows noninvasive evaluation of immune cell modulation in esophageal dysplasia. Mol Imaging. 2014;13(3):1–11. [PMC free article] [PubMed] [Google Scholar]
- Balducci A, et al. A novel probe for the non-invasive detection of tumor-associated inflammation. Oncoimmunology. 2013;2(2):e23034. doi: 10.4161/onci.23034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, et al. Plasmacytoid dendritic cells induce NK cell-dependent, tumor antigen-specific T cell cross-priming and tumor regression in mice. J Clin Invest. 2008;118(3):1165–1175. doi: 10.1172/JCI33583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadeiba H, et al. CCR9 expression defines tolerogenic plasmacytoid dendritic cells able to suppress acute graft-versus-host disease. Nat Immunol. 2008;9(11):1253–1260. doi: 10.1038/ni.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M, et al. A BALB/c murine lung alveolar carcinoma used to establish a surgical spontaneous metastasis model. Clin Exp Metastasis. 2004;21(4):363–369. doi: 10.1023/b:clin.0000046176.33867.c5. [DOI] [PubMed] [Google Scholar]
- Watkins SK, Zhu Z, Watkins KE, Hurwitz AA. Isolation of immune cells from primary tumors. J Vis Exp. 2012. p. e3952. [DOI] [PMC free article] [PubMed] [Retracted]
- Shafer-Weaver KA, et al. Cutting Edge: Tumor-specific CD8+ T cells infiltrating prostatic tumors are induced to become suppressor cells. J Immunol. 2009;183(8):4848–4852. doi: 10.4049/jimmunol.0900848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear AW, Kumar A, Dow S, Ryan EP. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J Immunol Methods. 2014;405:97–108. doi: 10.1016/j.jim.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Stewart JC, Villasmil ML, Frampton MW. Changes in fluorescence intensity of selected leukocyte surface markers following fixation. Cytometry A. 2007;71(6):379–385. doi: 10.1002/cyto.a.20392. [DOI] [PubMed] [Google Scholar]
- Hackstein H, et al. Heterogeneity of respiratory dendritic cell subsets and lymphocyte populations in inbred mouse strains. Respir Res. 2012;13:94. doi: 10.1186/1465-9921-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S. Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother. 2010;59(10):1593–1600. doi: 10.1007/s00262-010-0855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107(9):4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]


