Abstract
Activation of the visual pigment rhodopsin is caused by 11-cis to -trans isomerization of its retinal chromophore. High-resolution solid-state NMR measurements on both rhodopsin and the metarhodopsin II intermediate show how retinal isomerization disrupts helix interactions that lock the receptor off in the dark. We made 2D dipolar-assisted rotational resonance NMR measurements between 13C-labels on the retinal chromophore and specific 13C-labels on tyrosine, glycine, serine, and threonine in the retinal binding site of rhodopsin. The essential aspects of the isomerization trajectory are a large rotation of the C20 methyl group toward extracellular loop 2 and a 4- to 5-Å translation of the retinal chromophore toward transmembrane helix 5. The retinal–protein contacts observed in the active metarhodopsin II intermediate suggest a general activation mechanism for class A G protein-coupled receptors involving coupled motion of transmembrane helices 5, 6, and 7.
Gprotein-coupled receptors (GPCRs) are a large superfamily of membrane receptors that have seven transmembrane (TM) helices and respond to a wide array of signaling ligands. Similarities in the sequences of these receptors have led to the idea that they share a common activation mechanism, whereas sequence diversity is correlated with the specificity of different receptors for different ligands and G proteins. With the exception of the visual pigment rhodopsin (1, 2), the structures of GPCRs are unknown. The rhodopsin crystal structure shows that the TM helices are locked in an inactive conformation by interhelical interactions involving conserved amino acids. The general model of GPCR activation that has emerged over the past few years is that ligand-binding, or retinal isomerization in the case of the visual pigments, disrupts these interactions and drives a rigid body movement of one or more of the TM helices (3, 4).
Helix–helix interactions in rhodopsin are mediated by two sets of conserved amino acids. The highly conserved signature amino acids have sequence identities of >80% across the family of class A GPCRs. These amino acids are generally highly polar, aromatics, or proline. The group-conserved amino acids are small and/or weakly polar (Gly, Ala, Ser, Thr, and Cys) (5). They have low individual sequence identities but are highly conserved (>80%) when considered as a group. These amino acids are involved in mediating close helix contacts and facilitating interhelical hydrogen bonding (6).
The location of the group-conserved amino acids largely in the interfaces of TM helices H1–H4 of rhodopsin has suggested that these helices are locked in a stable structure that does not change significantly upon receptor activation (5). There are fewer group-conserved amino acids in H5–H7. H5 and H7 are unusual in containing conserved prolines that facilitate interactions with the H1–H4 core by exposing the i-4 backbone carbonyl for interhelical hydrogen bonding. H6 is unusual in that the TM region is composed of conserved aromatic and large hydrophobic amino acids that appear to interact with the H1–H4 core only by means of van der Waals contacts (5). However, despite the vast amount of sequence and mutational data on class A GPCRs and the high-resolution crystal structure of rhodopsin, it has not yet been established how ligand binding or retinal isomerization disrupts the interactions that lock these receptors in their inactive state.
Rhodopsin is a prototypical class A GPCR. For studies on GPCR activation, rhodopsin has the advantage of containing a covalently bound retinal chromophore that acts as an inverse agonist in the dark and is switched to an agonist upon absorption of light. To determine how retinal isomerization is coupled to receptor activation, we incorporated 13C labels into both the protein (7) and the retinal and identified close 13C...13C contacts by 2D solid-state NMR spectroscopy. The strategy is to first determine 13C...13C retinal–protein contacts in the inactive state of rhodopsin where their internuclear distances are known independently from the refined 2.6-Å rhodopsin crystal structure (1). This approach allows us to confirm that the NMR measurements are providing reliable retinal–protein contacts before converting the samples to metarhodopsin II (meta II), the active intermediate of rhodopsin.
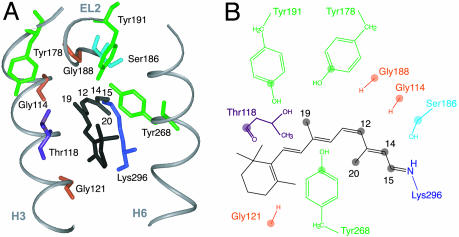
Fig. 1A shows the crystal structure of the retinal binding pocket in rhodopsin. In this study, we have incorporated 13C-labels into tyrosine, serine, glycine, and threonine of the apoprotein opsin and regenerated the rhodopsin pigment with 11-cis retinals that have been 13C-labeled at the C12, C14, C15, C19, and C20 carbons (Fig. 1B). In meta II, we can determine the location of the retinal chromophore and the direction of retinal isomerization by identifying specific retinal–protein contacts. We find that there is a significant translation of the retinal toward H5 upon receptor activation and a large rotation of the C20 methyl group toward extracellular loop 2 (EL2). We discuss the implications of the observed trajectory of the retinal on the motion of TM helices H5, H6, and H7, and the mechanism of rhodopsin activation.
Fig. 1.
Retinal binding pocket of rhodopsin. (A) Crystal structure of the retinal binding pocket showing the tyrosines, glycines, serines, and threonines within 7 Å of the retinal. (B) Schematic representation of the 11-cis retinal chromophore and the amino acids that are 13C-labeled in this study. The 13C-labeled atoms on the retinal and protein are represented by filled circles.
Methods
Expression and Purification of 13C-Labeled Rhodopsin. Rhodopsin was expressed in stable tetracycline-inducible HEK293S cells (8) containing the wild-type opsin gene (9). FBS was dialyzed against 20 liters of buffer A (137 mM NaCl/2.7 mM KCl/1.8 mM KH2PO4/10 mM Na2HPO4, pH 7.2) three times (7). The cells were grown in DMEM formulation (10) prepared from cell culture-tested components (Sigma). The suspension growth medium was supplemented with specific 13C-labeled amino acids (Cambridge Isotope Laboratories, Andover, MA) and 10% dialyzed FBS/0.1% Pluronic F-68/50 mg/liter heparin/100 units/ml penicillin/100 μg/ml streptomycin (7, 11). The cells were induced on day 5 with 2 mg/liter tetracycline (8) and harvested on day 7.
HEK293S cells were harvested, and the cell pellets were resuspended in buffer A (40 ml/liter cell culture). We added 11-cis retinal to a final concentration of 20 mM. Buffer B [(buffer A/1% n-dodecyl maltoside (DM)] was used to solubilize the cells (40 ml/liter cell culture) for 4 h at room temperature.
Purification by affinity chromatography with the 1D4 antibody (National Cell Culture Center, Minneapolis) (9) was done by washing with 25 column volumes of buffer C (buffer A/0.05% DM), followed by elution using buffer D (2 mM NaH2PO4/Na2HPO4, pH6/0.02% DM) containing the last nine C-terminal amino acids of rhodopsin as the antibody epitope. After elution, samples were concentrated in Centricon cones (Amicon) with a 10-kDa cutoff to a volume of 1 ml.
Synthesis of 13C-Labeled Retinals and Regeneration into Rhodopsin. Retinals with specific 13C-labels were synthesized by using standard methods (12). The 11-cis isomer was purified from an irradiated mixture of isomers by isocratic HPLC with a dried solution of 96% hexane/4% ethyl acetate at 8 ml/min on an Econosphere 10-μl silica column (Alltech).
The rhodopsin pigments in DM micelles were regenerated with 13C-labeled retinals by illumination of concentrated samples in the presence of a 2:1 retinal/protein molar ratio. Regeneration was typically >80%, as determined by recovery of the 500-nm visible absorption band. The added retinal was first dissolved in ethanol to a volume <1% of the total sample volume. We found that excess ethanol alters the ability to trap the meta II intermediate. This effect was noticeable only by NMR chemical shifts, which were not characteristic of meta II. The regenerated sample was then concentrated to a volume of ≤100 μl by water evaporation using a stream of argon gas. Samples were then transferred into a 4-mm magic angle spinning (MAS) rotor and frozen at -80°C until NMR data acquisition.
Trapping of the Meta II Intermediate. NMR measurements were first made on rhodopsin (≈250 nmol) in the dark. After thawing, the samples were illuminated by using a 400-W lamp with a >495-nm cutoff filter for 20 sec at room temperature. The rotor was then recapped and placed in the NMR probe with the probe stator warmed to room temperature. Under slow spinning (2 kHz), the heater was turned off and the sample was frozen within 3 min by using N2 gas cooled to -80°C.
NMR Spectroscopy. The solid-state NMR experiments were run on a 600-MHz Avance spectrometer (Bruker, Billerica, MA) with 4-mm MAS probes. The MAS spinning rate for each sample was selected to avoid overlap of 13C cross peaks with MAS side bands. Ramped amplitude cross-polarization (13) contact times were 2 ms in all experiments, and two pulse phase-modulated (14) proton decoupling was used during the evolution and acquisition periods. The decoupling field strength was typically 90 kHz. We referenced 13C chemical shifts to external tetramethylsilane. The samples were maintained at -80°C.
For dipolar-assisted rotational resonance (DARR) experiments (15), mixing times of 600 ms to 1 s were used to maximize homonuclear recoupling between 13C labels (16). The 1H radio-frequency field strength during mixing was matched to the MAS speed to satisfy the n = 1 condition for each sample. Each 2D data set represents 4,068–6,144 scans in each of 64 rows in the f1 dimension. We used 10 Hz of exponential line broadening in the f2 dimension, and a cosine multiplication was used in the f1 dimension along with a 32-coefficient forward linear prediction.
Results
Low-Temperature Trapping of the Meta II Intermediate. The first step in the current study was to demonstrate that we can trap the meta II intermediate at low temperature as a stable, homogeneous species. Meta II forms within milliseconds of light absorption and decays via meta III to opsin and free retinal within minutes at room temperature (17). The decay of the intermediate can be slowed significantly by low temperature (18, 19). For the experiments described below, we illuminated the sample in the NMR rotor and then lowered the temperature of the rotor rapidly (≈3 min) (see Methods).
Meta II occurs in equilibrium with meta I. The meta II intermediate is favored at low pH and in lipids with unsaturated acyl chains (20). We compared the ability to convert rhodopsin to meta II by using native rod outer-segment membranes, rhodopsin reconstituted into unsaturated lipids, and rhodopsin solubilized in DM detergent micelles (data not shown). The conversion was monitored by following the 13C chemical shifts of the 14-13C and 15-13C labels on the retinal chromophore. These chemical shifts are sensitive to the isomerization state of the retinal as well as to the protonation state of the retinal–Lys-296 Schiff base linkage (21, 22). DM detergent was the only environment that led to full conversion to meta II in the optically dense samples needed for solid-state NMR. It is known that there is a large conformational change associated with the conversion to meta II, and consequently, the DM detergent may provide a more flexible environment than lipids to accommodate this change in our concentrated samples.
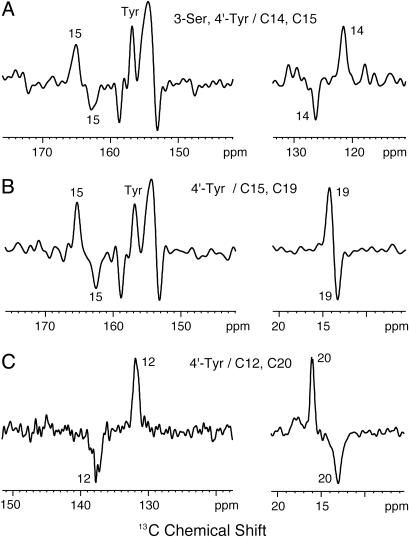
Fig. 2 shows difference spectra between rhodopsin (positive peaks) and meta II (negative peaks) in DM micelles. In Fig. 2 A, the C14 and C15 resonances observed at 121.6 and 165.1 ppm in rhodopsin shift to 126.3 and 162.4 ppm in meta II, respectively. The C14 and C15 chemical shifts in meta II are similar to those in the all-trans retinal unprotonated Schiff base model compound (21). The conversion to meta II is seen also in other samples following the characteristic shifts of the C12, C15, C19, and C20 resonances that are shown in Fig. 2 B and C. We observe a slight loss of intensity (≈10%) upon conversion to meta II. The intensity loss may be due to the formation of opsin and free retinal before the sample reaches the temperature needed to trap the meta II intermediate. It cannot be attributed to the presence of rhodopsin intermediates with an all-trans protonated Schiff base (e.g., meta I or meta III), which would generate C15 resonances of 164–166 ppm. By monitoring retinal resonances in meta II as a function of time, we have confirmed that the intermediate is stable for weeks at temperatures below -80°C and then converts rapidly to opsin and free retinal when the sample is brought to room temperature (data not shown).
Fig. 2.
Low-temperature trapping of meta II. Difference spectra between rhodopsin (positive peaks) and meta II (negative peaks) are shown for rhodopsin containing 3-13C serine, 4′-13C tyrosine, and 14,15-13C retinal (A); 4′-13C tyrosine and 15, 19-13C retinal (B); and 4′-13C tyrosine and 12, 20-13C retinal (C). Photoconversion of each sample was determined by the complete shifts of the positive retinal peaks in rhodopsin to their corresponding negative peaks in meta II.
The difference spectra shown in Fig. 2 A and B also show the chemical shift changes in the 4′-13C tyrosine resonances between 153 and 159 ppm. There are two (positive) rhodopsin tyrosine resonances at 154.3 and 156.8 ppm, and two (negative) meta II tyrosine resonances at 153.1 and 158.7 ppm. These differences between rhodopsin and meta II result from changes in hydrogen bonding rather than deprotonation. The 4′-13C chemical shift of tyrosinate is ≈165 ppm (23), which is not observed. Based on the 2D NMR correlation experiments (see below), we can assign the chemical shifts of the tyrosines that are in close proximity to the retinal. The chemical shifts of Tyr-268 (155. 2 ppm) and Tyr-178 (156.1 ppm) fall between the two maxima in the tyrosine difference spectra, suggesting that these tyrosines do not exhibit significant changes in chemical shift in meta II. Tyr-268 on H6 is part of a hydrogen-bonding network involving amino acids on EL2 (Glu-181, Ser-186, Cys-187, Tyr-191, and Tyr-192), which we propose remains intact in meta II. The lack of changes in the chemical shifts of Tyr-178 and Tyr-268 is consistent with rigid body motion of H6 that occurs on the cytoplasmic side of Pro-267 (3). Mutational data indicate that EL2 is important for the stability of the retinal binding site in rhodopsin and meta II (24, 25). Recent computational studies suggest that the Cys-110–Cys-187 disulfide bridge along with several residues on EL2 and at the ends of H3 and H4 are part of a stable folding core in rhodopsin (26).
Establishing Retinal–Protein Contacts in the Retinal Binding Site of Rhodopsin and Meta II. The strategy for establishing retinal–protein contacts in the retinal binding site is to incorporate 13C labels into both the protein and the retinal and to identify close 13C contacts by 2D solid-state NMR (16). The 2D NMR spectra are obtained by using MAS and DARR (15). MAS yields high-resolution spectra in solid-state NMR experiments of membrane proteins. DARR restores the dipolar couplings between 13C nuclei and gives rise to cross peaks between the corresponding recoupled 13C resonances.
Fig. 1 shows the retinal binding pocket based on the rhodopsin crystal structure. The retinal is attached to H7 and is bracketed largely by H3 and H6. The binding pocket is closed on the extracellular side of the protein by EL2, which connects TM helices H4 and H5. There are three tyrosines that are close to the retinal (Tyr-268, Tyr-178, and Tyr-191), as well as three glycines (Gly-114, Gly-121, and Gly-188), a single threonine (Thr-118), and a single serine (Ser-186). Establishing the retinal–protein contacts in both rhodopsin and meta II provides a way to determine the direction of retinal isomerization.
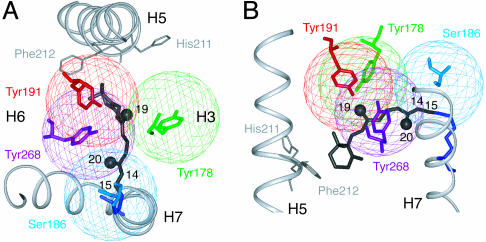
The cross-peak patterns seen in the 2D NMR spectra can be interpreted most easily by using Fig. 3 A and B, which show the crystal structure of the retinal binding pocket of rhodopsin with spheres around the 13C labels in Ser-186 (EL2), Tyr-178 (EL2), Tyr-191 (EL2), and Tyr-268 (H6). The radii of the spheres correspond to the ≈5.5-Å distance observable in the DARR experiment for mixing times of 600 ms to 1 s. The C19 and C20 retinal carbons are shown as solid black spheres. Cross peaks are generated in the 2D spectrum when the retinal 13C labels are within 5.5 Å of the 13C-labeled amino acids (i.e., within the colored spheres).
Fig. 3.
Distance range of the DARR NMR experiment. The 13C-labeled sites on tyrosine and serine in the retinal binding site of rhodopsin are highlighted. The spheres around the labeled atoms are drawn with a 5.5-Å radius, corresponding to the approximate detection limit of the DARR experiment. The 13C-labeled C19 and C20 retinal methyl carbons are indicated by black spheres.
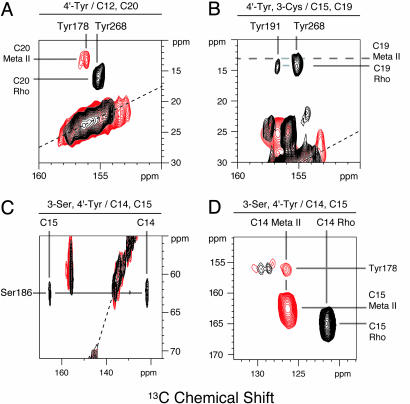
Fig. 4 shows regions of the 2D NMR spectra of rhodopsin (black) and meta II (red), showing specific retinal–protein contacts. Fig. 4A presents the 2D NMR spectrum of rhodopsin labeled with 4′-13C tyrosine and regenerated with 12,20-13C retinal. A single tyrosine-to-C20 cross peak in rhodopsin (black) is observed at 155.2 ppm. According to the crystal structure, the only 4′-13C tyrosine label within 8 Å of C20 is on Tyr-268 at a distance of 4.4 Å. Therefore, this cross peak is due to the C20–Tyr-268 contact. Importantly, upon conversion to meta II, the 155.2-ppm cross peak disappears and a new C20-Tyr cross peak appears at 156.1 ppm.
Fig. 4.
The 2D DARR NMR of rhodopsin and meta II. The expanded regions of the full 2D NMR spectra of rhodopsin (black) and meta II (red) shown contain cross peaks between the retinal and protein 13C-labels. The protein and retinal labels are as follows: 4′-13C tyrosine and 12, 20-13C retinal (A); 4′-13C tyrosine, 3-13C Cys and 15, 19-13C retinal (B); and 3-13C Ser, 4′-13C tyrosine and 14,15-13C retinal (C and D). The intense resonances along the dotted lines correspond to MAS side bands. The unlabeled cross peaks shown in B centered at 25 and 157 ppm correspond to cysteine–tyrosine correlations. The unlabeled cross peaks shown in C centered at 60 and 155 ppm correspond to serine–tyrosine correlations. The unlabeled cross peaks shown in D correspond to correlations between 4′-13C tyrosine and natural abundance 13C carbons on the tyrosine rings. The rhodopsin and meta II spectra were obtained at -80°C and a spinning speed of 10 kHz (A), 9.5 kHz (B), or 11 kHz (C and D). See the supporting information, which is published on the PNAS web site, for 1D MAS and full 2D DARR spectra.
Fig. 4B presents the 2D NMR spectrum of 4′-13C tyrosine rhodopsin regenerated with 15,19-13C retinal. In the rhodopsin spectrum, two tyrosine–retinal cross peaks are observed. One cross peak is at 155.2 ppm, the chemical shift of Tyr-268. According to the crystal structure, Tyr-268 is ≈4.8 Å from C19. The less intense cross peak at 156.7 ppm is due to Tyr-191 at 5.1 Å from C19. However, upon conversion to meta II, no C19-tyrosine cross peaks are observed despite longer data accumulations, indicating that C19 methyl group has moved away from all of the tyrosines in the retinal binding site.
Translation of the Retinal Toward H5 in the Formation of Meta II. The results presented imply that the C19 and C20 methyl groups do not simply rotate from one side of the retinal binding site to the other. First, C19 no longer exhibits cross peaks to any tyrosines in the retinal binding pocket in meta II. Second, C20 moves away from Tyr-268 and closer to a tyrosine other than Tyr-268 or Tyr-191. The observation of a single cross peak at 156.1 ppm argues that the C20 methyl group has rotated toward EL2 and is within 5.5 Å of Tyr-178, which is the only other tyrosine in the binding pocket. These observations can be explained by a translation of the retinal toward H5 in meta II.
To confirm that the retinal translates toward H5 in the binding site, we obtained 2D NMR spectra of rhodopsin regenerated with 14,15-13C retinal and labeled with 3-13C-serine and 4′-13C-tyrosine. In the dark, the 14,15-13C carbons exhibit cross peaks to Ser-186 (Fig. 4C) but not to any of the tyrosines in the binding pocket (Fig. 4D), consistent with the crystal structure. In meta II, the C14–Ser-186 and C15–Ser-186 cross peaks disappear and new cross peaks are observed between C14 and Tyr-178 (Fig. 4D) and between C15 and Tyr-178 (data not shown). Considering the detection range of the NMR experiment (see Fig. 3), these data indicate that the retinal translates 4–5 Å from its position in rhodopsin toward H5 in meta II.
Rotation of the C20 Methyl Group in the Formation of Meta II. Here, we further characterize the position of the C19 and C20 methyl groups in meta II. We obtained 2D NMR spectra on rhodopsin labeled with 2-13C glycine and 1-13C threonine and regenerated with either 19-13C or 20-13C retinal (data not shown). The α-carbons of Gly-188 and Gly-114 are 4.8 and 6.8 Å away from the C19 methyl group in rhodopsin, respectively. All other glycines are >8 Å away from C19. We observe a C19–Gly-188 contact in rhodopsin consistent with the crystal structure. The chemical shift of Gly-188 at 41.3 ppm, derived from this cross peak, is consistent with the β-sheet structure of EL2. The C19–Gly-188 cross peak disappears upon conversion to meta II, in agreement with a ≈4- to 5-Å translation of the retinal toward H5.
In rhodopsin, the C20 methyl group is roughly equidistant from Gly-114 (7.0 Å) and Gly-188 (6.3 Å) (see Fig. 1 A), and therefore, there are no contacts detected between the C20 and glycine labels. In meta II, we observe a C20–glycine cross peak at a frequency of 45.7 ppm. This chemical shift is different from that of Gly-188 and is consistent with a glycine in helical secondary structure, which is the case for Gly-114 or Gly-121 on H3. However, no Thr-118–C20 contacts are observed in either rhodopsin or meta II, as would be expected if the C20 methyl group were near Gly-121. We, therefore, assign the C20–glycine contact in meta II to Gly-114 on H3 in agreement with the correlation of C20 to Tyr-178 in EL2 and the absence of a C20 correlation to Thr-118.
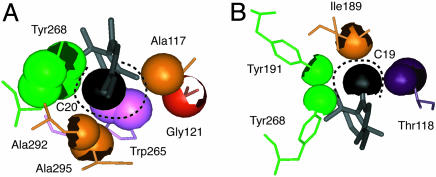
These observations imply that the C20 methyl group rotates >90° upon conversion of rhodopsin to meta II. Fig. 5A shows that the C20 methyl group is occluded predominantly by Trp-265 and Tyr-268 on H6, as well as by Ala-292, Ala-295, and Lys-296 on H7. C20 is packed more loosely in the direction of Ala-117 on H3, consistent with the conclusion that the C20 methyl group rotates toward the nonoccluded volume adjacent to Ala-117 (i.e., toward EL2 in the direction of H3). It is known that retinals lacking the C20 methyl group can be used to regenerate rhodopsin. They form stable pigments and are capable of receptor activation upon light absorption (27), implying that C20–protein steric interactions are not important for activation. However, mutation of Ala-117 to phenylalanine is predicted to occlude the space between Trp-265 and the side chain of Lys-296. In the A117F mutant, meta II forms very slowly (28), presumably because the rotation of the C20 methyl group is blocked.
Fig. 5.
Packing of the C19 and C20 retinal methyl groups in rhodopsin. The C20 (A) and C19 (B) retinal methyl carbons (black spheres), along with neighboring atoms (excluding hydrogen atoms) are rendered with their van der Waals radii. An occluded surface analysis of atomic packing (43) shows that the C20 methyl group is packed less tightly against H3 than C19.
In contrast, a significant rotation of C19 appears to be largely blocked by steric interactions in wild-type rhodopsin. Fig. 5B shows that the C19 methyl group is tightly constrained in the retinal binding site and packs against Ile-189 and Tyr-191 in EL2, Tyr-268 on H6 and Thr-118 on H3. However, C19 is not occluded by other residues along the axis of the retinal, and can be viewed as residing in a channel formed by H3, H6 and EL2. It was found that removal of the C19 methyl group prevents receptor activation (29, 30). These observations suggest that the C19 methyl group helps to direct translation of the retinal toward H5 by preventing significant rotation of the ionone ring end of the retinal.
Discussion
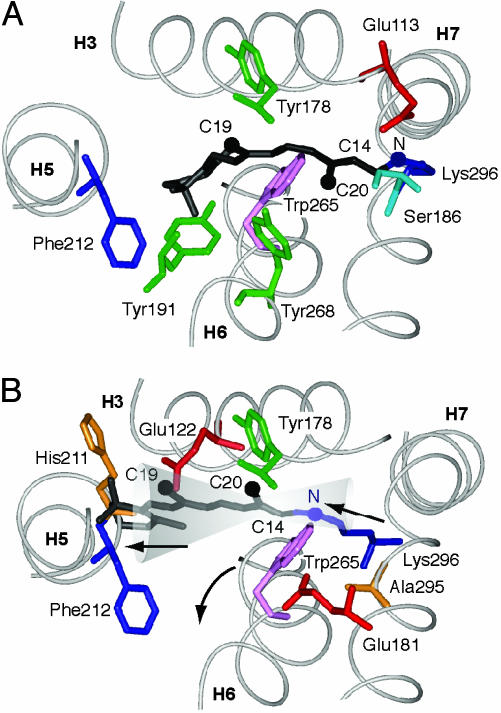
Implications of Retinal Rotation and Translation for Receptor Activation. Fig. 6 A and B show the position of the retinal in rhodopsin and meta II, respectively. Although the final position of the ionone ring has not yet been established, the data are consistent with the retinal translating roughly along the axis of the retinal polyene chain and increasing van der Waals contact on the face of H5 defined by His-211–Phe-212 (see Fig. 6B). The bulky ionone ring is tightly packed in rhodopsin and the proposed motion would minimize the change in conformation or orientation of the ring in meta II. Solid-state NMR measurements of the C16 and C17 methyl groups have indicated that the ionone ring does not significantly change conformation or environment in meta I (31). Below, we consider how the large C20 rotation and translation toward H5 disrupt the interactions of H5, H6, and H7 with the H1–H4 core of rhodopsin and lead to receptor activation.
Fig. 6.
Location of the retinal chromophore in meta II. (A) Location of 11-cis retinal in the crystal structure of rhodopsin. (B) A possible position of all-trans retinal in meta II. The NMR data constrain the C20 methyl group to be close to Gly-114 and Tyr-178 in meta II. These constraints place the Schiff base proton near Glu-181 in meta II. The position of the ionone ring is not tightly constrained by the current data. The cone indicates possible positions of the retinal consistent with the C20 and C14 carbons being near Tyr-178. The position shown is based on minimizing the movement of the ionone ring (31). The location of the retinal chromophore in meta II shown by Nakanishi and coworkers (44) is at one extreme of our estimated uncertainty.
Retinal–H5 Interactions. Retinal analogs that lack the ionone ring are not able to activate rhodopsin (32). Translation of the retinal in its binding pocket increases the contact of the ionone ring with H5 and explains the requirement of the ring for rhodopsin activation. The crystal structure of rhodopsin shows a hydrogenbonding contact between H3 and H5, namely, between Glu-122 and His-211. Because of the highly conserved Pro-215 at the i+4 position, the backbone carbonyl of His-211 is exposed and hydrogen bonds to the side chain of Glu-122. We have recently found that this interhelical interaction between H5 and the H1–H4 core is disrupted upon rhodopsin activation (A.B.P., E.C., P. Reeves, E. Getmanova, M.E., H. G. Khorana, and S.O.S. unpublished data). We propose that contact of the ionone ring with H5 in the region of His-211 moves this helix to an active orientation.
Ligand–H5 interactions are critical for activation in other class A GPCRs. For instance, in the β-adrenergic receptor, two serines (Ser-203 and Ser-207) hydrogen bond with hydroxyl groups on catecholamine agonists upon ligand binding and receptor activation (33, 34). Ser-207 is in the position equivalent to His-211 in rhodopsin, preceding a conserved proline by four amino acids. Ser-203 and Ser-207 are predicted to be in the H3–H5 interface and would mediate helix interactions in the absence of ligand. The SxxxS sequence has been identified as a common motif for mediating strong helix contacts (35). Together, these observations suggest that receptor-specific amino acids are designed to lock H5 in an inactive orientation and that ligand binding replaces helix–helix with helix–ligand interactions, which shift H5 to an active orientation.
Retinal–H6 Interactions. Trp-265 on H6 lies within the arc created by the 11-cis retinal and the Lys-296 side chain. Considering our proposed trajectory for the retinal, Trp-265 is the only amino acid in the retinal binding pocket that restricts retinal translation. The indole side chain is tightly packed between Gly-121 on H3 and Ala-295 on H7 (see Fig. 5). Translation of the retinal toward H5, as shown in Fig. 6B, would result in outward rotation of H6 by direct interaction between the tryptophan side chain and either the Lys-296 or Ala-295 side chains. The location of Trp-265 near highly conserved Pro-267 suggests that its large indole side chain acts as a lever for outward rotation of H6 by using the proline as a flexible hinge.
Rigid body motion of H6 has been shown to be a key element in the activation of rhodopsin (3). Preventing this motion by cross-linking H3 and H6 blocks activation (36). Mutation of Trp-265 to other aromatics or alanine decreases the activity of rhodopsin significantly (28). Trp-265 is part of a cluster of aromatic amino acids on the interior face of H6, which is thought to function as a conformational switch for activation of receptors in the amine and peptide subfamilies (37, 38). The conservation of Trp-265 and Pro-267 argues that these are common elements of the activation mechanism.
Retinal–H7 Interactions. The magnitude of retinal translation (4–5 Å) has significant implications for the structure of and the interactions involving H7. The 4- to 5-Å translation cannot be accommodated by extending the Lys-296 side chain alone. Because the retinal is covalently attached to H7, retinal translation implies there is a corresponding motion of this helix, at least in the region from Pro-290–Tyr-301. Motion of H7 toward H5 would result in increased steric contact between Ala-295 and Trp-265, which, as discussed above, would contribute to the outward rotation of H6.
In the rhodopsin crystal structure, H7 has two distinct regions that are separated near a kink at Pro-303. The sequence from Pro-290 to Tyr-301 appears to be ligand-specific and is conserved only within subfamilies of class A GPCRs. For instance, the retinal in the visual pigment subfamily is covalently linked to Lys-296. In contrast, the sequence from Asn-302 to Tyr-306 is highly conserved across the class A receptors, constituting the signature NPxxY motif. The NPxxY sequence, along with Asn-55 on H1 and Asp-83 on H2, are conserved across the class A GPCRs, even in the diverse olfactory receptor subfamily.
These observations suggest that ligand binding to the subfamily-specific end of H7 (or retinal isomerization) serves to alter H7 interactions with H6 and the H1–H4 core. Importantly, the side chains of the amino acids at positions 298 and 299 are subfamily-specific and are likely to be involved in ligand binding (39), whereas the backbone carbonyls at these sites can mediate hydrogen-bonding interactions with the signature asparagine and aspartic acid residues on H1 and H2, respectively. The carbonyl at position 299 is free because of the highly conserved Pro-303 in the NPxxY sequence and provides a bridge between the subfamily-specific amino acids and the conserved signature amino acids that are common to the activation mechanism. Interestingly, the P303A mutation in rhodopsin results in hyper-activity upon illumination (40), suggesting that the strong hydrogen bond between the Ala-299 carbonyl on H7 and Asn-55 on H1 is broken upon activation of wild-type rhodopsin.
Retinal–EL2 Interactions. The large rotation of the retinal C20 methyl group toward EL2 and translation of the retinal toward H5 is consistent with the counterion switch that has been proposed in the conversion to meta I (41). The protonated Schiff base proton, which is oriented toward Glu-113 on H3 in rhodopsin is thought to reorient toward Glu-181 on EL2 in meta I (see Fig. 6). Meta I has a blue-shifted absorption maximum relative to rhodopsin, which indicates a stronger interaction between the Schiff base proton and a negative protein counterion than in rhodopsin. The retinal trajectory shown in Fig. 6B would place the Schiff base proton into close proximity of Glu-181. An electrostatic interaction between the Schiff base proton and Glu-181 may help to guide the translation of the retinal during the photoconversion to meta I.
In conclusion, despite the ground-breaking high-resolution crystal structure of rhodopsin reported in 2000 (2), it has not been possible to establish how retinal isomerization leads to receptor activation. The results reported here provide a comprehensive picture for how retinal isomerization and translation are coupled to the structural changes in H5, H6, and H7 that were observed in EPR studies of Hubbell et al. (42). Importantly, our proposed mechanism explains the role of many of the signature and group-conserved amino acids that are common elements in the class A GPCR family.
Acknowledgments
We thank Martine Ziliox for assistance with the NMR experiments and critical reading of the manuscript; Philip Reeves and H. Gobind Khorana for the HEK293S cells for expression of isotope-labeled rhodopsin; and Michel Groesbeek for synthesis of the 15,19-13C retinal. The NMR facilities in the Center for Structural Biology at Stony Brook University are supported by the W. M. Keck Foundation. This work was supported by National Institutes of Health Grant GM-41412 (to S.O.S.) and National Institutes of Health National Science Foundation Instrumentation Grants S10 RR13889 and DBI-9977553. M.S. holds the Katzir-Makineni professorial chair in chemistry.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DARR, dipolar-assisted rotational resonance; DM, n-dodecyl maltoside; EL2, extracellular loop 2; GPCR, G protein-coupled receptor; MAS, magic angle spinning; meta, metarhodopsin; TM, transmembrane.
References
- 1.Okada, T., Fujiyoshi, Y., Silow, M., Navarro, J., Landau, E. M. & Shichida, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 3.Farrens, D. L., Altenbach, C., Yang, K., Hubbell, W. L. & Khorana, H. G. (1996) Science 274, 768-770. [DOI] [PubMed] [Google Scholar]
- 4.Gether, U. (2000) Endocr. Rev. 21, 90-113. [DOI] [PubMed] [Google Scholar]
- 5.Liu, W., Eilers, M., Patel, A. B. & Smith, S. O. (2004) J. Mol. Biol. 337, 713-729. [DOI] [PubMed] [Google Scholar]
- 6.Eilers, M., Patel, A. B., Liu, W. & Smith, S. O. (2002) Biophys. J. 82, 2720-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eilers, M., Reeves, P. J., Ying, W. W., Khorana, H. G. & Smith, S. O. (1999) Proc. Natl. Acad. Sci. USA 96, 487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeves, P. J., Kim, J. M. & Khorana, H. G. (2002) Proc. Natl. Acad. Sci. USA 99, 13413-13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves, P. J., Thurmond, R. L. & Khorana, H. G. (1996) Proc. Natl. Acad. Sci. USA 93, 11487-11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulbecco, R. & Freeman, G. (1959) Virology 8, 396-397. [DOI] [PubMed] [Google Scholar]
- 11.Eilers, M., Ying, W. W., Reeves, P. J., Khorana, H. G. & Smith, S. O. (2002) Methods Enzymol. 343, 212-222. [DOI] [PubMed] [Google Scholar]
- 12.Lugtenburg, J. (1985) Pure Appl. Chem. 57, 753-762. [Google Scholar]
- 13.Metz, G., Wu, X. & Smith, S. O. (1994) J. Magn. Reson. A 110, 219-227. [Google Scholar]
- 14.Bennett, A. E., Rienstra, C. M., Auger, M., Lakshmi, K. V. & Griffin, R. G. (1995) J. Chem. Phys. 103, 6951-6958. [Google Scholar]
- 15.Takegoshi, K., Nakamura, S. & Terao, T. (2001) Chem. Phys. Lett. 344, 631-637. [Google Scholar]
- 16.Crocker, E., Patel, A. B., Eilers, M., Jayaraman, S., Getmanova, E., Reeves, P. J., Ziliox, M., Khorana, H. G., Sheves, M. & Smith, S. O. (2004) J. Biomol. NMR 29, 11-20. [DOI] [PubMed] [Google Scholar]
- 17.Farrens, D. L. & Khorana, H. G. (1995) J. Biol. Chem. 270, 5073-5076. [DOI] [PubMed] [Google Scholar]
- 18.Hubbard, R., Brown, P. K. & Kropf, A. (1959) Nature 183, 442-446. [PubMed] [Google Scholar]
- 19.Yoshizawa, T. & Wald, G. (1963) Nature 197, 1279-1286. [DOI] [PubMed] [Google Scholar]
- 20.Straume, M., Mitchell, D. C., Miller, J. L. & Litman, B. J. (1990) Biochemistry 29, 9135-9142. [DOI] [PubMed] [Google Scholar]
- 21.Shriver, J. W., Mateescu, G. D. & Abrahamson, E. W. (1982) Methods Enzymol. 81, 698-703. [DOI] [PubMed] [Google Scholar]
- 22.Albeck, A., Livnah, N., Gottlieb, H. & Sheves, M. (1992) J. Am. Chem. Soc. 114, 2400-2411. [Google Scholar]
- 23.Herzfeld, J., Das Gupta, S. K., Farrar, M. R., Harbison, G. S., McDermott, A. E., Pelletier, S. L., Raleigh, D. P., Smith, S. O., Winkel, C., Lugtenburg, J. & Griffin, R. G. (1990) Biochemistry 29, 5567-5574. [DOI] [PubMed] [Google Scholar]
- 24.Doi, T., Molday, R. S. & Khorana, H. G. (1990) Proc. Natl. Acad. Sci. USA 87, 4991-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janz, J. M. & Farrens, D. L. (2003) Vision Res. 43, 2991-3002. [DOI] [PubMed] [Google Scholar]
- 26.Rader, A. J., Anderson, G., Isin, B., Khorana, H. G., Bahar, I. & Klein-Seetharaman, J. (2004) Proc. Natl. Acad. Sci. USA 101, 7246-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kropf, A., Whittenberger, B. P., Goff, S. P. & Waggoner, A. S. (1973) Exp. Eye Res. 17, 591-606. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama, T. A. & Khorana, H. G. (1991) J. Biol. Chem. 266, 4269-4275. [PubMed] [Google Scholar]
- 29.Vogel, R., Fan, G. B., Sheves, M. & Siebert, F. (2000) Biochemistry 39, 8895-8908. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, C. K., Bohme, M., Ockenfels, A., Gartner, W., Hofmann, K. P. & Ernst, O. P. (2000) J. Biol. Chem. 275, 19713-19718. [DOI] [PubMed] [Google Scholar]
- 31.Spooner, P. J. R., Sharples, J. M., Goodall, S. C., Seedorf, H., Verhoeven, M. A., Lugtenburg, J., Bovee-Geurts, P. H. M., DeGrip, W. J. & Watts, A. (2003) Biochemistry 42, 13371-13378. [DOI] [PubMed] [Google Scholar]
- 32.Jager, F., Jager, S., Krutle, O., Friedman, N., Sheves, M., Hofmann, K. P. & Siebert, F. (1994) Biochemistry 33, 7389-7397. [DOI] [PubMed] [Google Scholar]
- 33.Liapakis, G., Ballesteros, J. A., Papachristou, S., Chan, W. C., Chen, X. & Javitch, J. A. (2000) J. Biol. Chem. 275, 37779-37788. [DOI] [PubMed] [Google Scholar]
- 34.Strader, C. D., Candelore, M. R., Hill, W. S., Sigal, I. S. & Dixon, R. A. (1989) J. Biol. Chem. 264, 13572-13578. [PubMed] [Google Scholar]
- 35.Dawson, J. P., Weinger, J. S. & Engelman, D. M. (2002) J. Mol. Biol. 316, 799-805. [DOI] [PubMed] [Google Scholar]
- 36.Sheikh, S. P., Zvyaga, T. A., Lichtarge, O., Sakmar, T. P. & Bourne, H. R. (1996) Nature 383, 347-350. [DOI] [PubMed] [Google Scholar]
- 37.Shi, L., Liapakis, G., Xu, R., Guarnieri, F., Ballesteros, J. A. & Javitch, J. A. (2002) J. Biol. Chem. 277, 40989-40996. [DOI] [PubMed] [Google Scholar]
- 38.Singh, R., Hurst, D. P., Barnett-Norris, J., Lynch, D. L., Reggio, P. H. & Guarnieri, F. (2002) J. Pept. Res. 60, 357-370. [DOI] [PubMed] [Google Scholar]
- 39.Chanda, P. K., Minchin, M. C. W., Davis, A. R., Greenberg, L., Reilly, Y., McGregor, W. H., Bhat, R., Lubeck, M. D., Mizutani, S. & Hung, P. P. (1993) Mol. Pharmacol. 43, 516-520. [PubMed] [Google Scholar]
- 40.Fritze, O., Filipek, S., Kuksa, V., Palczewski, K., Hofmann, K. P. & Ernst, O. P. (2003) Proc. Natl. Acad. Sci. USA 100, 2290-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan, E. C. Y., Kazmi, M. A., Ganim, Z., Hou, J. M., Pan, D. H., Chang, B. S. W., Sakmar, T. P. & Mathies, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9262-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hubbell, W. L., Altenbach, C., Hubbell, C. M. & Khorana, H. G. (2003) Adv. Protein Chem. 63, 243-290. [DOI] [PubMed] [Google Scholar]
- 43.Pattabiraman, N., Ward, K. B. & Fleming, P. J. (1995) J. Mol. Recognit. 8, 334-344. [DOI] [PubMed] [Google Scholar]
- 44.Borhan, B., Souto, M. L., Imai, H., Shichida, Y. & Nakanishi, K. (2000) Science 288, 2209-2212. [DOI] [PubMed] [Google Scholar]