Abstract
Neutrophil Extracellular Traps (NETs) have been recently identified as part of the neutrophil’s antimicrobial armamentarium. Apart from their role in fighting infections, recent research has demonstrated that they may be involved in many other disease processes, including cancer progression. Isolating purified NETs is a crucial element to allow the study of these functions.
In this video, we demonstrate a simplified method of cell free NET isolation from human whole blood using readily available reagents. Isolated NETs can then be used for immunofluorescence staining, blotting or various functional assays. This enables an assessment of their biologic properties in the absence of the potential confounding effects of neutrophils themselves.
A density gradient separation technique is employed to isolate neutrophils from healthy donor whole blood. Isolated neutrophils are then stimulated by phorbol 12-myristate 13-acetate (PMA) to induce NETosis. Activated neutrophils are then discarded, and a cell-free NET stock is obtained.
We then demonstrate how isolated NETs can be used in an adhesion assay with A549 human lung cancer cells. The NET stock is used to coat the wells of a 96 well cell culture plate O/N, and after ensuring an adequate NET monolayer formation on the bottom of the wells, CFSE labeled A549 cells are added. Adherent cells are quantified using a Nikon TE300 fluorescent microscope. In some wells, 1000U DNAse1 is added 10 min before counting to degrade NETs
Keywords: Immunology, Issue 98, Neutrophil, neutrophil extracellular traps, inflammation, bacteria, infection, cancer, metastasis, adhesion
Introduction
Neutrophil extracellular traps (NETs) are newly discovered neutrophil-derived elements composed of strands of extracellular DNA decorated by hundreds of proteins and histones. They are secreted by neutrophils in the context of infection and inflammation following specific stimuli and they were initially recognized to be part of the neutrophil’s defense mechanisms against infections. They were first shown to promote trapping of various microbial invaders including bacteria, viruses and fungi 1-3. Trapping of the microbe would then lead to its destruction both directly by NET-associated proteins 3,4 and indirectly through local recruitment of phagocytic cells 5,6. The role of NETs however does not seem to be limited to host defense. More recently, they have been shown to play an important role in autoimmune diseases 7,8, thrombosis 9, pregnancy-related disorders 10 and even cancer progression 11,12. Accordingly, it appears that NETs play an important role in a large number of diverse physiologic processes. However, given the novel nature of such research, much remains to be elucidated with respect to the mechanisms by which NETs exert their effects. Herein, we present a simplified method for the isolation of NETs in the absence of neutrophils.
In the following video, we demonstrate a simplified and easy technique to isolate NETs from human whole blood. Cell-free NETs can then be used in a number of in vitro experiments including staining, imuunofluorescence, blotting as well as a number of assays such as adhesion, proliferation and migration. Other protocols exist 13, 19, however they are usually lengthy, complex, expensive, and often, low yield 13. This simplified protocol uses basic reagents and decreases the number of steps required to isolate neutrophils, therefore minimizing the length of the procedure while maximizing yield.
The following method combines different techniques for neutrophil isolation previously described in the literature to obtain a simple protocol yielding a very pure sample of live neutrophils. As suggested in the literature, venous blood is collected in EDTA tubes and used within 10 min to avoid neutrophil activation 14. We use Lymphocyte Separation Media (LSM) density gradient centrifugation to isolate granulocytes and red blood cells from heparinized whole blood, a method adapted from the Ficoll density centrifugation technique initially described by Boyum et al 15. LSM is a modification of the Boyum formulation that substitutes sodium diatrizoate for the sodium metrizoate and has been used successfully in many studies to isolate neutrophils 16-18.
Following differential density centrifugation, monocytes and lymphocytes are discarded; red blood cells are then sedimented using a 6% Dextran solution 14 and the remaining RBCs are lysed to obtain a population of pure neutrophils, which is verified with Trypan blue and Methylene blue staining.
Numerous agents have been used to induce NETosis both in vitro and in vivo, including lipopolysaccharide (LPS), and phorbol myristate acetate (PMA) and interleukins (IL-8) 3,5,6. In the following protocol, isolated neutrophils are stimulated with 500 nM PMA for 4 hr, which has been shown to be an adequate concentration to allow consistent and reliable NET formation without promoting apoptosis 19,20.
After isolation, we demonstrate how cell-free NETs obtained from this protocol can be used in an adhesion assay. DNAse1 is used to digest and degrade NETs as previously described and thus serves as a control 2,3,11. Other options include the use of Neutrophil elastase inhibitor (NEi) to inhibit NET formation, which is an acceptable alternative when the goal is to inhibit the NET formation rather than effect their degradation 11. Although NEi has a number of varied functions, it has been previously shown that NET deposition can be inhibited by NEi through blockage of chromatin decondensation, nuclear degranulation and neutrophil death 12
Protocol
NOTE: All experiments were conducted in accordance with local institutional ethical guidelines.
1. Blood Drawing
Through antecubital venipucture, collect 14 tubes of blood from a healthy volunteer into green top (heparinized) tubes and invert each tube before settling on ice. Collect blood within 10 - 15 min of experiment to ensure optimal neutrophil yield.
2. Neutrophil Isolation from Whole Blood
Wash each green top tube of blood with 5 ml of PBS without Ca and Mg to dilute the blood. In each of 4 x 50 ml conical tubes, add 15 ml of Lymphocyte Separation Media (LSM) at RT. Using a 18 G needle mounted on a 60 ml syringe, carefully layer the diluted blood onto the LSM, creating a sharp LSM-blood interface. NOTE: Avoid mixing of the layers as much as possible.
Centrifuge at 800 x g for 30 min at 21 °C without break. NOTE: Sudden breaks can cause mixing of the different layers.
Observe the erythrocytes and neutrophils sediment at the bottom. Aspirate and discard the top 2 layers (plasma and LSM) making sure to get rid of the interface between these layers which contains lymphocytes and mononuclear cells, leaving only the bottom red layer.
To each tube, add 20 ml of Phosphate Buffer Saline (PBS) and 20 ml of 6% Dextran solution. Gently invert each tube to mix with the layer of erythrocytes and neutrophils and allow to sit at RT for 30 min. NOTE: This will allow red blood cells (RBCs) to sediment at the bottom.
After 30 min, transfer the neutrophil rich supernatant into fresh tubes and discard the RBC pellets. Centrifuge supernatants at 450 x g at 4 °C for 5 min. After centrifugation, discard supernatant. The pellet will contain mostly neutrophils and few RBCs.
Prepare a lysing solution by adding 0.5 ml of lysing buffer to 4.5 ml of sterile water. Lyse remaining RBC with the prepared 5 ml of lysing solution, transferring all resuspended pellets into one tube. Allow lysis at RT in the dark for 10 min.
Centrifuge at 450 x g at 4 °C for 5 min. Discard supernatant and wash pellet with 5 ml PBS without Ca and Mg and centrifuge again at 450 x g at 4 °C to get rid of any remaining lysing solution. The pellet obtained at this point contains the neutrophils. Resuspend the pellet in 30 ml of cold 3% RPMI and put on ice. NOTE: 3% RPMI is made by supplementing RPMI media with 3% Fetal Bovine Serum (FBS)
Verify purity of the sample using Methylene blue staining. >95% of cells should be granulocytes with multi-lobar nuclei. Use Trypan blue staining to verify >95% viability of cells and count the final neutrophil yield. Dilute neutrophils in ice cold 3% RPMI to obtain a final concentration of 5 x 106 neutrophils/ml.
3. NETs Generation
Stimulate neutrophils with 500 nM of PMA (per 30 ml of neutrophil solution) and incubate on a 150 x 25mm flat tissue culture dish with 20 mm grid for 4 hr at 37 °C 5% CO2. This will allow NETosis.
After 4 hr of stimulation, gently aspirate and discard the media, leaving the layer of NETs and neutrophils adhered at the bottom. Do not disrupt this layer.
Using a total of 15 ml of cold PBS without Ca and Mg per dish, wash the bottom of each dish by pipetting 15 ml of PBS on the bottom of the dish in order to lift off all adherent material from the bottom.
Collect solution obtained from washing each dish (step 3.3) in a 15 ml conical tube and centrifuge for 10 min at 450 x g at 4 °C. Neutrophils and any remaining cells will pellet at the bottom, leaving a cell-free NET-rich supernatant.
Divide supernatant into 1.5 ml micro-centrifuge tubes and spin for 10 min at 18,000 x g at 4 °C. This will allow all DNA to pellet. NOTE: Centrifugation in larger tubes is easier and less time consuming if such high speeds are attainable on the regular centrifuge, otherwise will need to divide supernatants in smaller tubes in order to use high speed micro-centrifuge.
Discard supernatant and resuspend all pellets obtained together in ice cold PBS to a concentration corresponding to 2 x 107 neutrophils per 100 μl of PBS. This will yield the cell-free NET stock that can be used for subsequent experiments.
Measure DNA concentration in the sample obtained using spectrophotometry or alternate DNA quantification tool. An adequate concentration in the sample should range between 140 - 180 ng/μl.
4. NET-Cancer Cell Static Adhesion Assay
Add 100 μl of the previously obtained NET stock per well in a 96 well flat bottom plate and allow to coat wells O/N at 4 °C in the dark.
Between 12 and 20 hr later, verify formation of a uniform monolayer of cell free NETs at the bottom of the wells under the microscope (Figure 1). At this point, gently aspirate all non-adherent material out of the wells, making sure not to disrupt the NET monolayer at the bottom.
Add 100 μl of 1% Bovine Serum Albumin (BSA) blocking solution to each well and leave for 1 h at RT.
Detach A549 cancer cells from a T-75 flask of using 2 ml of 0.25% Trypsin solution. Once detached, add 10 ml of A549 media to trypsinized cells and centrifuge at 450 x g at 4 °C for 5 min. Discard supernatant and resuspend cells in media to obtain a concentration of 2 x104 cancer cells per 100 μl media. NOTE: A549 cells were grown separately. Briefly, cells were cultured and maintained in DMEM F12 media containing 10% FBS and 1% Penicillin Streptomycin and incubated at 37 °C 5% CO2. Once 70 - 80% cell confluence was reached, they were detached using 0.25% Trypsin-EDTA and resuspended in the same media described above.
Stain cancer cells using CFSE by adding 1 μl of CFSE per ml of media and leave to stain at RT for 10 min. After staining, centrifuge down at 450 x g at 4 °C for 5 min then discard supernatant and resuspend cells in initial volume of media to obtain a concentration of 2 x104 cancer cells per 100 μl media.
After 1 hr of NETs blocking, gently aspirate blocking solution and add 2 x104 cancer cells in media, which is equivalent to 100 μl, per well over the NET monolayer and allow to adhere for 90 min at 37 °C 5% CO2. Gently aspirate the cells and add 100 μl of PBS to each well to wash any non-adherent cells.
In some wells, add 1000U of DNAse1 per well for 10 min before washing to degrade NETs. This will serve as negative control. In other wells, add 100 μl of sterile water per well for 10 min, which will serve as the vehicle control (VC). NOTE: 3 replicates per condition are usually performed to increase sample size.
Aspirate and discard all solution in the wells leaving only NETs and adherent cancer cells at the bottom. Add 100 μl of 4% formaldehyde solution per well to fix adherent cancer cells to NETS and read assay under the fluorescence microscope (Figure 1). Plot and analyze the results (Figure 3).
Representative Results
Successful achievement of a static adhesion assay with NETs requires adequate coating of the wells with a monolayer of NETs before the addition of cancer cells (Figure 1). All subsequent steps must be done with care not to disrupt that layer. When an adequate layer of NETs is formed, it is expected to see significant cancer cells adhesion to NETs as seen in Figure 2A and 2C. This effect is abrogated by the addition of DNAse1, which degrades NETs as seen in Figures 2B and 2D. Conversely, there should be no significant decrease in A549 adhesion to NETs in the presence of the vehicle control (Figure 3). To obtain a good estimation of mean cancer cell adhesion per high power field (hpf), it is recommended to count at least 4 random hpfs per well. Fields that are not well coated in NETs due to technical issues should not be taken into consideration for counting as these might falsify results.
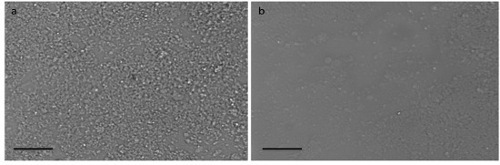 Figure 1. NET Monolayer. Light microscopy images of the 96 wells plate wells coated with NETs showing (A) uniform monolayer of NETs throughout well as opposed to (B) inadequate coating of the well with an interrupted layer of NETs. Scale bars represent 40 μm. Please click here to view a larger version of this figure.
Figure 1. NET Monolayer. Light microscopy images of the 96 wells plate wells coated with NETs showing (A) uniform monolayer of NETs throughout well as opposed to (B) inadequate coating of the well with an interrupted layer of NETs. Scale bars represent 40 μm. Please click here to view a larger version of this figure.
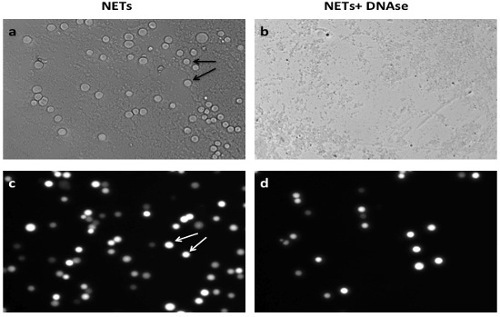 Figure 2. A549 Cancer Cell Adhesion to NETs In Vitro. (A) A549 cancer cells (arrows) adhere to monolayer of NETs in vitro. (B) Addition of DNAse1 decreases significantly the level of cancer cell adhesion; (C) shows a representative image of cancer cell adhesion on NETs under fluorescent light under fluorescent microscopy (Nikon TE300) before (C) and after (D) addition of DNAse1. Scale bars represent 40 μm. Please click here to view a larger version of this figure.
Figure 2. A549 Cancer Cell Adhesion to NETs In Vitro. (A) A549 cancer cells (arrows) adhere to monolayer of NETs in vitro. (B) Addition of DNAse1 decreases significantly the level of cancer cell adhesion; (C) shows a representative image of cancer cell adhesion on NETs under fluorescent light under fluorescent microscopy (Nikon TE300) before (C) and after (D) addition of DNAse1. Scale bars represent 40 μm. Please click here to view a larger version of this figure.
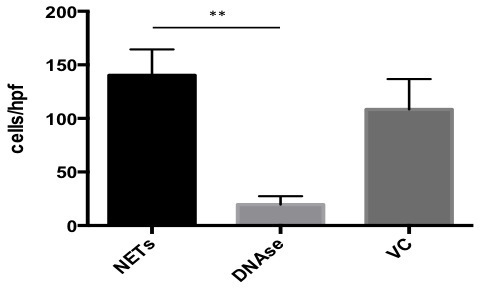 Figure 3. Results for In Vitro Adhesion Assay. GraphPad software was used to plot and analyze results from the A549 cancer cell adhesion assay to NETs. In the presence of DNAse, adhesion was 13.90% compared to untreated NETs. In the presence of vehicle control (VC), adhesion was 77.26% compared to untreated NETs. Data is presented as mean +/- SEM. Significance was determined using Kruskall Wallis test with **p<0.001.
Figure 3. Results for In Vitro Adhesion Assay. GraphPad software was used to plot and analyze results from the A549 cancer cell adhesion assay to NETs. In the presence of DNAse, adhesion was 13.90% compared to untreated NETs. In the presence of vehicle control (VC), adhesion was 77.26% compared to untreated NETs. Data is presented as mean +/- SEM. Significance was determined using Kruskall Wallis test with **p<0.001.
Discussion
The Neutrophil Extracellular Trap isolation protocol demonstrated in this video combines different techniques used in the literature for neutrophil isolation and NET formation. It simplifies a fairly complex and variable process and offers a very reliable and replicable way to isolate purified cell-free NETs with fewer steps than other protocols. Furthermore, readily available reagents are employed adding to the protocols simplicity and significantly reducing its cost. The application of NETs towards a static adhesion assay serves to demonstrate the flexibility afforded by this isolation technique, as NET concentration can easily be manipulated to suit a particular goal (17). Accordingly, NETs isolated by the demonstrated technique can be used for various types of assays including dynamic adhesion assays, migration assays, proliferation assays immunofluorescence and confocal imaging, electron microscopy, flow cytometry as well as traditional Western blotting.
There are a number of steps in this protocol that are critical for its success. First, neutrophils may be accidentally activated during the process of isolation leading to apoptosis. It is therefore important to perform all isolation steps under sterile conditions under the fume hood, avoid aggressive mixing of tubes, keep all samples on ice after the RBC lysis step, and avoid long wait times between steps. Second, if the goal of the assay is to coat wells for the purpose of assessing adhesion, it is important to respect the suggested NET concentration per well as well as the proposed final DNA concentration in the “NET stock” as this will ensure an even and consistent monolayer of NETs. Lower concentration may produce only patchy coating of the well and therefore less reliable results. Finally, it is also important to handle the NET monolayer very carefully while performing the assay to prevent its disruption. This can be facilitated by adjusting the intensity of the suction and pipetting on the sides of the wells.
Limitations of this technique include the fact that it still requires a significant amount of time to produce NETs, mostly caused by the 4 hr PMA stimulation step. In addition, a significant amount of blood is required to isolate a sufficient number of neutrophils required for one experiment. There definitely is loss of a significant amount of NETs at the first centrifugation step (step 3.4.) when discarding neutrophils. Modifications to minimize NET loss at this point could significantly increase the final NET yield. Also, isolated NETs must be used within 12 - 24 hr of their isolation, which requires careful experiment planning and time management.
Disclosures
The authors have nothing to disclose
Acknowledgments
The authors would like to acknowledge Dr. Paul Kubes for his guidance and mentoring that were central in the construction of this work.
References
- Saitoh T, et al. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe. 2012;12(1):109–116. doi: 10.1016/j.chom.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Bruns S, et al. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin RodA. PLoS Pathog. 2010;6(4):e1000873. doi: 10.1371/journal.ppat.1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Zychlinsky A. NETs: a new strategy for using old weapons. Trends Immunol. 2009;30(11):513–521. doi: 10.1016/j.it.2009.07.011. [DOI] [PubMed] [Google Scholar]
- McDonald B, Urrutia R, Yipp BG, Jenne CN, Kubes P. Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host Microbe. 2012;12(3):324–333. doi: 10.1016/j.chom.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Pilsczek FH, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol. 2010;185(12):7413–7425. doi: 10.4049/jimmunol.1000675. [DOI] [PubMed] [Google Scholar]
- Villanueva E, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. 2011;187(1):538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandpur R, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5(178):178ra140. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers M, et al. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc Natl Acad Sci U S A. 2012;109(32):13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Giaglis S, Hoesli I, Hasler P. Neutrophil NETs in reproduction: from infertility to preeclampsia and the possibility of fetal loss. Front Immunol. 2012;3:362. doi: 10.3389/fimmu.2012.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools-Lartigue J, et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest. 2013. [DOI] [PMC free article] [PubMed]
- Cools-Lartigue J, Spicer J, Najmeh S, Ferri L. Neutrophil extracellular traps in cancer progression. Cell Mol Life Sci. 2014. [DOI] [PMC free article] [PubMed]
- Brinkmann V, Laube B, Abu Abed U, Goosmann C, Zychlinsky A. Neutrophil extracellular traps: how to generate and visualize them. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Maqbool M, Vidyadaran S, George E, Ramasamy R. Optimisation of laboratory procedures for isolating human peripheral blood derived neutrophils. Med J Malaysia. 2011;66(4):296–299. [PubMed] [Google Scholar]
- Boyum A. Isolation of lymphocytes, granulocytes and macrophages. Scand J Immunol. Suppl. 1976;5:9–15. [PubMed] [Google Scholar]
- Calado RT, et al. Sex hormones, acting on the TERT gene, increase telomerase activity in human primary hematopoietic cells. Blood. 2009;114(11):2236–2243. doi: 10.1182/blood-2008-09-178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Qu X, Tan M, Meng YG, Ferrara N. Characterization and regulation of bv8 in human blood cells. Clin Cancer Res. 2009;15(8):2675–2684. doi: 10.1158/1078-0432.CCR-08-1954. [DOI] [PubMed] [Google Scholar]
- Wu YJ, et al. In vivo leukocyte labeling with intravenous ferumoxides/protamine sulfate complex and in vitro characterization for cellular magnetic resonance imaging. Am J Physiol Cell Physiol. 2007;293(5):C1698–C1708. doi: 10.1152/ajpcell.00215.2007. [DOI] [PubMed] [Google Scholar]
- Saffarzadeh M, et al. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 2012;7(2):e32366. doi: 10.1371/journal.pone.0032366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]


