Abstract
Longevity regulatory genes include the Forkhead transcription factor FOXO and the NAD-dependent histone deacetylase silent information regulator 2 (Sir2). Genetic studies demonstrate that Sir2 acts to extend lifespan in Caenorhabditis elegans upstream of DAF-16, a member of the FOXO family, in the insulin-like signaling pathway. However, the molecular mechanisms underlying the requirement of DAF-16 activity in Sir2-mediated longevity remain unknown. Here we show that reversible acetylation of Foxo1 (also known as FKHR), the mouse DAF-16 ortholog, modulates its transactivation function. cAMP-response element-binding protein (CREB)-binding protein binds and acetylates Foxo1 at the K242, K245, and K262 residues, the modification of which is involved in the attenuation of Foxo1 as a transcription factor. Conversely, Sir2 binds and deacetylates Foxo1 at residues acetylated by cAMP-response element-binding protein-binding protein. Sir2 is recruited to insulin response sequence-containing promoter and increases the expression of manganese superoxide dismutase and p27kip1 in a deacetylase-activity-dependent manner. Our findings establish Foxo1 as a direct and functional target for Sir2 in mammalian systems.
The mammalian FOXO family of forkhead transcription factors, Foxo1 (also known as FKHR), Foxo3a (also known as FKHRL1), and Foxo4 (also known as AFX) plays a key role in transmitting insulin signaling downstream of protein kinase B (also called Akt). In response to insulin, FOXO proteins are phosphorylated by protein kinase B, resulting in their nuclear exclusion (1–3) and subsequent degradation through ubiquitination (4). Previous studies have established the diverse functions of FOXO proteins, including glucose metabolism (5–9), cell-cycle regulation (10–14), apoptosis (1, 11, 15–17), and oxidative stress resistance (18, 19). Notably, accumulating evidence from works in Caenorhabditis elegans has uncovered that genetic determinants of longevity involve DAF-16, the nematode FOXO ortholog, downstream of the insulin/insulin-like growth factor I signaling pathway (20–23).
A second longevity regulatory gene is the evolutionarily conserved histone deacetylase silent information regulator (Sir2) (24, 25). Sir2 activity depends on NAD (nicotinamide adenine dinucleotide, oxidized form) (26) and links to various biological functions in budding yeast through transcriptional silencing of the silent mating-type loci, telomeres, and ribosomal DNA repeats (27). Moreover, Sir2 increases longevity by suppressing ribosomal DNA homologous recombination (28), and calorie restriction (CR), which extends lifespan in a wide variety of species, requires the NAD-dependent Sir2 activity in yeast (25). Interestingly, genetic analysis demonstrated that, similar to its yeast counterpart, Sir2 extends lifespan in the nematode by negatively regulating a signaling pathway involving insulin-like hormones upstream of DAF-16, a member of the FOXO family (24). However, the molecular mechanisms underlying the requirement of FOXO activity in Sir2-mediated longevity remain unclear.
Here we show that Sir2 binds Foxo1 in vivo and in vitro and deacetylates it with specificity for K242, K245, and K262 residues, which are involved in transcriptional attenuation by cAMP-response element-binding protein-binding protein (CBP)-induced acetylation. Furthermore, Sir2 enhances Foxo1-mediated transcription in a deacetylase-activity-dependent manner. These findings indicate that Sir2 serves as not a transcriptional silencer of gene(s) for the insulin-signaling pathway but rather as a transcriptional coactivator of Foxo1 that in turn increases antioxidant gene expression.
Experimental Procedures
Plasmids and Antibodies. The mutants of Foxo1 phosphorylation and acetylation sites were generated by site-directed mutagenesis. GST-Foxo1 deletion mutants were made by digesting the full-length cDNAs with appropriate enzymes or PCR-based subcloning into pGEX-5X (Amersham Pharmacia). p3×IRS-MLP-luc was generated by inserting three copies of the insulin-response sequence derived from the insulin-like growth factor-binding protein 1 promoter into the luciferase reporter gene vector containing an adenovirus major late promoter. GAL4 DNA-binding domain-fused Foxo1 plasmid was constructed in pGBT9 vector and subcloned into the pcDNA3 vector. Mouse Sir2 cDNA in the pUSEamp vector was purchased (Upstate Biotechnology, Lake Placid, NY), and a hemagglutinin (HA) tag was inserted at the C terminus of Sir2 cDNA. A catalytically inactive mutant of Sir2, H355A, was generated by site-directed mutagenesis. The following antibodies were used: anti-HA (12CA5, Roche, Gipf-Oberfrick, Switzerland), anti-FLAG (M2, Sigma), anti-manganese superoxide dismutase (MnSOD) (Stressgen Biotechnologies, Victoria, Canada), anti-p27kip1 (Transduction Laboratories, Lexington, KY), anti-β-actin (Sigma), anti-acetylated lysine, anti-Sir2, and anti-CBP-CT (Upstate Biotechnology). The anti-Foxo1 antibody was as described (8). Rabbit polyclonal antibody specific for Foxo1 acetylated at both K242 and K245 was raised against the Foxo1 peptide GKSGKSPRRR.
Cell Culture, Transfections, and Luciferase Assays. HepG2, HEK293, and HEK293T cells were cultured in DMEM supplemented with 10% FBS. To establish FK-1 and GM-1 cells, HEK293 cells were stably transfected with 3×IRS-MLP-luc and 5×UAS-MLP-luc plasmids, respectively. Transfections were performed by using FuGENE-6 (Roche). After transfection, cells were incubated in DMEM with 10% FBS for 24 h. Whereas GM-1 cells were continuously incubated for 18 h, FK-1 cells were serum-starved for 18 h. Luciferase and β-galactosidase assays were performed in triplicate, and representative experiments in the figures depict the average of three experiments with standard deviations indicated. The experiments in stably transfected cells were replicated in three independent clones to avoid clonal artifacts.
Immunoprecipitation and Western Blotting. HepG2 cells were serum-starved for 18 h and lysed in lysis buffer A (10 mM Hepes, pH 7.5/100 mM KCl/0.1% Nonidet P-40/protease inhibitors). The lysates were immunoprecipitated with anti-Foxo1 or anti-CBP-CT antibodies. To detect Foxo1 acetylation in vivo, HEK293T cells expressing FLAG-Foxo1 were lysed and immunoprecipitated in lysis buffer B (50 mM Hepes, pH 7.5/150 mM KCl/1% Triton X-100/10% glycerol/5 mM sodium butyrate/protease inhibitors) and followed by Western blotting. For preparation of CBP or Sir2 proteins, HEK293T cells transfected with CBP-HA or Sir2-HA were lysed in radioimmunoprecipitation assay buffer (10 mM Hepes, pH 7.2/150 mM NaCl/1% Nonidet P-40/0.1% SDS/1 mM EDTA/protease inhibitors) and immunoprecipitated with anti-HA antibody. After washing the beads, the immunoprecipitated CBP or Sir2 was used for in vitro assays.
GST Pull-Down Assays. GST fusion proteins were expressed in Escherichia coli strain BL-21 by using the pGEX vector system. In vitro binding assays were performed by incubating cell extracts from transfected HEK293T cells with various GST-fused proteins immobilized on glutathione-Sepharose in lysis buffer A. After incubation for 4 h at 4°C, the beads were washed four times with the same buffer, and proteins were analyzed by Western blotting.
In Vitro Acetylation Assays. Two micrograms of various GST-Foxo1 fusion proteins were incubated at 30°C for 1 h with immunoprecipitated CBP and 0.5 μl of [14C]acetyl-CoA [25 nCi (1 Ci = 37 GBq); Amersham Pharmacia Biotech] in the reaction buffer (50 mM Tris·HCl, pH 8.0/100 mM NaCl/10% glycerol/0.1 mM EDTA/1 mM DTT/1 mM PMSF/5 mM sodium butyrate). Reaction products were analyzed by Coomassie brilliant blue staining and autoradiography.
In Vitro Deacetylation Assays. GST-FHD and GST-C1 were first acetylated in vitro by using a GST-CBP-histone acetyltransferase (HAT) and 0.1 mM acetyl-CoA in the acetylation buffer. After washing the beads, the acetylated Foxo1s were incubated with purified Sir2, 1 mM NAD (Sigma), 5 mM nicotinamide (NIA) (Wako Biochemicals, Osaka), or 1 μM trichostatin A (Wako Biochemicals) as indicated at 30°C for 1 h. The reactions were performed in a buffer containing 50 mM Tris·HCl (pH 9.0), 50 mM NaCl, 4 mM MgCl2, 0.5 mM DTT, 0.2 mM PMSF, 0.02% Nonidet P-40, and 5% glycerol. The proteins were analyzed by Western blotting and Ponceau-S staining.
Chromatin Immunoprecipitation Assays. Chromatin immunoprecipitation assays were performed with a previously described protocol (8). Briefly, chromatin from crosslinked HEK293 cells was sheared by sonication and incubated overnight with anti-Foxo1, anti-Sir2, and anti-CBP-CT antibodies or normal rabbit IgG followed by incubation with protein G-Sepharose saturated with salmon sperm DNA. Precipitated DNAs were analyzed by PCR using specific primers for human MnSOD and p27kip1 promoters and the β-actin coding region.
Results
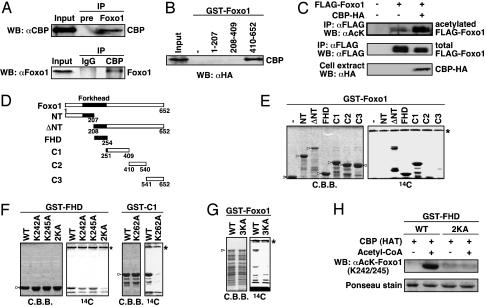
CBP Binds and Acetylates Foxo1 both in Vivo and in Vitro. Because we previously identified Foxo1 as an interactant of CBP by the yeast two-hybrid screen, we tried to confirm the interaction in mammalian cells. The coimmunoprecipitation assay revealed that endogenous Foxo1 and CBP could associate in HepG2 cells (Fig. 1A). This result is consistent with the data demonstrating the interaction with DAF-16 and CBP in a mammalian two-hybrid system (29). We further determined which portions of Foxo1 are important for the interaction with CBP. HA-CBP was transfected into HEK293T cells, and GST-Foxo1 protein mutants with a series of deletions were bacterially expressed. As shown in Fig. 1B, CBP exclusively bound the C terminus of Foxo1, which has been identified as the transactivation domain of Foxo1 (30).
Fig. 1.
CBP binds and acetylates Foxo1. (A) Interaction of endogenous CBP and Foxo1. Cell extracts from serum-starved HepG2 cells were immunoprecipitated (IP) with anti-Foxo1 or anti-CBP antibodies and analyzed by Western blotting (WB). The input lanes represent 5% of the total volume of whole-cell extracts used for the binding assay. (B) In vitro interaction of CBP and Foxo1. Cell extracts from HEK293T cells transfected with HA-CBP WT were incubated with GST or various GST-Foxo1 deletion mutants. (C) Foxo1 is acetylated by CBP. Cell extracts from HEK293T cells transfected with indicated plasmids were immunoprecipitated with anti-FLAG antibody and probed with anti-acetylated lysine or anti-FLAG antibodies. The expression of CBP-HA in the cell extract was shown by Western blotting. (D–G) Identification of Foxo1 acetylation sites. Schematics of Foxo1 deletion mutants are shown in D. The GST-Foxo1 proteins, indicated by arrowheads, were subjected to in vitro acetylation assays with immunoprecipitated CBP. Reaction products were analyzed by Coomassie brilliant blue staining and autoradiography (14C). The asterisk shows the autoacetylated CBP. (H) The anti-acetylated Foxo1 (K242/K245) antibody specifically recognizes the acetylated K242 and K245 residues in Foxo1. The GST-FHD WT or -FHD 2KA (KK242, 245AA) proteins were incubated with GST-CBP (HAT) protein in the presence or absence of acetyl-CoA (0.1 mM) followed by immunoblotting with anti-acetylated-Foxo1 (K242/K245) antibody and staining with Ponceau-S solution.
Next, because several examples of direct transcription factor acetylation by CBP have been described (31), we investigated whether Foxo1 is acetylated by CBP in a cellular condition. HEK293T cells were cotransfected with FLAG-Foxo1 and HA-CBP, and immunoprecipitated Foxo1 was probed with an anti-acetyl lysine antibody. Consistent with previous data in erythroid progenitor cells (32), Foxo1 was acetylated in HEK293T cells, and the extent of its acetylation prominently increased with the coexpression of CBP (Fig. 1C). Furthermore, to identify the acetylation sites of Foxo1 by CBP, a series of GST-fusion Foxo1 deletion mutants were used for in vitro acetylation assays (Fig. 1D). An immunoprecipitated CBP was able to acetylate Foxo1-ΔNT, FHD, and C1 regions, but not NT, C2, or C3 (Fig. 1E). In the forkhead domain, the four lysine residues are present, encompassing amino acids 208–254, and the alanine substitution at the latter two lysine residues, both K242A and K245A, completely abolished acetylation, whereas each mutation still allowed acetylation to a lower extent (Fig. 1F Left). Next, to identify the acetylated lysine residue(s) in the C1 region, we compared its sequence pattern with the CBP/p300-mediated acetylation sites of histone H2B (33) and focused on K262, which is present between alanine and serine residues (AKS). Actually, K262A substitution significantly decreased the acetylation by CBP (Fig. 1F Right). Finally, GST-Foxo1 3KA mutant was not acetylated (Fig. 1G), suggesting that the three lysine residues of Foxo1, K242, K245, and K262, can serve as acetylation sites by CBP in vitro. These results were further confirmed by generating polyclonal antisera specific for both acetylated K242 and K245 (Fig. 1H). Notably, the two acetylated lysine residues, K242 and K245, are addressed in the basic region of the forkhead domain and conserved in its related proteins, Foxo4 and Foxo3a, indicating that modification of the forkhead domain by acetylation may be highly conserved throughout the FOXO family members.
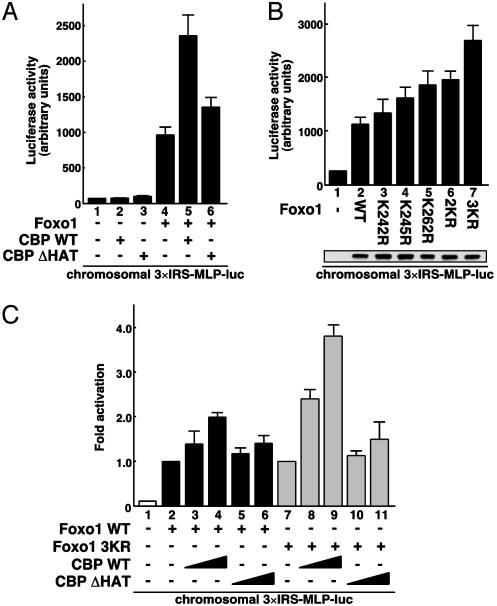
Acetylation of Foxo1 Mitigates Its Transcriptional Activity. To evaluate the effect of CBP on Foxo1-mediated transcription through its acetyltransferase activity, we performed luciferase assays in FK-1 cells, which were HEK293 cell clones chromosomally integrating the 3×IRS-MLP-luc plasmid. As shown in Fig. 2A, coexpression of wild-type CBP (CBP WT) significantly augmented Foxo1-mediated transcription (lanes 4 and 5). On the other hand, coexpression of a HAT inactive mutant (CBP ΔHAT) was less effective on the CBP-mediated transactivation (lane 6), suggesting that CBP serves as a transcriptional coactivator for Foxo1 by acetylating histones and/or Foxo1 on the chromosomal promoter. We next assessed the effect of Foxo1 acetylation by generating various acetylation-deficient Foxo1 mutants (Lys to Arg). Surprisingly, substitutions of the individual lysine residues at K242, K245, and K262 to arginine induced transcriptional activities by 1.5- to 2.0-fold compared with that of Foxo1 WT, and moreover, the triple substitution (3KR) of Foxo1 stimulated the promoter activity by 3.0-fold (Fig. 2B). No differences were observed in the levels of ectopically expressed WT and mutant Foxo1 (Fig. 2B Lower). This finding clearly indicates that acetylation of Foxo1 at the three lysine residues mitigates its transactivating function. Finally, to distinguish between the two substrates of an acetyltransferase CBP, namely histones or Foxo1, the WT or 3KR mutant of Foxo1 was cotransfected with CBP WT or ΔHAT in FK-1 cells. As shown in Fig. 2C, CBP WT exhibited a marked increase in the Foxo1 3KR-mediated transcription compared with Foxo1 WT (lanes 2–4 versus lanes 7–9), suggesting that CBP-induced acetylation of Foxo1, but perhaps not of histones, attenuates its transactivation function. Also, similar to the effect on Foxo1 WT (lanes 5 and 6), CBP ΔHAT was impaired in activating the transcription of Foxo1 3KR (lanes 10 and 11). Thus, we concluded that CBP coactivates Foxo1-mediated transcription, presumably by acetylating chromosomal histones around the insulin response sequence (IRS)-containing promoter and thereby forming the preinitiation complex, but subsequent acetylation of Foxo1 leads in turn to its transcriptional attenuation.
Fig. 2.
CBP regulates Foxo1-mediated transcription through its acetyltransferase activity on the chromosomal reporter. (A) Effect of CBP acetyltransferase activity on Foxo1-mediated transcription. FK-1 cells were transfected with Foxo1 together with either CBP WT or CBP ΔHAT, and the luciferase activity was measured. (B) KR mutations alter Foxo1 transactivation function. FK-1 cells were transfected with the indicated Foxo1 mutants, and the luciferase activity was measured. (Lower) Equal amounts of FLAG-Foxo1 proteins were detected by Western blotting. (C) Coactivation of Foxo1 WT and 3KR by CBP. FK-1 cells were transfected with Foxo1 WT (black boxes) or 3KR (gray boxes) together with CBP, and the luciferase activity was presented as fold induction above the activity obtained by Foxo1 WT or 3KR alone.
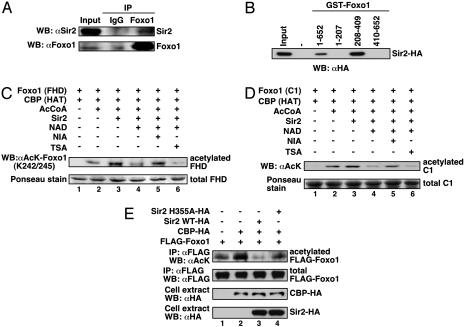
Sir2 Binds and Deacetylates Foxo1. Considering the data discussed above along with the genetic correlation between FOXO/DAF-16 and Sir2 in longevity (24), it was possible that Foxo1 functionally associates with Sir2 as a deacetylase. To test this possibility, we first investigated whether endogenous Foxo1 interacts with endogenous Sir2 in mammalian cells. HepG2 cells were serum-starved, a condition under which Foxo1 accumulates in the nucleus, and a coimmunoprecipitation assay was performed by using an anti-Foxo1 antibody. As expected, the immunoprecipitated Foxo1 complex contained a slight but detectable amount of Sir2 (Fig. 3A). The low efficiency of the coimmunoprecipitation seems to imply a condition, in response to which the binding of Foxo1 and Sir2 would increase. Next, we sought to determine which portions of Foxo1 are important for the interaction with Sir2 in vitro. Sir2 tightly bound immobilized GST-Foxo1 (full length) and its middle region (208–409 aa), which is around the forkhead domain, including the three acetylatable lysine residues (Fig. 3B). To investigate further whether Sir2 could deacetylate Foxo1, we acetylated GST-Foxo1 (FHD, containing the CBP-dependent acetylated-lysine residues K242 and K245) and GST-Foxo1 (C1, containing the CBP-dependent acetylated-lysine residues K262) (Fig. 1D) and performed in vitro deacetylation assays by using an immunoprecipitated Sir2. As shown in Fig. 3 C and D, Sir2 efficiently deacetylated Foxo1 in an NAD-dependent manner. Moreover, the Sir2-mediated deacetylation of Foxo1 was significantly inhibited by NIA, an inhibitor of Sir2 (34), but not by trichostatin A, an inhibitor of class I and II histone deacetylases. These results indicate that Sir2 can preferentially deacetylate Foxo1 at K242, K245, and K262 in vitro. To verify the role for Sir2 in deacetylating Foxo1 in vivo, we transfected Foxo1 and CBP together with either Sir2 WT or a catalytically inactive mutant (H355A) in HEK293T cells. A great deal of Foxo1 acetylation was observed when Foxo1 and CBP were transfected; however, the acetylation level was substantially abrogated by expression of Sir2 WT (Fig. 3E, lanes 1–3). In contrast, Sir2 H355A had no obvious effect on Foxo1 deacetylation (lane 4). These findings demonstrate that Sir2 is a bona fide deacetyltransferase for Foxo1.
Fig. 3.
Sir2 binds and deacetylates Foxo1. (A) Interaction of endogenous Sir2 and Foxo1. HepG2 cells were coimmunoprecipitated (IP) with anti-Foxo1 antibody. The input lanes represent 5% of the total volume of whole-cell extracts used for the binding assay. WB, Western blotting. (B) Association between Sir2 and Foxo1 in vitro. Cell extracts from HEK293T cells transfected with HA-Sir2 were incubated with GST or various GST-Foxo1 deletion mutants. (C and D) Sir2 deacetylates Foxo1 in vitro. Acetylated GST-Foxo1 (FHD) (C) or (C1) (D) proteins were incubated with immunoprecipitated Sir2. NAD (50 μM), NIA (5 mM), and/or trichostatin A (TSA, 1 μM) were added as indicated, and the reactions were analyzed by Western blotting (Upper) and Ponceau-S stain (Lower). (E) Sir2 deacetylates Foxo1 in vivo. Cell extracts from HEK293T cells were transfected with FLAG-Foxo1, HA-CBP, HA-Sir2 WT, or HA-Sir2 H355A as indicated and analyzed by immunoprecipitation and Western blotting.
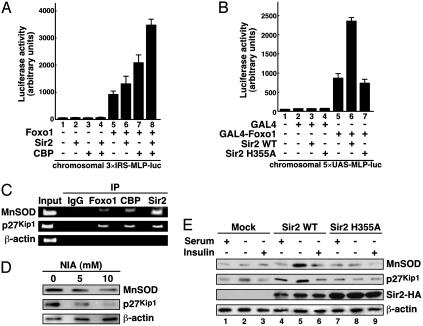
Sir2 Up-Regulates FOXO-Targeted Gene Expression Through Its Deacetylase Activity. To test whether Sir2 changes the properties of Foxo1, transcriptional activities were evaluated in FK-1 cells in which Sir2 and/or CBP expression plasmids were transfected. Although potent transcriptional repression by Drosophila Sir2 has been shown in in vitro transcription systems (35), expression of Sir2, interestingly, had a slight activation on Foxo1-mediated transcription in FK-1 cells (Fig. 4A, lanes 5 and 6). Moreover, coexpression of both CBP and Sir2 together with Foxo1 potentiated the reporter activity 2-fold compared with CBP alone (lanes 7 and 8). Given that the overexpression of CBP augments the level of acetylated Foxo1 (Figs. 1C and 3E), the hypothesis could be drawn that Sir2 might increasingly deacetylate Foxo1, and it therefore up-regulates the transcriptional activity of Foxo1. To gain a better understanding of the consequence of Sir2-dependent deacetylation in Foxo1-mediated transcription, we established stable HEK293 cell clones (referred to as GM-1 cells) containing chromosomally integrated luciferase reporter plasmids with the GAL4 recognition sites (5×UAS-MLP-luc) and carried out a GAL4 mammalian one-hybrid assay. As shown in Fig. 4B, GAL4-Foxo1 stimulated the chromosomal GAL4-luciferase reporter, and coexpression of Sir2 WT significantly enhanced the reporter activity (lanes 5 and 6). Importantly, Sir2 H355A was completely impaired in the synergistic effect on Foxo1-mediated transcription (lane 7). These results implicate Sir2 as a positive cofactor for Foxo1-dependent transactivation through its deacetylase activity.
Fig. 4.
Sir2 coactivates Foxo1-mediated transcription. (A) Sir2 and CBP facilitates Foxo1 transcription in chromatin context. FK-1 cells were transfected with either empty vector or Foxo1 WT together with or without Sir2 and/or CBP as indicated, and the luciferase activity was measured. (B) Sir2 potentiates GAL4-Foxo1-mediated transcription via its deacetylase activity. GM-1 cells were cotransfected with GAL4-Foxo1 together with or without Sir2 WT or H355A as indicated. (C) Recruitments of Foxo1, CBP, and Sir2 onto the MnSOD and p27kip1 promoters. Chromatin immunoprecipitation assays were performed with the indicated antibodies in HEK293 cells. Immunoprecipitated (IP) DNA was analyzed by PCR using specific primer sets. (D) NIA reduces Foxo1-mediated gene expression. HEK293 cells were treated with NIA (0, 5, and 10 mM) for 24 h in serum-starved conditions. (E) Overexpression of Sir2 affects Foxo1-mediated gene expression. HEK293 cells stably transfected with Sir2 WT or Sir2 H355A were cultured with or without 10% FBS or insulin (100 nM) for 18 h.
To verify the direct effect of Sir2 on FOXO-targeted gene expression including MnSOD (18) and p27kip1 (16), we performed chromatin immunoprecipitation assays in HEK293 cells. Endogenous Foxo1, CBP, and Sir2 were recruited to Foxo1-binding sites of the MnSOD and p27kip1 promoters but not on a β-actin coding region (Fig. 4C). Next, to investigate whether the expression of MnSOD and p27kip1 proteins are regulated by deacetylase activity of Sir2, HEK293 cells were treated with NIA in serum-starved conditions. As shown in Fig. 4D, the expression levels of both MnSOD and p27kip1 were reduced in the NIA-treated cells in a dose-dependent manner. To examine further the effect of the enzymatic activity of Sir2, we stably transfected Sir2 WT or H355A into HEK293 cells and evaluated the expression patterns of the FOXO-targeted genes. Compared with mock transformants, HEK293 cells expressing Sir2 WT increased the amounts of MnSOD and p27kip1 by serum deprivation, and the addition of insulin, which inactivates Foxo1 transactivation function by nuclear exclusion, abolished these expressions (Fig. 4E, lanes 1–6). On the other hand, cells expressing Sir2 H355A impaired the induction of these protein amounts along with serum deprivation (lanes 7–9). Together, these results indicate that deacetylase activity of Sir2 actually mediates Foxo1-mediated transcription in mammalian cells.
Discussion
Here we tried to elucidate the mechanisms of how Foxo1 and Sir2 communicate in mammalian cells. We found that Foxo1 is acetylated by CBP and that this acetylation is counteracted by Sir2. Furthermore, we demonstrated that Sir2 coactivates the transcriptional function of Foxo1 in a deacetylase-activity-dependent manner. Based on these results, a speculative model would be that (i) formation of the Foxo1–CBP complex leads to histone acetylation and the recruitment of preinitiation complex containing RNA polymerase II to the target promoter, but (ii) the induced transcription could be attenuated by subsequent Foxo1 acetylation by CBP, after which (iii) Sir2 may restore Foxo1 function through deacetylase activity, and thereby Foxo1-mediated transcription could be sustained further. Such a model would be supported by the seminal findings that Drosophila Sir2 is associated with numerous euchromatic, constitutively active genes (36) and that Sir2-null mice provide no evidence for failure of gene silencing (37, 38). Alternatively, more recent reports have shown that deacetylase activity of histone deacetylases is required for signal transducer and activator of transcription-dependent gene expressions (39–41). These findings lead us to conjecture that deacetylase enzymes may be generally involved in the transactivating mechanism.
Our present findings provide a direct and functional correlation between two genetic determinants of longevity, FOXO and Sir2, at the molecular level. Thus far, a plausible explanation of Sir2-dependent longevity in C. elegans has been the Sir2-mediated silencing of genes for components of the insulin-signaling pathway; namely, Sir2 represses the expression of genes upstream of FOXO/DAF-16, which in turn attenuates the insulin signals and oppositely stimulates FOXO/DAF-16 activity (24). In this study, however, we present a molecular mechanism in which opposing enzymatic activities of CBP (acetylation) and Sir2 (deacetylation) modulate the transcriptional ability of Foxo1 and propose that Sir2-mediated coactivation of FOXO/DAF-16 might account for its effect on longevity in C. elegans.
CR promotes longevity in a wide spectrum of organisms. Although it has been shown to require NAD and Sir2 in yeast (25), the molecular mechanism by which CR extends lifespan is largely exclusive (42). More recently, Lin et al. (43) demonstrated that CR reduces NADH levels, and the NADH is a competitive inhibitor of Sir2. Given that CR decreases the blood insulin level, our findings provide a model indicating that two major indicators of intercellular (insulin level) and intracellular (NAD/NADH ratio) energy states converge to FOXO transcriptional regulation through multiple modifications: phosphorylation and acetylation, respectively. It is possible that CR efficiently enhances FOXO activity by down-regulating the insulin-signaling pathway and by up-regulating deacetylase activity of Sir2 and hence leads diverse organisms toward longevity.
As established previously (44–46), Sir2 also deacetylates p53 and down-regulates its transactivation function on damage-response target genes including proapoptotic factor BAX. Interestingly, while this article was under review, Motta et al. (47) and Brunet et al. (48) found that SIRT1, the human Sir2 ortholog, repressed the proapoptotic target genes for FOXO such as BIM and FAS ligand. These studies indicate that Sir2 could serve to resist stress-induced cell death cooperatively by repressing both p53- and FOXO-dependent apoptosis. In the present study, however, we demonstrated that Sir2 potentiates FOXO-dependent expression of an antioxidant gene, MnSOD. This finding suggests that an increased ability to detoxify reactive oxygen species mediated by Sir2 and FOXO slows oxidative damage and promotes endurance. Although additional study will be required to elucidate the mechanism underlying the opposite effect of Sir2 on FOXO activity, our findings together with recent reports imply that Sir2 may modulate the dual function of FOXO, cell death and survival, and consequently prolong lifespan.
Acknowledgments
We thank the Fukamizu Laboratory members for helpful discussion. This work was supported by the 21st Century Center of Excellence Program; Grant-in-Aid for Scientific Research (A); Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Technology of Japan; “Research for the Future” Program (the Japan Society for the Promotion of Science, Grant RFTF 97L00804); the Research Grant for Cardiovascular Diseases (11C-1); Comprehensive Research on Aging and Health from the Ministry of Health, Labour, and Welfare; TAKEDA Science Foundation; and the Cell Science Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Sir, silent information regulator; CR, calorie restriction; CBP, cAMP-response element-binding protein-binding protein; HA, hemagglutinin; MnSOD, manganese superoxide dismutase; HAT, histone acetyltransferase; NIA, nicotinamide.
References
- 1.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857-868. [DOI] [PubMed] [Google Scholar]
- 2.Kops, G. J., de Ruiter, N. D., De Vries-Smits, A. M., Powell, D. R., Bos, J. L. & Burgering, B. M. (1999) Nature 398, 630-634. [DOI] [PubMed] [Google Scholar]
- 3.Rena, G., Guo, S., Cichy, S. C., Unterman, T. G. & Cohen, P. (1999) J. Biol. Chem. 274, 17179-17183. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki, H., Daitoku, H., Hatta, M., Tanaka, K. & Fukamizu, A. (2003) Proc. Natl. Acad. Sci. USA 100, 11285-11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayala, J. E., Streeper, R. S., Desgrosellier, J. S., Durham, S. K., Suwanichkul, A., Svitek, C. A., Goldman, J. K., Barr, F. G., Powell, D. R. & O'Brien, R. M. (1999) Diabetes 48, 1885-1889. [DOI] [PubMed] [Google Scholar]
- 6.Schmoll, D., Walker, K. S., Alessi, D. R., Grempler, R., Burchell, A., Guo, S., Walther, R. & Unterman, T. G. (2000) J. Biol. Chem. 275, 36324-36333. [DOI] [PubMed] [Google Scholar]
- 7.Hall, R. K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R. & Granner, D. K. (2000) J. Biol. Chem. 275, 30169-30175. [DOI] [PubMed] [Google Scholar]
- 8.Daitoku, H., Yamagata, K., Matsuzaki, H., Hatta, M. & Fukamizu, A. (2003) Diabetes 52, 642-649. [DOI] [PubMed] [Google Scholar]
- 9.Nakae, J., Biggs, W. H., III, Kitamura, T., Cavenee, W. K., Wright, C. V., Arden, K. C. & Accili, D. (2002) Nat. Genet. 32, 245-253. [DOI] [PubMed] [Google Scholar]
- 10.De Ruiter, N. D., Burgering, B. M. & Bos, J. L. (2001) Mol. Cell. Biol. 21, 8225-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahl, M., Dijkers, P. F., Kops, G. J., Lens, S. M., Coffer, P. J., Burgering, B. M. & Medema, R. H. (2002) J. Immunol. 168, 5024-5031. [DOI] [PubMed] [Google Scholar]
- 12.Medema, R. H., Kops, G. J., Bos, J. L. & Burgering, B. M. (2000) Nature 404, 782-787. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura, N., Ramaswamy, S., Vazquez, F., Signoretti, S., Loda, M. & Sellers, W. R. (2000) Mol. Cell. Biol. 20, 8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka, M., Kirito, K., Kashii, Y., Uchida, M., Watanabe, T., Endo, H., Endoh, T., Sawada, K., Ozawa, K. & Komatsu, N. (2001) J. Biol. Chem. 276, 15082-15089. [DOI] [PubMed] [Google Scholar]
- 15.Tang, E. D., Nunez, G., Barr, F. G. & Guan, K. L. (1999) J. Biol. Chem. 274, 16741-16746. [DOI] [PubMed] [Google Scholar]
- 16.Dijkers, P. F., Medema, R. H., Pals, C., Banerji, L., Thomas, N. S., Lam, E. W., Burgering, B. M., Raaijmakers, J. A., Lammers, J. W., Koenderman, L., et al. (2000) Mol. Cell. Biol. 20, 9138-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijkers, P. F., Birkenkamp, K. U., Lam, E. W., Thomas, N. S., Lammers, J. W., Koenderman, L. & Coffer, P. J. (2002) J. Cell. Biol. 156, 531-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kops, G. J., Dansen, T. B., Polderman, P. E., Saarloos, I., Wirtz, K. W., Coffer, P. J., Huang, T. T., Bos, J. L., Medema, R. H. & Burgering, B. M. (2002) Nature 419, 316-321. [DOI] [PubMed] [Google Scholar]
- 19.Nemoto, S. & Finkel, T. (2002) Science 295, 2450-2452. [DOI] [PubMed] [Google Scholar]
- 20.Ogg, S., Paradis, S., Gottlieb, S., Patterson, G. I., Lee, L., Tissenbaum, H. A. & Ruvkun, G. (1997) Nature 389, 994-999. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., Li, H. & Kenyon, C. (2003) Nature 424, 277-283. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. S., Kennedy, S., Tolonen, A. C. & Ruvkun, G. (2003) Science 300, 644-647. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, A. L., Murphy, C. T. & Kenyon, C. (2003) Science 300, 1142-1145. [DOI] [PubMed] [Google Scholar]
- 24.Tissenbaum, H. A. & Guarente, L. (2001) Nature 410, 227-230. [DOI] [PubMed] [Google Scholar]
- 25.Lin, S. J., Defossez, P. A. & Guarente, L. (2000) Science 289, 2126-2128. [DOI] [PubMed] [Google Scholar]
- 26.Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. (2000) Nature 403, 795-800. [DOI] [PubMed] [Google Scholar]
- 27.Guarente, L. (2000) Genes Dev. 14, 1021-1026. [PubMed] [Google Scholar]
- 28.Kaeberlein, M., McVey, M. & Guarente, L. (1999) Genes Dev. 13, 2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasrin, N., Ogg, S., Cahill, C. M., Biggs, W., Nui, S., Dore, J., Calvo, D., Shi, Y., Ruvkun, G. & Alexander-Bridges, M. C. (2000) Proc. Natl. Acad. Sci. USA 97, 10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomizawa, M., Kumar, A., Perrot, V., Nakae, J., Accili, D., Rechler, M. M. & Kumaro, A. (2000) J. Biol. Chem. 275, 7289-7295. [DOI] [PubMed] [Google Scholar]
- 31.Sterner, D. E. & Berger, S. L. (2000) Microbiol. Mol. Biol. Rev. 64, 435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmud, D. L., G-Amlak, M., Deb, D. K., Platanias, L. C., Uddin, S. & Wickrema, A. (2002) Oncogene 21, 1556-1562. [DOI] [PubMed] [Google Scholar]
- 33.Schiltz, R. L., Mizzen, C. A., Vassilev, A., Cook, R. G., Allis, C. D. & Nakatani, Y. (1999) J. Biol. Chem. 274, 1189-1192. [DOI] [PubMed] [Google Scholar]
- 34.Bitterman, K. J., Anderson, R. M., Cohen, H. Y., Latorre-Esteves, M. & Sinclair, D. A. (2002) J. Biol. Chem. 277, 45099-45107. [DOI] [PubMed] [Google Scholar]
- 35.Parsons, X. H., Garcia, S. N., Pillus, L. & Kadonaga, J. T. (2003) Proc. Natl. Acad. Sci. USA 100, 1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Steensel, B., Delrow, J. & Henikoff, S. (2001) Nat. Genet. 27, 304-308. [DOI] [PubMed] [Google Scholar]
- 37.McBurney, M. W., Yang, X., Jardine, K., Bieman, M., Th'ng, J. & Lemieux, M. (2003) Mol. Cancer Res. 1, 402-409. [PubMed] [Google Scholar]
- 38.McBurney, M. W., Yang, X., Jardine, K., Hixon, M., Boekelheide, K., Webb, J. R., Lansdorp, P. M. & Lemieux, M. (2003) Mol. Cell. Biol. 23, 38-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nusinzon, I. & Horvath, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 14742-14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rascle, A., Johnston, J. A. & Amati, B. (2003) Mol. Cell. Biol. 23, 4162-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, M., Nie, L., Kim, S. H. & Sun, X. H. (2003) EMBO J. 22, 893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koubova, J. & Guarente, L. (2003) Genes Dev. 17, 313-321. [DOI] [PubMed] [Google Scholar]
- 43.Lin, S. J., Ford, E., Haigis, M., Liszt, G. & Guarente, L. (2004) Genes Dev. 18, 12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L. & Gu, W. (2001) Cell 107, 137-148. [DOI] [PubMed] [Google Scholar]
- 45.Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. & Weinberg, R. A. (2001) Cell 107, 149-159. [DOI] [PubMed] [Google Scholar]
- 46.Langley, E., Pearson, M., Faretta, M., Bauer, U. M., Frye, R. A., Minucci, S., Pelicci, P. G. & Kouzarides, T. (2002) EMBO J. 21, 2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motta, M. C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma, Y., McBurney, M. & Guarente, L. (2004) Cell 116, 551-563. [DOI] [PubMed] [Google Scholar]
- 48.Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E., Mostoslavsky, R., Cohen, H. Y., et al. (2004) Science 303, 2011-2015. [DOI] [PubMed] [Google Scholar]