Abstract
Next-generation sequencing of environmental samples can be challenging because of the variable DNA quantity and quality in these samples. High quality DNA libraries are needed for optimal results from next-generation sequencing. Environmental samples such as water may have low quality and quantities of DNA as well as contaminants that co-precipitate with DNA. The mechanical and enzymatic processes involved in extraction and library preparation may further damage the DNA. Gel size selection enables purification and recovery of DNA fragments of a defined size for sequencing applications. Nevertheless, this task is one of the most time-consuming steps in the DNA library preparation workflow. The protocol described here enables complete automation of agarose gel loading, electrophoretic analysis, and recovery of targeted DNA fragments.
In this study, we describe a high-throughput approach to prepare high quality DNA libraries from freshwater samples that can be applied also to other environmental samples. We used an indirect approach to concentrate bacterial cells from environmental freshwater samples; DNA was extracted using a commercially available DNA extraction kit, and DNA libraries were prepared using a commercial transposon-based protocol. DNA fragments of 500 to 800 bp were gel size selected using Ranger Technology, an automated electrophoresis workstation. Sequencing of the size-selected DNA libraries demonstrated significant improvements to read length and quality of the sequencing reads.
Keywords: Environmental Sciences, Issue 98, environmental samples, next-generation sequencing, DNA libraries, Ranger Technology, gel size selection, metagenomics, water samples
Introduction
Metagenomics involves the sequencing of all the genetic material in a sample to characterize the microbial communities present. It is a complex and expensive process which involves the conversion of extracted nucleic acids into DNA libraries followed by next-generation sequencing. High quality libraries are essential for maximal data output and accurate metagenomics analysis. Environmental samples, such as water samples, often pose significant challenges to generating high quality libraries, due to low amounts of DNA that may also be degraded1-3 and the presence of inhibitors of PCR4-6.
High quality libraries ideally consist of longer segments of DNA within a narrow range of lengths. In order to maximize the amount of useful data generated per sequencing run, the length of the DNA in the library should be at least as long as the maximum read length of the sequencing method being used. When using a sequencing-by-synthesis technology such as the Illumina MiSeq, the size of the DNA fragments affects the efficiency at which clusters are generated on the flow cell. For instance, when a library contains both shorter and longer DNA fragments, the shorter ones will be over-represented in the sequencing data7,8. In contrast, a library with similarly sized DNA fragments will be proportionally represented in the sequencing data. Many library preparation kits use ligation-based methods to add adapters to the DNA fragments and size selection is necessary to remove adapter dimers that do not contain an insert9,10. There are numerous methods11,12 to achieve this but the one technique that gives the most consistent results is the electrophoretic separation of DNA followed by the recovery of the desired lengths of DNA13,14. This process can be performed manually for a small number of samples, but when faced with processing hundreds of samples, automated solutions are required. The currently available platforms for automated gel size selection are low throughput and new platforms are needed to process large numbers of samples for sequencing. The Ranger Technology can be integrated with existing liquid handling workstations to enable the use of agarose gel electrophoresis for size selection and analytical purposes on a scale that satisfies today’s high throughput environment.
Protocol
1. Water Collection and Filtration
Collect freshwater samples from various sites (Figure 1). Pass samples through a series of filters: 1 µm filter, 0.2 µm filter, and 30 kDa cutoff tangential flow filter to systematically separate eukaryotic, bacterial and viral sized particles, respectively. Note: Only the purification and analysis of the material recovered on the 0.2 µm filters (free-living bacteria) is described in this report, but similar approaches can be used for the other particles recovered from the filters.
2. Bacterial Concentration, DNA Extraction and Purification
Remove the 0.2 µm membrane filter(s) from the water filtration system (schematic diagram, Figure 2). Fold the 0.2 µm disc filter in half and place it into a 50 ml tube with the open side up. Add 5 ml of 1x phosphate buffered solution (PBS) and 0.01% Tween, pH 7.4 into the tube. If more than one filter is used during the process employ a different tube per filter. Store tube(s) with filter at 4 °C until processed. Otherwise, proceed to step 2.2.
- Remove the 0.2 µm membrane filter from the 50 ml tube with sterile forceps. Place filter(s) on a petri dish (leave the PBS buffer in the tube).
- With sterile forceps and sterile scissors, cut the filter into small strips (1 cm x 1 cm). Disinfect forceps and scissors by soaking in 70% isopropanol, wiping with a DNA surface decontaminant and rinsing with ultrapure water of type 1 between different samples.
Place the filter cut into strips back into the same 50 ml tube. Add 10 ml of 1x PBS and 0.01% Tween, pH 7.4 to the filter so there is a total of 15 ml of buffer in the 50 ml tube.
Add 6 clean tungsten beads (3 mm diameter) to each 50 ml tube, cover the tube with Parafilm, and vortex vigorously for 20 min (use 50 ml tubes vortex adaptor). If multiple filters are processed from one sample, transfer and pool all the homogenate into a clean 50 ml tube. Centrifuge the tubes at 3,300 x g for 15 min at 4 °C.
Resuspend the pellet and aliquot bacterial cells (~1 ml aliquots) into 1.7 ml microcentrifuge tubes. Note that there will be 5 microcentrifuge tubes per sample.
Spin down the microcentrifuge tubes at 10,000 x g for 10 min, and remove most of the supernatant down to ~200 µl mark.
Store the samples at -80 °C, or proceed to extract bacterial DNA by using a commercially available DNA isolation kit as per the manufacturer’s instructions. During bacterial DNA extraction use either PBS or water as a negative extraction control, and E. coli as a positive extraction control.
Because DNA from multiple tubes are being extracted, pool parallel sub-aliquots from the same sample into one 2 ml tube. Then conduct an O/N precipitation by using a solution comprised of 0.1 volumes of 3 M sodium acetate, 2 volumes of 100% ethanol, and 5 µl of 5 µg/µl linear acrylamide. Mix well and store samples at -80 °C.
Centrifuge at maximum speed (17,000 x g) for 30 min at 4 °C. Wash pellet with 1 ml of ice-cold 70% ethanol, air dry in a biosafety cabinet for no more than 5 min and resuspend DNA pellet in 34 µl of 10 mM Tris-Cl, pH 8.5. Note: Linear acrylamide will help to visualize DNA pellet after the O/N precipitation.
Determine DNA quantity and quality using a high sensitivity fluorescent nucleic acid quantitation assay and a microvolume spectrophotometer, respectively, following the manufacturers’ instructions (See Table of Materials). Note: If multiple samples are prepared at the time, an ultrasensitive fluorescent nucleic acid assay for quantifying dsDNA and plate-based fluorometer/spectrophotometer can be used instead.
3. DNA Library Preparation
Normalize genomic bacterial DNA extracted from water samples to a concentration of ~0.2 ng/μl.
Enzymatically fragment one nanogram of bacterial DNA using a transposon-based method (tagmentation). Add adapter and index sequences onto fragmented DNA following the manufacturer’s instructions. Subject tagmented samples to 12 cycles of PCR as indicated in the manufacturer’s instructions, at which point the sequencing indexes are incorporated.
4. Automated Gel Size Selection
- Skip standard post-PCR clean-up step described in the manufacturer’s instructions (See Table of Materials). Size select post-PCR samples directly using an automated gel-based size selection platform.
- Add 3.5 μl of loading buffer to each sample.
- Following the manufacturer's protocol, load the samples onto the electrophoresis workstation, specifying a size-selection target of 500-800 bp, in a 300 μl extraction volume, using a pre-cast 1.5% agarose cassette.
- Concentrate the raw output material (300 μl) down to 25 μl via ethanol precipitation. Note: Alternatively, use the electrophoresis workstation to automate concentration for larger numbers of samples via filter plate and vacuum manifold.
- Precipitate the raw elution volume using 0.1 volumes of 3 M sodium acetate, 2 volumes of 100% ethanol and 5 μl of 5 µg/µl linear acrylamide. Mix well, place samples at -80 °C O/N. Centrifuge samples at 17,000 x g for 30 min.
- Discard supernatants and proceed to wash DNA pellet with 1 ml of ice-cold 70% ethanol.
- Centrifuge samples again at 17,000 x g for 30 min, discard supernatants, air dry, and finally resuspend DNA pellet in 25 μl of 10 mM Tris-Cl, pH 8.5.
(Optional) Use 1 μl of DNA libraries size selected with the automated gel size selection platform to make a five-fold dilution using Tris-EDTA buffer solution, pH 8.0. Analyze libraries using a chip-based capillary electrophoresis instrument to analyze dsDNA quality according to manufacturer’s instructions.
5. Library Normalization and Sequencing
- Proceed to normalize samples using library normalization beads (included in the transposon-based library preparation kit) as described in the manufacturer’s preparation guide.
- Alternatively, measure dsDNA libraries using a high sensitivity fluorescent nucleic acid quantitation assay, followed by pooling at equimolar ratios to reach a final dsDNA concentration of 2 nM. Proceed to denature libraries using 10 μl of pooled libraries and 10 μl of 0.2 N NaOH, incubate for 5 min at RT. Dilute denatured libraries using hybridization buffer (included with the transposon-based preparation kit) to a final volume of 1 ml and concentration of 11.5 pM.
Load samples in a next-generation sequencing platform by following the manufacturer’s standard sequencing protocols.
Representative Results
DNA Concentration, and Quality Assessment
Bacterial DNA concentrations ranged from 0.01 to 0.11 ng/ml per water sample of the different watershed sites (Table 1). DNA isolated from bacterial fraction had A260/280 and A260/230 ratios of 1.4 to 1.8 and 0.3 to 1.6, respectively. While some of the A260/230 ratios were relatively low, no apparent inhibition was observed in the prepared library and other downstream applications. These applications included PCR and qPCR tests targeting 16S rRNA and cpn60, and subsequent amplicon sequencing (data not shown).
Ranger Technology
DNA libraries prepared with a transposon-based method and gel size selected with a high-throughput automated platform yielded a tight range of DNA fragments between 500-800 bp (Figure 3). This alternative protocol yielded concentrations ranging from 0.3 to 3.4 ng/μl (Table 1). Using an average of 650 bp of DNA fragment size, average concentration for this library was 3.81 nM of input material. Libraries were consistently size selected in all DNA libraries prepared from multiple watershed locations.
DNA Sequencing QC Parameters
DNA sequencing of this library on the Illumina MiSeq generated a cluster density of 1,198 K/mm2. The percentage of cluster passing filters was 84.2% with an estimated yield of 9.2 Gb. Moreover, 7.4 Gb or 82.8% of the reads had a quality score ≥30. Utilization of an automated gel size selection platform consistently resulted in suitable yields and minimized the clustering of unwanted fragments under 200 bp.
 Figure 1.
Water samples were collected from urban (A) and agriculturally (B) influenced watersheds.
Figure 1.
Water samples were collected from urban (A) and agriculturally (B) influenced watersheds.
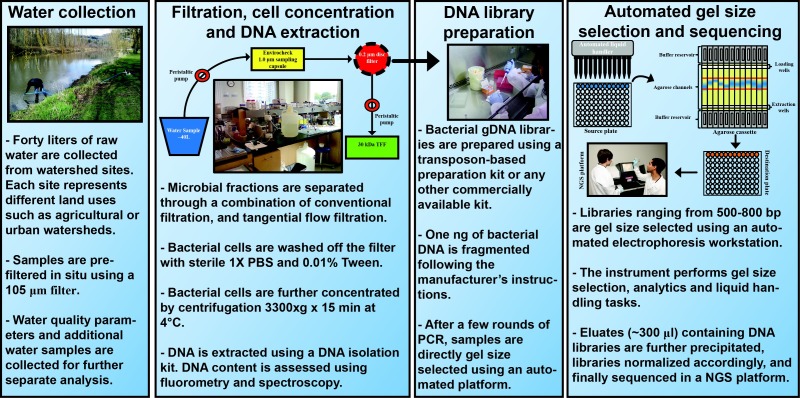 Figure 2.
Schematic representation of methods used to generate transposon-based DNA libraries from freshwater samples.
Please click here to view a larger version of the figure.
Figure 2.
Schematic representation of methods used to generate transposon-based DNA libraries from freshwater samples.
Please click here to view a larger version of the figure.
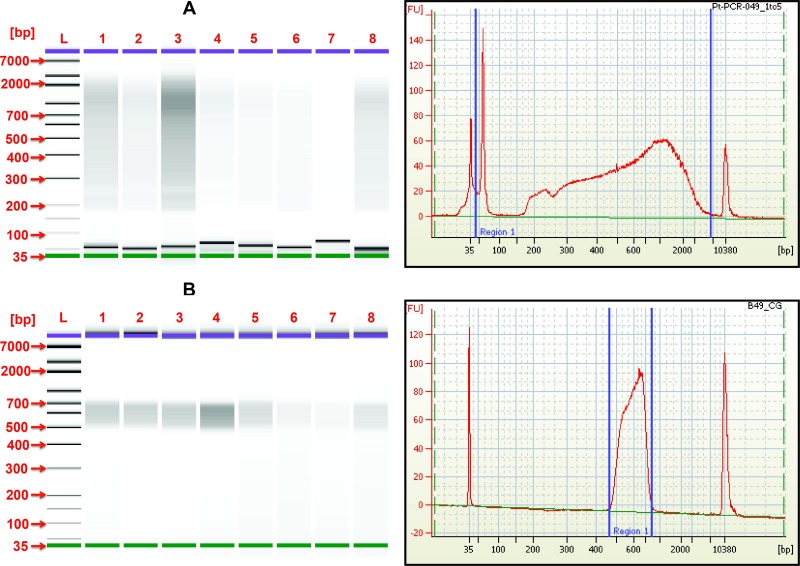 Figure 3.Electropherograms of the DNA libraries generated from freshwater samples: (A) Post-PCR DNA libraries prior to size selection and (B) DNA libraries size selected using the automated gel size selection platform (target range: 500-800 bp). All samples were diluted five-fold with TE buffer.
Figure 3.Electropherograms of the DNA libraries generated from freshwater samples: (A) Post-PCR DNA libraries prior to size selection and (B) DNA libraries size selected using the automated gel size selection platform (target range: 500-800 bp). All samples were diluted five-fold with TE buffer.
| Environmental sample | Bacterial DNA concentration (ng/ml)* | A260/280 | A260/230 | DNA concentration (ng/µl) before size selection, volume: 25 μl | DNA concentration (ng/µl) after size selection, volume: 25 μl |
| 1 | 0.11 (0.16) | 1.8 | 1.4 | 27 | 1.2 |
| 2 | 0.11 (0.20) | 1.8 | 1.3 | 27.4 | 3.3 |
| 3 | 0.10 (0.37) | 1.8 | 1.6 | 19.5 | 3.4 |
| 4 | 0.04 (0.04) | 1.5 | 0.5 | 24.4 | 1.6 |
| 5 | 0.11 (0.13) | 1.7 | 1.0 | 26.1 | 1.1 |
| 6 | 0.05 (0.03) | 1.5 | 0.7 | 15.2 | 0.4 |
| 7 | 0.01 (0.01) | 1.4 | 0.3 | 15.1 | 0.3 |
| 8 | 0.03 (0.05) | 1.6 | 0.7 | 17 | 0.9 |
| *Values indicate dsDNA concentration using a high sensitivity fluorescent nucleic acid quantitation assay. Number in parentheses represents concentration using a microvolume spectrophotometer. |
Table 1. Quality and quantity assessment of DNA extracted from environmental freshwater samples, and transposon-based libraries before and after automated gel size selection.
Discussion
The presence of adapter dimers and clustering of small insert sizes preferentially being sequenced in current platforms represent a decrease in useable yields and under-utilization of the equipment’s capacity15. The use of bead beating methods combined with a transposon-based approach to prepare libraries may result in more DNA shearing compared to other methods of extraction4,5. Nevertheless, all methods of extraction and library preparation potentially introduce biases to the distribution of sequencing reads8,16. Experimental observations using an automated nucleic acid extraction platform, combined with a ligation-based method, did not seem to remove the excess of adapter dimers from shotgun libraries (data not shown). This study employed a modified protocol to prepare libraries from environmental samples by size selecting DNA fragments within a very specific range, thereby minimizing carryover of adapter dimers or small DNA fragments. The use of an automated gel size selection platform generated reproducible traces of DNA libraries within a specific range of 500-800 bp (Figure 3). While a larger number of raw reads (~1.5 million) were obtained using standard treatment (paramagnetic beads at 0.5x ratio) compared to other cleanups (i.e., 2 times at 0.5x ratio) and the modified protocol here described, only 55% of those reads per sample were greater than 225 bp. The percentage of reads per sample using the modified protocol generated at least 75% of reads greater than 225 bp. In the standard treatment, the large number of reads indicated an over representation of short reads in the library. Fragment sizes selected with paramagnetic beads (standard treatment) have been reported to be variable as compared to gel size selection approaches17. This modified protocol generated more informative reads than paramagnetic beads. The use of gel selection to build libraries has also indicated a reduction in the incidence of chimeras from ~5% to 0.02%9.
Critical steps in the present protocol include considerations for normalization of input DNA, amount of DNA in the libraries, and chemistry of the PCR master mix. Depending on the starting input DNA concentration, it may be time-consuming to measure and dilute the DNA down to an amount of 1 ng required by the transposon-based method used here and thus faster automatable bead-based approaches to normalize input DNA can also be considered in the initial sample preparation18. Following tagmentation and incorporation of indexes, samples were directly size selected using the electrophoresis workstation described in this study. A critical step in this procedure is the amount of DNA loaded to the system. The minimum sample DNA concentration for optical detection depends on the nature of the sample (amplicon versus shotgun sequencing). Intrinsic recovery yields have been shown to hold up with low-input samples and recovery efficiencies >75%. The lowest DNA amount that can be detected by the automated gel size selection system described here is 1.5 ng of total DNA (Nesbitt, M. and Slodoban, J., 2014, unpublished data). Nevertheless, optical detection of the sample is not required to conduct size selection, as dual dye markers are used to determine where to size select a specific target. Before libraries prepared from environmental samples were loaded into the platform, DNA concentration was quantified using a fluorometer. A relatively narrow range of DNA concentrations was detected in post-PCR samples (Table 1). Although these samples contained a mixture of salts, dimers, DNA polymerase, amplified environmental DNA, and undisclosed reagents19, these concentrations represented at least 252-fold higher than the reported lowest amount of DNA detectable by the electrophoresis workstation. Normally, size selection of DNA fragments would be carried out in a buffer solution instead of the PCR master mix. Even though information regarding PCR master mix used in this protocol was undisclosed from the manufacturer, preliminary tests were conducted to track migration of dual dye markers in the mix (data not shown). A high ionic strength buffer in the PCR master mix will alter the electrophoretic mobility of the sample, and thus this variable needs to be accommodated for. The automated gel selection platform described in this study exposes an option to indicate if a sample is in high ionic strength. When this option is used, then altered electrophoretic mobility curves and delayed band tracking parameters are employed. Therefore, it is possible that the chemistry of the PCR mix will have an impact on the loading buffer and markers, and affects migration of DNA fragments in the gel.
There are a few systems commercially available for automated or semi-automated gel size selection. These instruments also provide separation, recovery, and documentation in less than 1 hr. Nevertheless, there are limitations when it comes to number of samples that these systems can handle, and the costs associated with consumables17,20. In this context, the technique described here handles aspects of both agarose gel size selection and analytics. Up to 96 samples can be size selected in a single run, while up to 192 samples can be characterized with high resolution electrophoresis. Furthermore, the instrument can also complete the generic liquid handling tasks users expect from an automated workstation.
Raw elution volume of this protocol ranges from 100 to 400 μl, depending on the target range (25 bp to 40 kb). While this aspect of the instrument may be perceived as a limitation, it must be noted that DNA libraries eluted with a volume of 300 μl would have an average concentration of 301 pM (data not shown). This resultant value represents a 26-fold higher than the required final concentration for sequencing in a MiSeq platform (~11.5 pM). In the present study, ethanol precipitation represented an additional step in terms of time employed in library preparation. Currently, the elution volume can be further reduced by using an on-deck vacuum filtration step that uses a commercial filter plate and a vacuum manifold that fits on the deck (upgrade). Using this setup, the raw elution volume is transferred into the filter plate, concentrated and then resuspended in 25 μl (Slodoban, J., 2014, personal communication). For this study, DNA precipitation was conducted to keep the workflow consistent with the input volume needed for normalization sample step as described in the transposon-based kit.
This modified technique can be applicable to DNA or cDNA libraries prepared from other environmental samples such as seawater, wastewater treatment plant, sediment, soil, or microbial mats. The automated gel size selection platform reported here can be applied with minimum modifications not only to Illumina sequencing platforms, but also to other next-generation sequencing platforms including Roche 454, Life Technologies, and Pacific BioSciences. Special considerations to adapt this technology to other platforms include loading well capacity (up to 51 μl), sizing references or markers, and percentage of agarose gel cassette (dependent on target fraction). This high-throughput automated gel size selection technique is a discriminating form of purification and is adequate for low cost electrophoretic characterizations. Other applications that can benefit from this technology include RNA-Seq projects, long fragment sequencing (including functional metagenomics), gene synthesis, mate pair library construction, de novo and complex genome sequencing, restriction digest analysis, and genotyping.
Disclosures
Jared R. Slobodan and Matthew J. Nesbitt both hold shares of Coastal Genomics, a privately owned British Columbia corporation offering the Ranger Technology.
Acknowledgments
This work was funded by Genome BC, Genome Canada, and Coastal Genomics. The authors thank Kirby Cronin and Michael Chan for their help in sample collection and processing. We also would like to acknowledge Thea Van Rossum and Dr. Fiona Brinkman for bioinformatics assistance.
References
- Siuda W, Chrost RJ. Concentration and susceptibility of dissolved DNA for enzyme degradation in lake water - some methodological remarks. Aquatic Microbial Ecology. 2000;21:195–201. [Google Scholar]
- Butler JM, Hill CR. Scientific Issues with Analysis of Low Amounts of DNA. 2010. Available from: http://www.promega.ca/resources/profiles-in-dna/2010/scientific-issues-with-analysis-of-low-amounts-of-dna.
- Ficetola GF, Miaud C, Pompanon F, Taberlet P. Species detection using environmental DNA from water samples. Biology letters. 2008;4:423–425. doi: 10.1098/rsbl.2008.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liles MR, et al. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Applied and environmental microbiology. 2008;74:3302–3305. doi: 10.1128/AEM.02630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey BS, Fichot EB, Dayama G, Decho AW, Norman RS. Extraction of high molecular weight DNA from microbial mats. BioTechniques. 2010;49:631–640. doi: 10.2144/000113486. [DOI] [PubMed] [Google Scholar]
- Tebbe CC, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Applied and environmental microbiology. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solonenko SA, et al. Sequencing platform and library preparation choices impact viral metagenomes. BMC genomics. 2013;14:320. doi: 10.1186/1471-2164-14-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome biology. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Swerdlow H, Turner DJ, et al. Improved protocols for the illumina genome analyzer sequencing system. Current protocols in human genetics / editorial board, Jonathan L. Haines .. [et al.] 2009;18:18. doi: 10.1002/0471142905.hg1802s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head SR, et al. Library construction for next-generation sequencing: overviews and challenges. BioTechniques. 2014;56:61–64, 66, 68. doi: 10.2144/000114133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk EL, Jaszczyszyn Y, Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Experimental cell research. 2014;322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Grunenwald H, Baas B, Caruccio N, Syed F. Rapid, high-throughput library preparation for next-generation sequencing. Nature Methods. 2010;7 [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. 3rd edn. Cold Spring, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Lee PY, Costumbrado J, Hsu CY, Kim YH. Agarose gel electrophoresis for the separation of DNA fragments. Journal of visualized experiments. 2012. [DOI] [PMC free article] [PubMed]
- Quail MA, et al. A large genome center's improvements to the Illumina sequencing system. Nature methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine R, et al. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Applied and environmental microbiology. 2011;77:8071–8079. doi: 10.1128/AEM.05610-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. Cost-effective high-throughput DNA sequencing libraries for multiplexed target capture. Genome research. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamble S, et al. Improved workflows for high throughput library preparation using the transposome-based nextera system. Bmc Biotechnology. 2013;13 doi: 10.1186/1472-6750-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome research. 2014. [DOI] [PMC free article] [PubMed]
- Rhodes J, Beale MA, Fisher MC. Illuminating Choices for Library Prep: A Comparison of Library Preparation Methods for Whole Genome Sequencing of Cryptococcus neoformans Using Illumina HiSeq. PloS One. 2014;9:e113501. doi: 10.1371/journal.pone.0113501. [DOI] [PMC free article] [PubMed] [Google Scholar]


