Abstract
The important anticancer drug Taxol (paclitaxel) binds to tubulin in a stoichiometric ratio and promotes its assembly into microtubules. The conformation of microtubule-bound drug has been the subject of intense study, and various suggestions have been made. In this work we present experimental and theoretical evidence that Taxol adopts a T-shaped conformation when it is bound to tubulin.
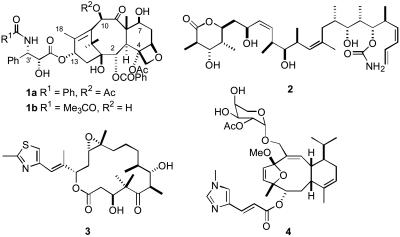
The diterpenoid natural product Taxol (1a) and its semisynthetic analog docetaxel (1b) are clinically important antitumor agents (1) whose clinical uses are still being expanded as various combination therapies are being explored (2, 3). They are known to exert their therapeutic effect, at least in part, by their ability to promote the assembly of tubulin into microtubules (4, 5). In recent years, several other natural products have been discovered that have a similar mechanism of action to Taxol, including discodermolide (2) (6), epothilone B (3) (7, 8), and eleutherobin (4) (9) (see Scheme 1). These compounds, together with several recently discovered analogs of Taxol, are all in preclinical development or in clinical trials as potential antitumor agents (10).
Scheme 1.
Structures of Taxol (1a), docetaxel (1b), discodermolide (2), epothilone B (3), and eleutherobin (4).
Taxol has been shown to bind to assembled microtubules and to stabilize them. It binds with an approximate stoichiometry of 1 mole of Taxol to 1 mole of tubulin dimer, stabilizes the polymer, and disrupts the equilibrium between tubulin and microtubules, leading to cell death by apoptosis (11, 12). Taxol also binds to Bcl-2, which then undergoes hyperphosphorylation (13), but it has been shown that this effect is linked to Taxol's tubulin-assembly activities. It has been proposed that Taxol-promoted microtubule assembly leads to Raf-1 activation and Bcl-2 phosphorylation, and thence to apoptosis (14). The tubulin-binding activity of Taxol (and, by implication, of other compounds that have similar effects) thus appears to be the key to its antitumor activity.
The interaction of Taxol with tubulin has been studied intensively by several methods. Thus, photoaffinity labeling has shown that a 3′-(p-azidobenzamido)Taxol derivative labels the N-terminal 31 amino acids of β-tubulin (15), whereas 2-(m-azidobenzoyl)Taxol labels residues 217–231 of β-tubulin (16), and a C-7 benzophenone derivative labeled Arg282 in β-tubulin (17). Fluorescence spectroscopy has yielded valuable information (18–20), but the most important results to date have come from the recently determined 3.7-Å structure of the αβ-tubulin–Taxol complex obtained by electron crystallography of zinc-induced tubulin sheets (21, 22). Although this structure shows the location of the binding site on β-tubulin, it does not enable the conformation of the ligand to be determined with precision.
A knowledge of the conformation of Taxol in its bound state on the microtubule has important ramifications. On one hand, it offers one piece of the puzzle that presently obscures the basis for the similar biological activity of the four very different chemical compounds 1-4. On the other hand, it can provide a conceptual model for the synthesis of simplified analogs that may well retain the full activity of the parent compound. Several experimental attempts to identify this conformation have been made by NMR measurement of internuclear distances within Taxol bound to microtubules (23, 24), and by the synthesis of various conformationally restricted Taxol analogs (25–30). In addition, studies have compared the conformations of Taxol and epothilone B by molecular modeling and other methods (29, 31–33). Separate investigations have proposed distinguishable conformations of the Taxol side chain, with a T-shaped conformation being favored on the basis of its fit with the election-density map of zinc-induced tubulin sheets (34, 35). In the present work, we provide experimental evidence that Taxol can be constrained to the T-conformation in solution, and that this form both stabilizes genuine microtubules and induces cell death.
Materials and Methods
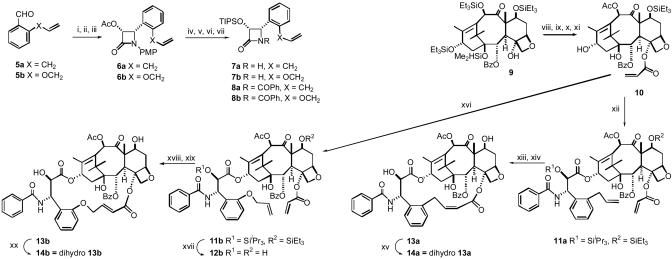
Synthesis of Open Chain Analogs 12b and Bridged Analogs 13a, 13b, 14a, and 14b. Fig. 1 outlines the synthesis of bridged Taxol derivatives 13a, 13b, 14a, and 14b, together with the open-chain analog 12b. Experimental details and characterization data for the intermediates are published as supporting information on the PNAS web site.
Fig. 1.
Synthetic scheme for the bridged paclitaxels 13a, 13b, 14a, and 14b. The reagents and conditions for the “a” series of β-lactams were similar to those below for the “b” series. Reagents and conditions for the “b” series β-lactams follow. i, 5b, p-MeOC6H4NH2, MgSO4, CH2Cl2, 100%. ii, CH3COOCH2COCl, Et3N, -78°C to room temperature (rt), 12 h, 85%. iii, Lipase (Amano PS), phosphate buffer, pH 7.2, CH3CN, 24 h, 98%. iv, 1M KOH, THF, 0°C, 100%. v, TIPSCl, imidazole, DMF, 94%. vi, CAN, CH3CN, -5°C, 62%. vii, PhCOCl, Et3N, DMAP, CH2Cl2, 95%. viii, LiHMDS, THF, 0°C, CH2—CHCOCl, 52%. ix, HF-pyridine, THF, 70%. x, CeCl3,Ac2O, THF, 96%. xi, Et3SiCl, imidazole, DCM, 72%. xii, 8a, NaH, THF, 0°C to rt, 24 h. xiii, (H2IMes)(PCy3)(CI)2Ru—CHPh, CH2Cl2, 3 h. xiv, HF-pyridine, 12 h. xv, H2,Pd/C(10%), 35 psi, 2.5 h. xvi, 8b, NaH, THF, 0°C to rt, 24 h, 50%. xvii, HF-Pyridine, THF, 81%. xviii, (H2IMes)(PCy3)(CI)2Ru—CHPh, CH2Cl2, 3 h, 64%. xix, HF-pyridine, 12 h, 98%. xx, H2, Pd/C(10%), 35 psi, 2.5 h, 96%.
Critical Concentration Determination. The critical concentration of tubulin in the presence of 10 μM Taxol or Taxol analog was determined from assembly experiments performed at 37°C with GDP-tubulin in PME buffer (100 mM Pipes/2 mM MgSO4/1 mM EGTA, pH 6.90) containing 4% DMSO. The extent of assembly was measured at different tubulin concentrations (0.5–6 μM) by light scattering (apparent absorption at 350 nm). Critical concentrations were calculated from the x intercepts of plots of apparent A350 vs. tubulin concentration.
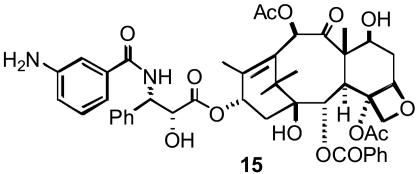
Competition Binding Experiments. The relative affinities of Taxol and derivatives for polymerized tubulin were assessed by competition assays. Crosslinked microtubules were prepared in glycerol assembly buffer (10 mM phosphate/1 mM EGTA/0.1 mM GTP/3.4 M glycerol, pH 6.9) as described by Andreu and Barasoain (37). Before use, the crosslinked microtubules were dialyzed against PME buffer for 16–18 h. Crosslinked microtubules (5 μM) and the fluorescent Taxol derivative N-AB-PT (15, 5 μM; Scheme 2) in PME buffer were incubated for 20 min at room temperature with 5 μM Taxol or the taxane. The fluorescence emission intensity at 412 nm of each sample was recorded by using a Jobin–Yvon Horiba Fluoromax-3 spectrofluorometer in a 2 mm × 10 mm quartz cell (λex = 320 nm) and compared to the emission intensity of the system in the absence of competitor.
Scheme 2.
Structure of the fluorescent Taxol derivative N-AB-PT (15).
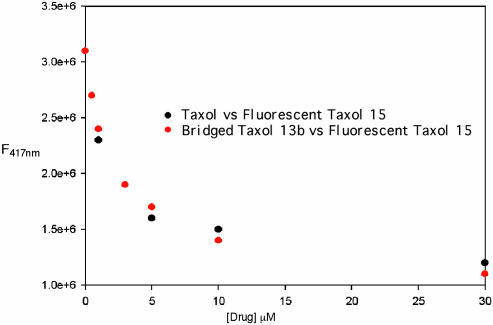
For the full competition experiment shown in Fig. 3, microtubules, N-AB-PT, and varying concentrations of Taxol or 13b (0–30 μM) were treated in the same manner, and their emission intensities were measured. The full competition experiment was repeated by using pure tubulin assembled by N-AB-PT, and identical results were obtained (data not shown).
Fig. 3.
Competition displacement of fluorescent Taxol 15 from tubulin by Taxol (black dots) and by compound 13b (red dots). Compound 15 was maintained at 5 μM, and increasing amounts of Taxol or compound 13b were added.
Conformational Analysis for 13b in Solution. The 400-MHz rotating-frame Overhauser effect spectroscopy (ROESY) analysis of 13b delivered 17 intramolecular distances. Monte Carlo conformational analysis using macromodel 6.5 (38) yielded 858 fully optimized conformations for 13b. Subsequent NMR/NAMFIS (NMR analysis of molecular flexibility in solution) treatment (35, 39) integrated the latter and the ROESY spectra to yield three conformations in CDCl3, two of which differ by torsions in the C-4 to C-3′ bridge but correspond to a total of 83% of 13b in the T-Taxol form. For details, see supporting information.
Results and Discussion
As summarized in the Introduction, a series of studies point to a T-shaped Taxol conformer bound to β-tubulin in microtubules as being the best fit to the experimental data. However, definitive confirmation of this hypothesis is still lacking. As a result, we sought experimental verification through independent but complementary methods: molecular design, synthesis, and tubulin-binding studies of constrained T-Taxol analogs 13a, 13b, 14a, and 14b, and NMR analysis of conformation in solution.
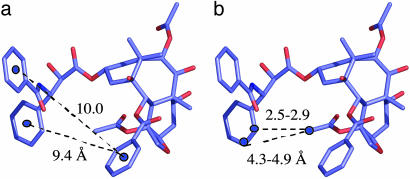
T-Taxol as a Design Template. The T-Taxol conformation, derived by docking experimentally based conformers of the ligand into the tubulin–Taxol electron crystallographic density (36), shows several unusual features. Unlike a number of earlier propositions concerning the conformation of bound Taxol (see ref. 36 for a summary of previous proposals), hydrophobic collapse between the C-2 benzoyl phenyl moiety and either of the phenyl rings emanating from C-3′ is not observed. Rather, both of the latter rings reside 9–10 Å from the C-2 substituent (Fig. 2a). The overall spatial disposition of the C-13 side chain in the T-Taxol conformation resembles that of the so-called “nonpolar” conformation in which the C-3′ benzamidophenyl has been shifted significantly away from the C-2 benzoyl center. Examination of the computationally refined tubulin binding site illustrates that His-227 resides between these two rings. This accounts for the fact that previous attempts to bridge the C-2 and C-3′ positions have delivered either inactive Taxol analogs (28) or compounds that are one or two orders of magnitude less active than Taxol itself (25, 26, 29, 30, 31). By contrast, inspection of T-Taxol reveals that the C-4 acetate methyl hydrogens are just 2.5–2.9 Å and 4.3–4.9 Å distant from the o- and m-hydrogens of the C-3′ phenyl, respectively (Fig. 2b), suggesting that a bridge between these centers would contribute to a reduction in conformational mobility while locking the structure into the T-conformer.
Fig. 2.
T-Taxol conformation. (a) The similar and extended ring-to-ring distances between the C-2 benzoyl phenyl and the C-3′ phenyl and benzamido phenyl centers, respectively. (b) H—H separations between the C-4 acetate methyl group and the ortho and meta postions of the C-3′ phenyl ring.
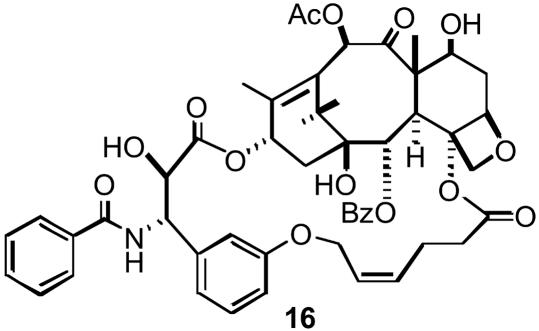
Consequently, we contemplated a number of bridging units from the C-4 methyl to the C-3′ phenyl that could be formed readily by ring-closing metathesis methodology. Two initial proof-of-principle targets connected the meta position of the C-3′ phenyl and the C-4 methyl with three- and five-atom bridges, e.g., 16 (Scheme 3). The compounds proved to be 30-fold and 10-fold less potent than Taxol, respectively, as microtubule stabilizers and cytotoxins (27). Therefore, we turned to ortho-bridged structures with three or four atoms separating the side chains, namely 13a and 13b and their bridge-saturated analogs 14a and 14b. In an attempt to quantify the SAR of the modifications, we used our most recently parameterized 3D-QSAR Taxol minireceptor (refs. 34 and 40 and M. Wang, M., A.S.L., and J.P.S., unpublished data) to evaluate the tubulin polymerization capacity of the structures. Both were predicted to show activity comparable to Taxol in complete accord with the subsequently measured biological quantities described below.
Scheme 3.
Structure of bridged Taxol 16.
Bridged Taxol Synthesis. The ultimate test of the binding conformation of Taxol would be to prepare a conformationally constrained derivative that shows better activity than Taxol itself. Our earlier modeling studies on the bridged analog 16 (27) revealed that the compound is seated higher than Taxol in the binding pocket of tubulin as a result of a close contact between the propene moiety of the m-phenol linked tether and Phe-270 of the protein. Structural analysis forecast that a tether linked to the ortho position of the 3′-phenyl would be pulled closer to the baccatin core and thereby minimize the ligand–protein interaction. Thus, we prepared a number of bridged Taxols with linkages from the ortho position of the 3′-phenyl group to the C-4 acetyl methyl group. The synthesis of the key compounds 13a and 13b is shown in Fig. 1. The double bond in the bridging linker of 13a was shown to be Z based on the NMR coupling constant of the α-proton (J = 11.5 Hz). No E isomer was detected in this case. Surprisingly, the double bond in the bridging linker of 13b was found to be E (J = 15.5 Hz). Hydrogenation provided the dihydro analogs 14a and 14b, whereas compound 12b was evaluated as an “open chain” analog of 13a and 13b to ensure that the αβ-unsaturated ester and 3′-phenyl substitutions alone were not responsible for any unusual activity.
Microtubule Assembly and Cytotoxicity Assessment. Compound 13a proved to be highly active in two cytotoxicity assays (Table 1). In the A2780 ovarian cancer cell line, it was approximately 20 times more potent than Taxol, and ≈300-fold more potent than our previous best analog, 16 (27). In the PC-3 prostate cell line, it was slightly more cytotoxic than Taxol. Compound 13b was equipotent with Taxol in the two cell lines. The dihydro compound 14a was more potent than Taxol in both cell lines, whereas the dihydro derivative 14b was slightly less potent when compared with the same standard. The “open chain” analog 12b was over three orders of magnitude less cytotoxic to A2780 cells and over two orders of magnitude less cytotoxic to PC-3 cells, demonstrating that the activity of 13a is not caused by the presence of the αβ-unsaturated ester at C-4 or to the ortho substituent on the phenyl ring.
Table 1. Bioactivity of Taxol and analogs 12–14.
| Compound | IC50(cp)/IC50(tx) A2780 | IC50(cp)/IC50(tx) PC3 | ED50, Tb polymerization,* μM | Critical Tb concentration,† μM | Inhibit binding F-Taxol, % |
|---|---|---|---|---|---|
| Taxol | —‡ | —§ | 0.50 ± 0.14 | 1.8 ± 0.30 | 26 |
| 12b | 1,190 | 150 | 1.02 ± 0.37 | ND | ND |
| 13a | 0.045 | 0.69 | 0.30 ± 0.09 | 0.53 ± 0.07 | 72 |
| 13b | 0.97 | 1.0 | 0.28 ± 0.11 | 1.2 ± 0.24 | 30 |
| 14a | 0.08 | 0.67 | 0.21 ± 0.09 | 0.35 ± 0.06 | 79 |
| 14b | 1.2 | 3.3 | 0.83 ± 0.19 | 1.3 ± 0.33 | 7 |
ND, not determined
Tubulin concentration, 5 μM
Taxoid concentration, 10 μM
Taxol has IC50 values of 6–15 nM in this assay
Taxol has an average IC50 value of 4 nM in this assay
A characteristic in vitro activity of Taxol is its ability to induce purified tubulin to assemble into microtubules. The analogs' capability to promote tubulin assembly was roughly parallel to their cytotoxicities: compounds less cytotoxic than Taxol were also less potent promoters of assembly in these assays. Conversely, compounds with cytotoxicities equal to or greater than Taxol were more effective polymerizing agents.
The induction of tubulin assembly by Taxol and related molecules is a function of both the affinity of the ligand for the Taxol binding site on tubulin and the effect of ligand binding on the conformation of the protein (41). These two parameters can be measured separately. The affinity of ligands for the Taxol binding site on microtubules can be determined by competition between the ligand in question and a radioactive or fluorescent derivative of Taxol (19, 37). Fig. 3 shows that the binding of the fluorescent Taxol derivative N-AB-PT (15) to stabilized microtubules was inhibited to the same extent by Taxol and 13b, demonstrating that they bind to the Taxol site on microtubules with equal affinity. Single point assays for inhibition of N-AB-PT binding to tubulin by other Taxoids indicate that the relative affinities of the molecules for the Taxol-binding site on tubulin are roughly parallel to their assembly promoting abilities and cytotoxicities.
Taxol binding to polymerized tubulin affects the conformation of the protein in a way that favors tubulin assembly, i.e., by increasing the equilibrium constant for polymer growth (Kp). The reciprocal of the critical concentration is a very close approximation of Kp (42, 43). Table 1 shows the critical concentration for tubulin assembly in the presence of Taxol and the conformationally restricted Taxol analogs. All four molecules are at least as active as Taxol in lowering the critical concentration of tubulin, indicating that they are all effective promoters of the assembly active conformation of tubulin.
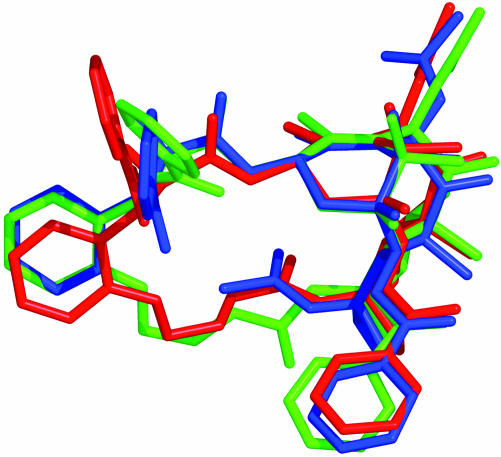
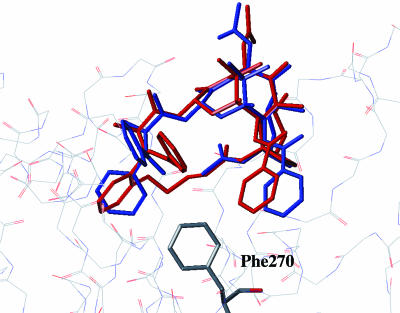
Ligand–Tubulin Structure: Solution Conformations by NMR/NAMFIS Analysis. In our initial approach to fitting the electron crystallographic density with conformational candidates, we reasoned that a data set of conformations determined by combining single crystal X-ray and solution NMR structures would prove to furnish more realistic binding candidates than a completely virtual docking approach. Thus, we used the NMR/NAMFIS method (39) to identify a low-population Taxol conformation (4%)¶ that matched the electron crystallographic density on tubulin, namely T-Taxol (36). The meta-bridged compound 16 with less than one-tenth the activity of Taxol was similarly determined to exist in solution to the extent of 5% (27). According to the NAMFIS analysis, the considerably more constrained ortho-bridged compound 13b in CDCl3 exhibits two related conformers with populations of 76 and 7%. As illustrated by Fig. 4, these conformations together comprise a total of 83% in the T-Taxol conformer proposed as the tubulin-bound form. Clearly, the decreased torsional freedom and reduced molecular volume of ortho-bridged 13b contributes to its equipotency with Taxol relative to the more flexible, larger and less active meta-bridged 16 (27). Fig. 5 illustrates that 13b seats itself in the tubulin-taxoid binding site in a manner almost identical with that of Taxol, escaping a steric clash with hydrophobic residues at the bottom of the ligand pocket as previously modeled for 16 (27). Compounds 13a and 14a with one less bridge atom between the ortho position of the C3′-phenyl and the 4-OAc methyl introduce additional molecular rigidification while maintaining the T-Taxol conformation as determined by conformational analysis. Accordingly, respective cytotoxicities exceed those of Taxol by a factor of up to 20.
Fig. 4.
NAMFIS-derived T-conformations for 13b (76% red, 7% green) superimposed on the tubulin-bound T-Taxol form (blue).
Fig. 5.
T-Taxol (blue) bound to β-tubulin as described in ref. 35. The ortho-bridged T-form of 13b (red, both ligand and protein binding site subjected to MD relaxation similar to T-Taxol) superimposed on Taxol avoids the steric contact with Phe-270 at the bottom of the hydrophobic taxoid pocket experienced by the meta-bridged analog 16 (27).
Summary, Conclusions, and Prospects. Analysis of the T-Taxol conformation has suggested a previously undescribed bridging strategy linking the C-4 OAc methyl and the C-3′ phenyl group that locks the molecule into the T-Taxol geometry. Minireceptor evaluation predicted the ortho-bridged unsaturated esters 13a and 13b and the corresponding saturated analogs 14b and 14b to be at least equipotent to Taxol's action as microtubule stabilizers. Subsequent synthesis taking advantage of the olefin metathesis approach has led to both compounds, the NMR/NAMFIS analysis for 13b demonstrating that >80% of the compound adopts the T-Taxol conformation in solution. Tubulin polymerization and cytotoxicity assays are complementary by demonstrating that bridged taxoids, namely 13b and 14b, are capable of showing equivalence to Taxol in their biological action. The outcomes highlight three important conclusions to be drawn from our work.
First, the electron crystallographic-based modeling study that identified T-Taxol as the bioactive conformation would appear to be independently substantiated. By constraining the two C-3′ and C-2 phenyl rings to be distant from one another (Fig. 2), the hydrophobically collapsed “polar” and “nonpolar” conformations are eliminated as viable binding forms.
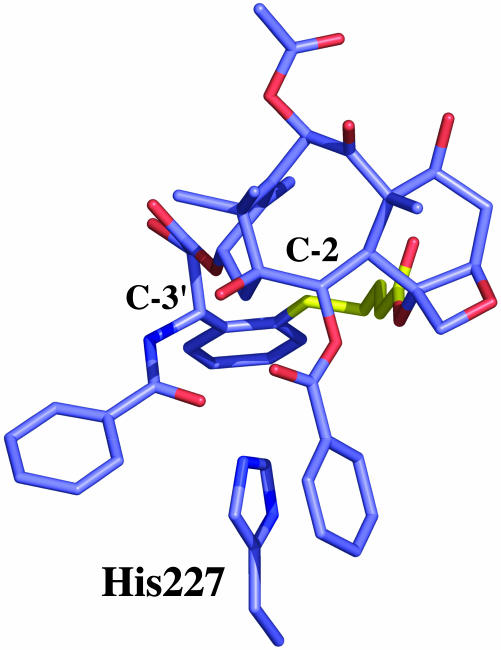
The second point follows, which is that the present results contrast sharply with those derived by bridging from either the C-3′ phenyl (25, 28, 29) or the benzamido (26, 30, 31) side chain directions to the C-2 position of the baccatin core; cases in which the activity is either nonexistent or at best one-tenth that of Taxol. The fundamental reason for activity in the present series and its absence in other bridging schemes is related to the location of β-tubulin's His-227 in the taxoid binding pocket. This protein residue is part of a three-ring stacking motif in which its imidazole ring resides between the C-2 benzoyl and C-3′ benzamido phenyl rings of Taxol (36). Consequently, most tethers connecting the C-3′ and C-2 centers are unable to achieve the necessary arrangement. Compounds 13a/b and 14a/b, on the other hand, not only accommodate the His-227-ligand interaction, but constrain the molecules to the bioactive conformation by bridging behind the stacked rings (Fig. 6).
Fig. 6.
A view of ortho-bridged 13b in which phenyl rings emanating from C-2 and C-3′ surround the imidazole of His-227 in a sandwich motif; the C-4 OAc to C-3′ phenyl bridge (yellow) avoids the latter while stabilizing the T-Taxol conformation.
Third, the electron crystallographic analyses (21, 22, 36) are derived from zinc-stabilized tubulin sheets in which the α,β-tubulin protofilaments are antiparallel. This contrasts with the parallel arrangement of protofilaments in cellular microtubules. It has been argued that the difference implies T-Taxol derived from the former is inapplicable to the latter (44). The present results obviate this argument and support the earlier inference that the Taxol-binding site in the tubulin dimer derived from zinc-stabilized sheets is unaltered in the microtubule structure (45–47, ∥).
Finally, we anticipate that the T-Taxol design strategy evolved here, short rigid bridges from C-4 to C-3′, will permit other bridged structures to likewise surpass the activity of parent Taxol. We also foresee that significantly modified or truncated compounds designed around this principle will provide novel classes of active and easily synthesized antitumor agents.
Acknowledgments
We thank Tom Glass and William Bebout in the Department of Chemistry, Virginia Polytechnic Institute and State University, for assistance with rotating-frame Overhauser effect spectroscopy (ROESY) NMR measurements and for mass spectroscopic determinations, respectively. We also thank Jennifer Schilling for some preliminary cytotoxicity determinations. This work was supported by National Institutes of Health Grant CA-69571, and we are grateful for this support.
Abbreviation: NAMFIS, NMR analysis of molecular flexibility in solution.
Footnotes
Taxol in either CDCl3 (35) or D2O/DMSO-d6 (J.P.S., N. Nevins, J. Jiménez-Barbero, D. Cicero, and J. M. Jansen, unpublished data) displays 8 and 14 conformers, respectively, the populations of which are solvent-dependent. T-Taxol appears in CDCl3 and D2macromodel 6.5O/DMSO-d6 with mole fractions of 0.04 (4%) and 0.02 (2%), respectively.
The orientation of T-Taxol in the tubulin binding site is different from what two of us proposed in an earlier paper (24) based on measurements performed on colchicinoid–tubulin complexes assembled with a fluorescent Taxol analog. The Taxol-binding site on the colchicine–tubulin–stathmin complex is now known to be different from that in the zinc-sheet structure of tubulin (47). The binding mode of Taxol we proposed (24) is therefore likely to be different from that of Taxol in the tubulin conformation found in microtubules and zinc-sheets.
References
- 1.Crown, J. & O'Leary, M. (2000) Lancet 355, 1176-1178. [DOI] [PubMed] [Google Scholar]
- 2.Nabholtz, J.-M., Tonkin, K., Smylie, M., Au, H.-J., Lindsay, M.-A. & Mackey, J. (2000) Exp. Opin. Pharmacother. 1, 187-206. [DOI] [PubMed] [Google Scholar]
- 3.Miller, K. D. & Sledge, G. W., Jr. (1999) Cancer Invest. 17, 121-136. [PubMed] [Google Scholar]
- 4.Schiff, P. B., Fant, J. & Horwitz, S. B. (1979) Nature 277, 665-667. [DOI] [PubMed] [Google Scholar]
- 5.Ringel, I. & Horwitz, S. B. (1991) J. Natl. Cancer Inst. 83, 288-291. [DOI] [PubMed] [Google Scholar]
- 6.ter Haar, E., Kowalski, R. J., Hamel, E., Lin, C. M., Longley, R. E., Gunasekera, S. P., Rosenkranz, H. S. & Day, B. W. (1996) Biochemistry 35, 243-250. [DOI] [PubMed] [Google Scholar]
- 7.Bollag, D. M., McQueney, P. A., Zhu, J., Hensens, O., Koupal, L., Liesch, J., Goetz, M., Lazarides, E. & Woods, C. M. (1995) Cancer Res. 55, 2325-2333. [PubMed] [Google Scholar]
- 8.Kowalski, R. J., Giannakakou, P. & Hamel, E. (1997) J. Biol. Chem. 272, 2534-2541. [DOI] [PubMed] [Google Scholar]
- 9.Lindel, T., Jensen, P. R., Fenical, W., Long, B. H., Casazza, A. M., Carboni, J. & Fairchild, C. R. (1997) J. Am. Chem. Soc. 119, 8744-8745. [Google Scholar]
- 10.Kavallaris, M., Verrills, N. M. & Hill, B. T. (2002) Drug Resist. Updat. 4, 392-401. [DOI] [PubMed] [Google Scholar]
- 11.Manfredi, J. J. & Horwitz, S. B. (1984) Pharmacol. Ther. 25, 83-125. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz, S. B. (1992) Trends Pharmacol. Sci. 13, 134-136. [DOI] [PubMed] [Google Scholar]
- 13.Scatena, C. D., Stewart, Z. A., Mays, D., Tang, L. J., Keefer, C. J., Leach, S. D. & Pietenpol, J. A. (1998) J. Biol. Chem. 273, 30777-30784. [DOI] [PubMed] [Google Scholar]
- 14.Blagosklonny, M. V., Giannakakou, P., El-Deiry, W. S., Kingston, D. G. I., Higgs, P. I., Neckers, L. & Fojo, T. (1997) Cancer Res. 57, 130-135. [PubMed] [Google Scholar]
- 15.Rao, S., Krauss, N. E., Heerding, J. M., Swindell, C. S., Ringel, I., Orr, G. A. & Horwitz, S. B. (1994) J. Biol. Chem. 269, 3132-3134. [PubMed] [Google Scholar]
- 16.Rao, S., Orr, G. A., Chaudhary, A. G., Kingston, D. G. I. & Horwitz, S. B. (1995) J. Biol. Chem. 270, 20235-20238. [DOI] [PubMed] [Google Scholar]
- 17.Rao, S., He, L., Chakravarty, S., Ojima, I., Orr, G. A. & Horwitz, S. B. (1999) J. Biol. Chem. 274, 37990-37994. [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., Edsall, J., R., Jagtap, P. G., Kingston, D. G. I. & Bane, S. (2000) Biochemistry 39, 616-623. [DOI] [PubMed] [Google Scholar]
- 19.Diaz, J. F., Strobe, R., Engelborghs, Y., Souto, A. A. & Andreu, J. M. (2000) J. Biol. Chem. 275, 26265-26276. [DOI] [PubMed] [Google Scholar]
- 20.Sengupta, S., Boge, T. C., Liu, Y., Hepperle, M., Georg, G. I. & Himes, R. H. (1997) Biochemistry 36, 5179-5184. [DOI] [PubMed] [Google Scholar]
- 21.Nogales, E., Wolf, S. G. & Downing, K. H. (1998) Nature 39, 199-203. [DOI] [PubMed] [Google Scholar]
- 22.Lowe, J., Li, H., Downing, K. H. & Nogales, E. (2001) J. Mol. Biol. 313, 1045-1057. [DOI] [PubMed] [Google Scholar]
- 23.Ojima, I., Inoue, T. & Chakravarty, S. (1999) J. Fluorine Chem. 97, 3-10. [Google Scholar]
- 24.Li, Y., Poliks, B., Cegelski, L., Poliks, M., Cryczynski, Z., Piszczek, G., Jagtap, P., G., Studelska, D. R., Kingston, D. G. I., Schaefer, J. & Bane, S. (2000) Biochemistry 39, 281-291. [DOI] [PubMed] [Google Scholar]
- 25.Ojima, I., Lin, S., Inoue, T., Miller, M. L., Borella, C. P., Geng, X. & Walsh, J. J. (2000) J. Am. Chem. Soc. 122, 5343-5353. [Google Scholar]
- 26.Geng, X., Miller, M. L., Lin, S. & Ojima, I. (2003) Org. Lett. 5, 3733-3736. [DOI] [PubMed] [Google Scholar]
- 27.Metaferia, B. B., Hoch, J., Glass, T. E., Bane, S. L., Chatterjee, S. K., Snyder, J. P., Lakdawala, A., Cornett, B. & Kingston, D. G. I. (2001) Org. Lett. 3, 2461-2464. [DOI] [PubMed] [Google Scholar]
- 28.Boge, T. C., Wu, Z.-J., Himes, R. H., Vander Velde, D. G. & Georg, G. I. (1999) Bioorg. Med. Chem. Lett. 9, 3047-3052. [DOI] [PubMed] [Google Scholar]
- 29.Ojima, I., Chakravarty, S., Inoue, T., Lin, S., He, L., Horwitz, S. B., Kuduk, S. C. & Danishefsky, S. J. (1999) Proc. Natl. Acad. Sci. USA 96, 4256-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Querolle, O., Dubois, J., Thoret, S., Roussi, F., Montiel-Smith, S., Gueritte, F. & Guenard, D. (2003) J. Med. Chem. 46, 3623-3630. [DOI] [PubMed] [Google Scholar]
- 31.Querolle, O., Dubois, J., Thoret, S., Dupont, C., Guéritte, F. & Guénard, D. (2003) Eur. J. Org. Chem. 2003, 542-550. [Google Scholar]
- 32.Giannakakou, P., Gussio, R., Nogales, E., Downing, K. H., Zaharevitz, D., Bollbuck, B., Poy, G., Sackett, D., Nicolaou, K. C. & Fojo, T. (2000) Proc. Natl. Acad. Sci. USA 97, 2904-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, L., Jagtap, P. G., Kingston, D. G. I., Shen, H.-J., Orr, G. A. & Horwitz, S. B. (2000) Biochemistry 39, 3972-3978. [DOI] [PubMed] [Google Scholar]
- 34.Wang, M., Xia, X., Kim, Y., Hwang, D, Jansen, J. M., Botta, M., Liotta, D. C. & Snyder, J. P. (1999) Org. Lett. 1, 43-46. [DOI] [PubMed] [Google Scholar]
- 35.Snyder, J. P., Nevins, N., Cicero, D. O. & Jansen, J. (2000) J. Am. Chem. Soc. 122, 724-725. [Google Scholar]
- 36.Snyder, J. P., Nettles, J. H., Cornett, B., Downing, K. H. & Nogales, E. (2001) Proc. Natl. Acad. Sci. USA 98, 5312-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreu, J. M. & Barasoain, I. (2001) Biochemistry 40, 11975-11984. [DOI] [PubMed] [Google Scholar]
- 38.Mohamadi, F., Richards, N. G. J., Guida, W. C., Liscamp, R., Lipton, M., Caufield, C., Chang, G., Hendrickson, T. & Still, W. C. (1990) J. Comput. Chem. 11, 440-467. [Google Scholar]
- 39.Cicero, D.O., Barbato, G. & Bazzo, R. (1995) J. Am. Chem. Soc. 117, 1027-1033. [Google Scholar]
- 40.Barboni, L., Datta, A., Dutta, D., Georg, G. I., Vander Velde, D., Himes, R. H., Wang, M. & Snyder, J. P. (2001) J. Org. Chem. 66, 3321-3329. [DOI] [PubMed] [Google Scholar]
- 41.Diaz, J. F., Menedez, M. & Andreu, J. M. (1993) Biochemistry 32, 10067-10077. [DOI] [PubMed] [Google Scholar]
- 42.Timasheff, S. N. (1981) in Protein–Protein Interactions, eds. Frieden, C. & Nichol, L. W. (Wiley, New York), pp. 315-336.
- 43.Andreu, J. M. & Timasheff, S. N. (1986) Methods Enzymol. 130, 47-59. [DOI] [PubMed] [Google Scholar]
- 44.Ivery, T. G. M. & Le, T. (2003) Oncol. Res. 14, 1-19. [DOI] [PubMed] [Google Scholar]
- 45.Nogales, E., Whittaker, M., Milligan, R. A. & Downing, K. H. (1999) Cell 96, 79-88. [DOI] [PubMed] [Google Scholar]
- 46.Li, H., DeRosier, D. J., Nicholson, W. V., Nogales, E. & Downing, K. H. (2002) Structure (London) 10, 1317-1328. [DOI] [PubMed] [Google Scholar]
- 47.Ravelli, R. B. G., Gigant, B., Curmi, P. A., Jourdain, I., Lachkar, S., Sobel, A. & Knossow, M. (2004) Nature 428, 198-202. [DOI] [PubMed] [Google Scholar]