Abstract
The neonate is exposed to the maternal vaginal microbiota during parturition, providing the primary source for normal gut colonization, host immune maturation, and metabolism. These early interactions between the host and microbiota occur during a critical window of neurodevelopment, suggesting early life as an important period of cross talk between the developing gut and brain. Because perturbations in the prenatal environment such as maternal stress increase neurodevelopmental disease risk, disruptions to the vaginal ecosystem could be a contributing factor in significant and long-term consequences for the offspring. Therefore, to examine the hypothesis that changes in the vaginal microbiome are associated with effects on the offspring gut microbiota and on the developing brain, we used genomic, proteomic and metabolomic technologies to examine outcomes in our mouse model of early prenatal stress. Multivariate modeling identified broad proteomic changes to the maternal vaginal environment that influence offspring microbiota composition and metabolic processes essential for normal neurodevelopment. Maternal stress altered proteins related to vaginal immunity and abundance of Lactobacillus, the prominent taxa in the maternal vagina. Loss of maternal vaginal Lactobacillus resulted in decreased transmission of this bacterium to offspring. Further, altered microbiota composition in the neonate gut corresponded with changes in metabolite profiles involved in energy balance, and with region- and sex-specific disruptions of amino acid profiles in the developing brain. Taken together, these results identify the vaginal microbiota as a novel factor by which maternal stress may contribute to reprogramming of the developing brain that may predispose individuals to neurodevelopmental disorders.
During parturition, the neonate is first exposed to a complex microbial milieu while passing through the birth canal. These microbiota colonize the neonate gut and assist in a wide variety of critical functions, including host immune maturation, metabolism, as well as extraction and availability of substrates necessary for growth (1–5). Disruptions to the vaginal ecosystem via perturbations in the prenatal environment, such as by maternal stress, could have significant and long-term consequences for the offspring (6–8). Gastrointestinal dysfunction and dysbiosis of gut microbes has been associated with exacerbated behavioral symptoms and severity in children with autism spectrum disorders, consistent with studies in rodent models showing that a normal gut microbiota can influence brain development and behavior (9–18). Although stress during pregnancy may disrupt the normal composition of the vagina, the potential involvement of these changes in the microbiome in neurodevelopmental reprogramming has not been considered (8, 19–22). Further, as the timing of neonate gut colonization overlaps with a critical period of neurodevelopment, shifts in composition of the colonizing microbiota due to maternal stress could impact nutrient metabolism and availability in the neonate (6).
The bacterial communities that dominate colonization of the newborn gut are limited to a consortium of taxa that closely resemble the maternal vaginal microbiota, such as Lactobacillus, Prevotella, and Snethia species (23, 24). Environmental factors including antenatal antibiotic exposure, mode of delivery, and stress can alter colonization of Lactobacillus in the neonate gut, with lasting consequences on immune function, metabolism, and behavior (25–32). These results are in line with emerging evidence that maternal loss of vaginal commensal bacteria parallel similar patterns in the offspring gut, although the impact of these environmental factors, such as stress, on maternal-offspring microbial transmission and downstream offspring consequences has not been examined (33–35). Because the developing brain exhibits a substantially high metabolic demand during this period, microbial colonization may be poised to influence the efficiency of nutrient extraction and availability, further impacting accessibility of circulating energy substrate (18, 36–38). Impaired availability or transport of substrates such as amino acids (AAs) into the brain can influence specific regions undergoing maturation during this critical developmental window and further mediate disease risk (39).
Therefore, to examine the potential contribution of changes in the maternal vaginal microbiota in offspring programming effects from maternal stress, we used our established mouse model of early prenatal stress (EPS), in which male, but not female, offspring demonstrate significant neurodevelopmental changes in hypothalamic and limbic circuits and in regulation of stress responsivity, cognitive dysfunction, and postpubertal growth (40–46). Because perturbations such as maternal stress increase neurodevelopmental disease risk, disruptions to the composition of the vaginal ecosystem could be a contributing factor in significant and long-term consequences for the offspring (47–49). Specifically, we hypothesize that maternal stress alters vaginal microbiota composition, and the vertical transmission of this dysbiotic community may promote distinct bacterial colonization patterns in the offspring gut, impairing the metabolism, availability and use of nutrients necessary for normal neurodevelopment in a sex-specific manner. To examine this, the maternal vaginal and neonate gut microbiota, in addition to neonate colon, plasma, and brain were examined using genomic, proteomic and metabolomic technologies. These outcomes were then coupled with multivariate modeling to identify programmatic changes resulting from maternal stress. Time series proteome profiling was made between the end of stress (embryonic d [E]7.5) and postpartum (postnatal d [PN]2) periods to assess long-term impacts of stress on the vaginal environment. In the neonate, offspring sex was included as a factor to discern between outcomes that correlate with programming of the EPS phenotype, which we have reported is only detected in male offspring (40–46). Because neurodevelopmental disorders have strong sex biases, including 4:1 males:females in autism spectrum disorders, identification of such sex differences in mechanisms related to brain development are of critical importance.
Materials and Methods
Experimental animals
Male C57BL/6J and female 129S1/SvImJ mice were obtained from The Jackson Laboratory and subsequently used as breeding stock to produce C57BL/6J:129S1/SvImJ hybrids (F1 hybrids). A total of F1 hybrid breeding pairs (n = 44) were used for these studies, and females were checked daily at 7 am for copulation plugs. Dams were housed under a 12-hour light, 12-hour dark photoperiod with a standard chow diet (28.1% protein, 59.8% carbohydrate, and 12.1% fat; Purina Rodent Chow) and ad libitum access to water. Noon on the day that the plug was observed was considered to be E0.5. Samples from one male and one female per litter were used for all subsequent analysis. All experiments were approved by the University of Pennsylvania Institutional Animal Care and Use Committee and performed in accordance with National Institutes of Health Animal Care and Use Guidelines.
Early prenatal stress
Administration of chronic variable stress was performed as previously described (40–46). Dams were randomly assigned to treatment groups to receive stress during days 1–7 of gestation (EPS; n = 21) or to a control (n = 23) nonstressed group. After confirmation of a copulation plug, pregnant mice assigned to the EPS group experienced each of the next stressors on different days: 60 minutes of fox odor exposure (1:5000, 2,4,5-trimethylthiazole; Acros Organics), 15 minutes of restraint (beginning at 1 pm) in a modified 50-mL conical tube, 36 hours of constant light, novel noise (White Noise/Nature Sound-Sleep Machine; Brookstone) overnight, 3 cage changes throughout the light cycle, saturated bedding (700 mL, 23°C water) overnight, and novel object (8 marbles of similar shape and color) exposure overnight. Stressors were selected to be nonhabituating and did not induce pain or directly influence maternal food intake, weight gain, behavior, litter size, or sex ratio (40–46).
Tissue collection
For assessing relationship between maternal and pup gut Lactobacillus, vaginal lavages (n = 8 stress and 13 control females) were obtained by triturating 125 μL of sterile saline in the vagina of the dam 2 days postpartum (PN2). Maternal vaginal lavage samples were collected at PN2 as lavage collection near the time of parturition would not only prematurely induce delivery, but may also alter the composition of microbiota to which offspring are exposed within the birth canal, thereby confounding the effect of stress on the maternal vaginal microbiota and subsequent neonate colonization. Maternal fecal pellets were collected at the same time point. Samples were snap frozen in liquid nitrogen and stored at −80°C until ready for analysis.
Vaginal proteomics
Temporal effects of EPS on vaginal mucosal function were assessed in a separate cohort (n = 5–6 females per treatment and time point). Vaginal lavage samples were collected at E7.5 and PN2 after the procedures as mentioned above. For proteomics analysis, 25 μL of vaginal lavage sample was denatured with dithiothreitol and protein was precipitated with a 4:1 acetone:water solution. Vaginal protein extracts were trypsin digested and analyzed using liquid chromatography-mass spectrometry methods to determine AA sequences. These sequences were then used to infer protein counts using Scaffold 3.0 (Proteome Software), which were then normalized to median per sample count, and then these values were used for subsequent analyses.
Colon and plasma metabolomics
PN2 distal colon (n = 5–6 per sex and treatment) was removed and snap frozen in liquid nitrogen and stored at −80°C until ready for analysis. PN2 trunk blood (n = 6–7 per sex and treatment) was collected at time of killing, centrifuged at 5000g at 4°C for 10 minutes and plasma was aspirated. Proteins were removed from the distal colon and plasma samples by precipitation. Subsequently, water soluble metabolites were extracted from colon and plasma with a dual methanol and 40:40:20 methanol:acetonitrile:water extraction methods. Fatty acid metabolites were extracted from plasma and colon using an ethyl-acetate extraction method. Both water-soluble and fatty acid extracts were sent to the University of Pennsylvania Diabetes Research Center Metabolomics Core for analysis using standard liquid chromatography-mass spectrometry methods and compared with a background profile of mass/charge ratios of known metabolites, as previously described (50, 51).
Brain AA HPLC analysis
PN2 brains (n = 8 per sex and treatment) were cryosectioned at −20°C. Using a 0.75 mm in diameter hollow needle (Ted Pella, Inc), brain regions were micropunched according to the atlas of the developing mouse brain as follows: paraventricular nucleus, 2 successive from 2 250-μm slices +3.27 mm to +3.75 mm anterior to bregma, atlas figures 64–68; paraventricular thalamus, 2 successive punches from 2 300-μm slices +3.15 mm to +3.39 mm, atlas figures 63–65; dorsal hippocampus, bilateral punches from 2 successive 300 μm slices from +3.39 mm to +3.87 mm anterior to bregma, atlas figures 65–69. PN2 hippocampus, Paraventricular nucleus of the thalamus-enriched (PvThal), and paraventricular nucleus of the hypothalamus (PVN)-enriched punches (n = 6 per sex/treatment/region) were plunged into a 0.5-mL tube, snap frozen in liquid nitrogen, and stored at −80°C until ready for analysis. The concentration of AAs was determined by the University of Pennsylvania Children Hospital Metabolomic Core using an Agilent 1260 Infinity LC system, using precolumn derivatization with o-phthalaldehyde, as previously described (52).
C-section surgeries
Donor and foster females (n = 5–10 females per group) were time-mated, and 24 hours before expected delivery (E18.5), vaginal fluid was collected from donor dams with 200 μL of sterile saline and the entire contents of the pipette were expelled into the tube, which remained at room temperature until neonatal gavage. Immediately after vaginal lavaging, cesarean deliveries of offspring were conducted under sterile conditions according to established protocols with modifications (53, 54). See Supplemental Materials and Methods.
Quantitative PCR
Extraction of genomic 16S rRNA from maternal vaginal lavage, maternal fecal pellet, and offspring distal gut was processed using the PSP Spin Stool DNA kit manufacturers protocol for difficult to lyse bacteria (Stratec Biomedical). Quantification of total bacteria count is as previously described (55). Previously published 16S rDNA-targeted primers were used for the identification and detection of bacteria. Supplemental Table 1 shows the genus-specific primers used for quantification of relative abundance in maternal vaginal lavage, maternal fecal pellets, and offspring colon. Samples were run in technical triplicates with a critical threshold standard deviation cut-off set at 0.5, and calculated relative bacterial abundance was normalized to total bacteria. See Supplemental Materials and Methods.
Statistical analysis
Given the high degree of interrelatedness of molecules detected in proteomics and metabolomics, multivariate statistical methods were chosen over more traditional univariate statistical approaches (56–60). Multivariate modeling was conducted in SIMCA-P+ (UMETRICS). Significance of these proteins or metabolites identified by multivariate modeling was verified using univariate statistical testing, with a P < .05. Gene Ontology (GO) enrichment analysis was conducted using proteins identified by multivariate modeling using the Function Annotation Chart using only Gene Ontology Biological Process “fat” terms in the Database for Visualization and Integrative Discovery (DAVID) (61). Selection criterion included more than or equal to 3 differentially altered proteins within a Gene Ontology Biological Process term and P < .05. PN2 plasma and colon metabolite set enrichment analysis (MSEA) was performed using MetaboAnalyst, a web-based analytical pipeline for high-throughput metabolomics studies (62, 63). ANOVA, t tests, and Tukey's Honest Significant Difference post hoc comparisons for group-level contrasts were performed within the R environment (version 2.14.2) using Bonferroni corrected α to control for multiple comparisons. GraphPad Prism version 5.02 (GraphPad Software) was used to create figures. Data are presented as mean ± SEM. See Supplemental Materials and Methods.
Results
Stress during pregnancy produces long-term effects on the vaginal mucosal environment
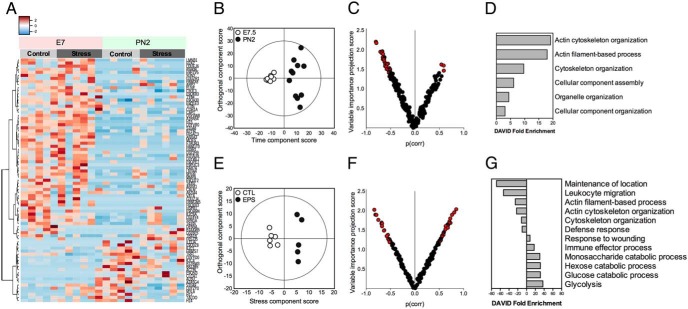
To establish the long-term impact of stress on the vaginal mucosal environment, pregnant dams were exposed to chronic variable stress or not during the first week of pregnancy (E1–E7). Vaginal samples were collected from the same control or stressed dams either immediately after stress exposure (E7.5) or in the early postpartum period (PN2). By proteomics, a total of 375 unique proteins were extracted for each sample at E7.5 and PN2. Hierarchical clustering analysis revealed transient shifts in protein profiles across pregnancy that interacted with stress (Figure 1A). To determine whether stress during the first week of pregnancy results in distinct proteomic signatures, multivariate models were constructed. Orthogonal partial least squares discriminant analysis (OPLS-DA) showed clear separation between protein profiles at E7.5 vs PN2, confirming dynamic shifts in the vaginal mucosal protein profiles across pregnancy (Q2) (cumulative) = 0.664, total amount of variance explained in the x matrix (R2X) (cumulative) = 0.238, and total amount of variance explained in the y matrix (R2Y) (cumulative) = 0.974 (P[CV-ANOVA] = 0.000996) (Figure 1B). According to this multivariate model, 39 proteins were down-regulated and 6 proteins were up-regulated at PN2 relative to E7.5 (Figure 1C and Supplemental Dataset 1). To determine whether these proteins clustered to distinct pathways, overrepresentation analysis using Gene Ontology terms were performed, and revealed significant associations with cytoskeletal organization and assembly (Figure 1D and Supplemental Dataset 2).
Figure 1.
Early pregnancy stress altered protein content of the maternal vaginal mucosal environment measured by proteomics assessment. A, Heat map of vaginal proteins with significantly different abundance levels between control and stress-exposed females at E7.5 and PN2, demonstrating changes in protein profiles across pregnancy and exposure to stress. B, OPLS-DA score plots showing significant separation of vaginal protein profiles at E7.5 (n = 10) vs PN2 (n = 11), indicating that the vaginal proteome was significantly different at these 2 time points. C, Volcano plot of vaginal proteins demonstrating the contribution that each protein made to the differences between time points. Highlighted in red are proteins with VIP scores above 1 and considered to be strong contributors to group separation, showing that most proteins are down-regulated at PN2 relative to E7.5. VIP, Variable Importance in the Projection. D, Differentially abundant proteins identified by volcano plots grouped into functional annotation categories using DAVID functional clustering tools, indicating that differentially abundant proteins cluster to pathways related to cytoskeletal reorganization. E, OPLS-DA scores plot showing clear separation between control (n = 6) and stress (n = 5) groups at PN2, suggesting that stress drives distinct vaginal protein profiles at this time. F, Volcano plot of vaginal proteins demonstrating the contribution that each protein makes to the differences between control and stress dams. Highlighted in red are proteins with VIP scores above 1 and considered to be strong contributors to group separation, showing 45 proteins differentially regulated by stress at PN2. G, Proteins differentially regulated by stress were grouped into functional annotation categories using the DAVID clustering tool. Negative DAVID enrichment scores correspond to significantly enriched pathways by proteins decreased by stress, whereas positive DAVID enrichment scores correspond to significantly enriched pathways by proteins increased by stress.
Further, the protein profiles at E7.5 projected toward a common central area in the first model, indicating that the protein profiles of control and stress females at E7.5 are relatively undifferentiated (Figure 1B). A subsequent OPLS-DA model confirmed no significant separation between control and stress females at E7.5, although subsequent univariate analysis adjusted for multiple comparisons revealed that stress significantly reduced levels of lactoferrin (t(14) = 4.604, P = .0004) (Supplemental Figure 1). In contrast, the significant spread along the orthogonal component (y axis) at PN2 indicated that another variable unrelated to time is contributing to this variance (Figure 1B). Modeling by OPLS-DA revealed clear separation between stress and control females at PN2 (Q2 [cumulative] = 0.661, R2X [cumulative] = 0.359, R2Y [cumulative] = 0.765; P[CV-ANOVA] = 0.02274) (Figure 1E). Stress exposure elevated 29 proteins while simultaneously decreasing 34 proteins relative to control females (Figure 1F and Supplemental Dataset 3). To determine whether these proteins clustered to distinct biological pathways, overrepresentation analysis using GO terms was performed and revealed significant involvement with altered defense response, response to wounding, leukocyte migration, and immune effector processes (Figure 1G and Supplemental Dataset 4).
Stress during pregnancy is associated with disruption of maternal vaginal and offspring gut microbiota composition
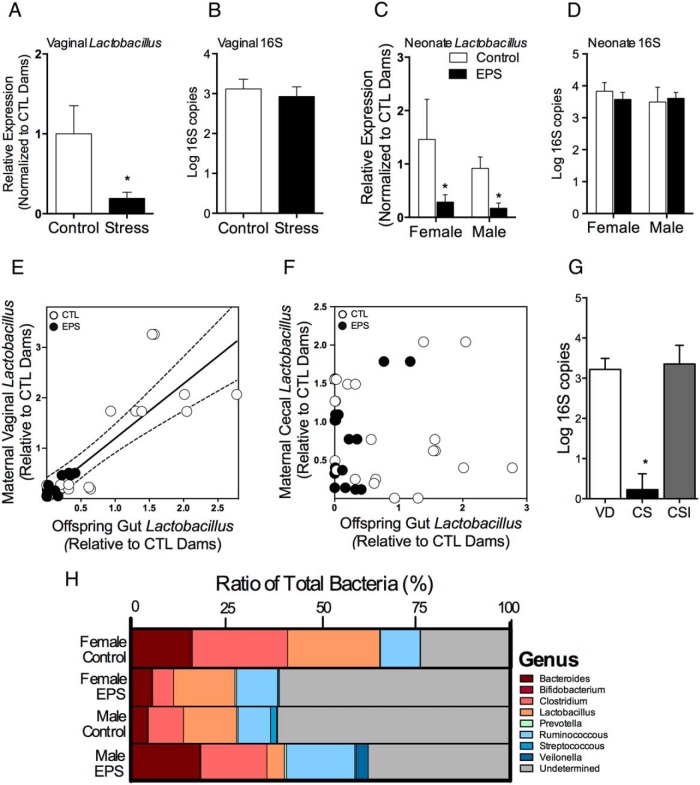
Because alterations in vaginal barrier defense and innate immune function are associated with dysbiosis to the vaginal microbiota, we next determined whether stress altered Lactobacillus abundance, the prominent taxa of the vaginal microbiota, using a quantitative PCR strategy. Vaginal Lactobacillus abundance was decreased in dams exposed to stress relative to control dams (t(15) = 1.880, P = .0398) (Figure 2A). To confirm that the suppression of vaginal Lactobacillus abundance was not related to effects of stress on the total bacterial load colonizing the vagina at PN2, we used a quantitative PCR strategy based on a small amplicon of 16S rRNA gene (55). The microbial census in stress and control dams showed no significant differences in bacterial counts in control and EPS females (t(30) = 0.54, P = .595) (Figure 2B), indicating that loss of Lactobacillus may be associated with outgrowth by other microbiota.
Figure 2.
Stress early in pregnancy altered maternal Lactobacillus transmission to offspring and more broadly disrupted neonate gut microbiota composition. A, Vaginal Lactobacillus abundance at PN2 was significantly decreased in dams exposed to EPS (n = 8) compared with control dams (n = 13). B, No difference in overall PN2 vaginal microbial census was noted. C, Comparison of Lactobacillus abundance in the PN2 neonate gut from control (n = 13) and offspring exposed to EPS (n = 8). Asterisks indicate a main effect of treatment by univariate analysis. D, No difference in PN2 neonate gut microbial census was noted. E, Maternal vaginal Lactobacillus levels were significantly positively correlated with neonate distal gut Lactobacillus levels. Solid line signifies the best-fit line from a linear regression, and the dotted lines signify the 95% confidence interval (CI) for that best fit line. F, No relationship was noted between the maternal fecal Lactobacillus and offspring gut Lactobacillus levels. G, Disrupting transmission of maternal vaginal microbiota by cesarean delivery significantly reduced total bacterial count in the PN2 neonate gut, whereas seeding cesarean delivered offspring with maternal vaginal fluid recovered total bacterial count compared with VD offspring. VD, vaginally delivered; CS, non-inoculated cesarean delivered; CSI, inoculated CS (n = 4–5 offspring/mode of delivery). Asterisks indicate a significant difference between CS offspring and all other groups determined by univariate analysis. H, Effect of EPS on selected bacterial genera in the PN2 neonate gut. In control offspring, a clear sex difference in microbiota composition was noted, whereas EPS disrupted microbiota composition disrupted this sex difference with male EPS offspring showing increased colonization by strict anaerobes and a microbiota profile more similar to control females (n = 8 per sex and treatment). *, P < .05.
To determine the effect of EPS on neonatal gut bacterial composition, genomic DNA was extracted from the distal colon of EPS and control male and female offspring at PN2. Lactobacillus abundance in the distal colon was decreased in EPS-exposed offspring (F1,36 = 5.29, P = .027), an effect that was independent of sex (F1,36 = 0.082, P = .77) (Figure 2C). Similar to their mothers, depletion of Lactobacillus was not related to differences in the total bacterial load colonized the offspring gut (P = .8, P =.58, and P =.51 for the main effect of EPS, sex, and sex × EPS interaction, respectively) (Figure 2D). Correlation analysis revealed that maternal vaginal Lactobacillus abundance was positively correlated with offspring's distal gut Lactobacillus abundance (r2 = 0.74, P < .0001) (Figure 2E), whereas comparison of maternal fecal Lactobacillus levels with offspring's distal gut Lactobacillus levels revealed no significant relationship (r2 = 0.028, P = .28) (Figure 2F).
To assess the impact of EPS on more broad changes in neonate gut microbial composition, 8 genus level 16S rRNA-targeted primers of selected intestinal bacteria were measured by quantitative polymerase chain reaction. The bacterial genera measured include Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, Prevotella, Ruminococcous, Streptococcus, and Veilonolla (primer sequences are listed in Supplemental Table 1). Bacterial profiles reveal distinct microbial patterns in the PN2 distal gut that were dependent on sex and EPS exposure (Figure 2G). For example, relative abundance of strict anaerobes Bacteroides and Clostridium in EPS males were increased compared with control males (18.3% vs 4.4% and 17.4% vs 9.3%, respectively), and the distal colon pattern of EPS males more closely resembled that of control females than control males (Figure 2H).
To confirm that offspring gut bacterial count is directly influenced by vertical transmission of maternal vaginal microbiota, we compared PN2 microbial census from a separate cohort of vaginally delivered offspring with offspring delivered by cesarean that were inoculated with maternal vaginal fluid. Total bacterial load was significantly decreased in noninoculated C-section offspring compared with vaginally delivered offspring (t(9) = 5.238, P = .0005) (Figure 2G). Total bacterial load between vaginally delivered and inoculated C-section offspring were similar, indicating that inoculation with maternal vaginal fluid recovered total bacterial count in offspring born by cesarean (P > .05) (Figure 2G).
EPS altered offspring gut microbial and host metabolism
To relate alterations in neonate gut microbiota composition to programmatic effects of EPS on metabolic function, we conducted global metabolomics profiling of distal colonic contents and plasma of PN2 offspring. We identified a total of 87 water-soluble metabolites and 44 fatty acids, and 112 water-soluble metabolites and 30 fatty acids in the PN2 distal colon and plasma, respectively. To determine whether EPS exposure results in distinct metabolite profiles in the distal colon and plasma, we performed similar supervised multivariate analyses detailed above. Because no effect of sex, EPS, or its interaction was observed for fatty acids in either the distal colon or plasma, these data were excluded from subsequent analysis.
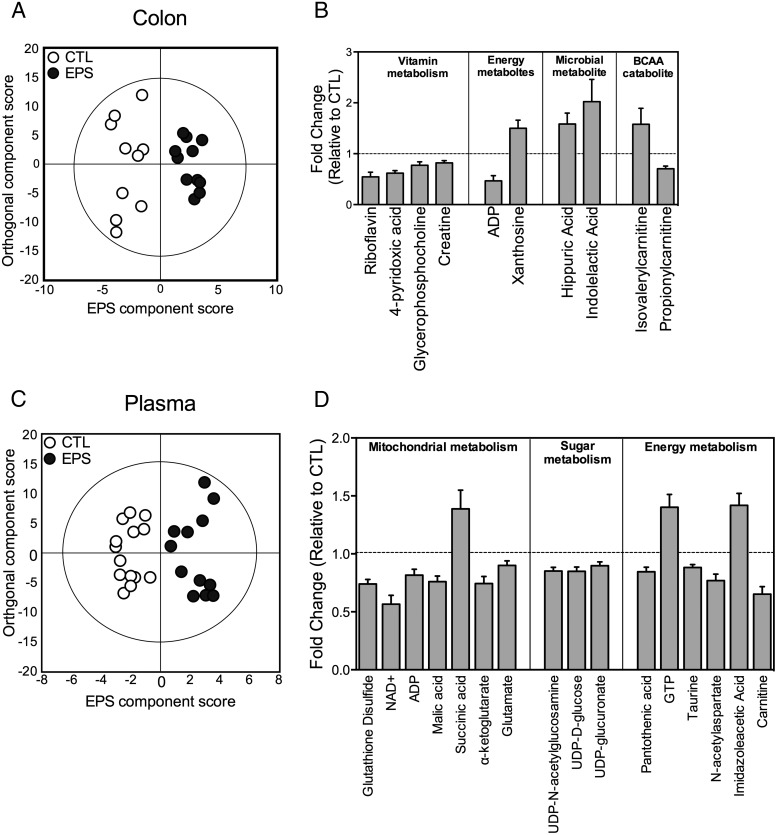
To determine whether EPS exposure resulted in distinct metabolite signatures in PN2 offspring, a series of multivariate models were constructed. Modeling by OPLS-DA revealed that colonic metabolite profiles clustered according treatment with clear separation among EPS and control offspring (R2X [cumulative] = 0.53, R2Y [cumulative] = 0.9, Q2 [cumulative] = 0.588; P[CV-ANOVA] = 0.037) (Figure 3A). This EPS-associated metabolite signature in the colon was driven by 18 metabolites, of which 12 were decreased and 6 were elevated in EPS offspring (Supplemental Dataset 5). To determine whether any processes or pathways were overrepresented by the altered metabolites, we performed a MSEA in MetaboAnalyst (62, 63). Pathways involved in RNA transcription, the urea cycle, glycolysis, gluconeogenesis, and the glucose-alanine cycle were significantly overrepresented in the metabolite signature associated with EPS (Supplemental Dataset 6 and Supplemental Figure 2). Of these colon metabolites identified by multivariate modeling, propionylcarnitine, isovalerylcarnitine, xanthosine, adenosine diphosphate, creatine, glycerophosphocholine, 4-pyridoxic acid, riboflavin, and the microbial cometabolites indolelactic acid and hippuric acid were significantly different in EPS offspring relative to control offspring (univariate P < .05) (Figure 3B).
Figure 3.
Stress alterations in neonate microbiota correlate with changes in colon and plasma metabolites as measured by metabolomics assessment. A, OPLS-DA plots showed clear separation between control (n = 10) and EPS (n = 11) offspring, indicating that EPS exposures resulted in distinct colon metabolite profiles already at PN2. B, Colon metabolites that were significantly increased or decreased in EPS offspring relative to controls were clustered into metabolic pathways identified by MSEA. Dotted lines signify mean value of control offspring. C, OPLS-DA plots showed clear separation between control (n = 14) and EPS (n = 12) offspring, indicating that EPS exposure resulted in distinct plasma metabolite profiles already at PN2. D, Plasma metabolites that were significantly increased or decreased in EPS offspring relative to controls clustered into metabolic pathways identified by MSEA.
Similarly, modeling by OPLS-DA revealed that plasma metabolite profiles clustered according to treatment with clear separation among EPS and control offspring (R2X [cumulative] = 0.555, R2Y [cumulative] = 0.877, Q2 [cumulative] = 0.603; P[CV-ANOVA] = 0.0038) (Figure 3C). This EPS-associated plasma metabolite signature was driven by 29 metabolites, of which 6 metabolites were increased and 23 metabolites were decreased relative to control offspring (Supplemental Dataset 7 and Supplemental Figure 2). MSEA revealed that these metabolites are involved in RNA transcription, glutamate metabolism, nucleotide sugars metabolism, the urea cycle, glycolysis, gluconeogenesis, starch and sucrose metabolism, mitochondrial electron transport chain, ammonia recycling, insulin signaling, malate-aspartate shuttle, citric acid cycle, and glutathione metabolism (Supplemental Dataset 8 and Supplemental Figure 2). Of these plasma metabolites identified by multivariate modeling, glutathione disulfide, nicotinamide adenine diphosphate (NAD+), adenosine diphosphate (ADP), malic acid, succinic acid, α-ketoglutarate, glutamate, Uridine diphosphate (UDP)-N-acetylglucosamine, UDP-D-glucose, UDP-glucuronate, pantothenic acid, guanosine-5′-triphosphate, taurine, N-acetylaspartate, imidazoleacetic acid, and carnitine were significantly different in EPS offspring relative to control offspring (univariate P < .05) (Figure 3D). Although no multivariate models using sex and EPS as class variables were significant, univariate analysis adjusted for multiple comparisons revealed a significant sex × treatment interaction for plasma histidine (F1,20 = 7.362, P = .013).
EPS altered AA availability in the developing brain
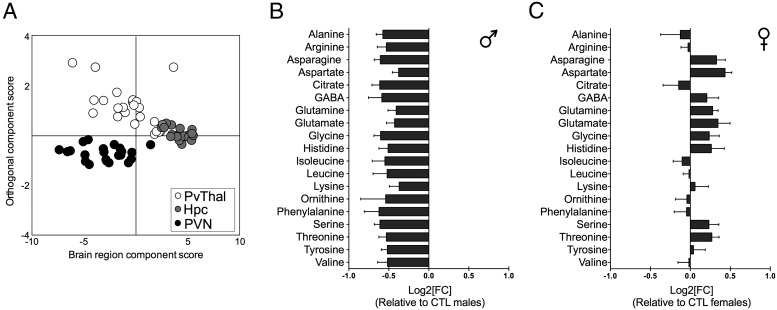
Because the developing brain comprises a significant component of the total nutritional and metabolic demand on the host, we determined whether EPS disruption of peripheral metabolism influences substrate availability in the developing brain. To identify region-specific metabolic signatures in the developing brain associated with EPS, we compared AA profiles in the PVN, PvThal and hippocampus of male and female PN2 offspring using HPLC. OPLS-DA models revealed that the PVT, PVN, and hippocampus exhibit distinct AA profiles (R2X [cumulative] = 0.895, R2Y [cumulative] = 0.674, Q2 [cumulative] = 0.615; P[CV-ANOVA] < 0.001) (Figure 4A), indicating that these regions may already exhibit metabolically differentiated states already at PN2.
Figure 4.
In accordance with changes in gut and plasma metabolites, EPS altered free AA levels in the PN2 brain in a region- and sex-specific manner. A, OPLS-DA scores plots showed significant brain region-specific differences in AA profiles of the hippocampus (Hpc), PvThal, and the PVN at PN2. B and C, AA differences with Log2 fold change in the PVN demonstrating that EPS altered the PVN AA profile in a sex-dependent manner. Stressed males (B) displayed down-regulation compared with control males, whereas stressed females (C) were more likely to display up-regulation of AAs compared with control females.
Owing to the small number of AAs detected by HPLC within each brain region at this early age, multivariate models were not able to detect treatment or sex differences. As a result, univariate analyses adjusting for multiple comparisons were used to determine the effect of EPS and sex on AA concentrations in the developing hippocampus, PvThal, and PVN (Supplemental Figures 3–5). In the PVN, a region dysregulated in our model of EPS, there was a main effect of sex for histidine (F1,18 = 8.346, P = .0098), citrate (F1,18 = 5.298, P = .034), aspartate (F1,18 = 9.717, P = .006), serine (F1,18 = 8.993, P = .008), asparagine (F1,18 = 9.298, P = .0069), histidine (F1,18 = 6.338, P = .022), glutamine (F1,18 = 7.892, P = .012), glutamate (F1,18 = 6.483, P = .02), and glycine (F1,18 = 9.182, P = .007) (Supplemental Figure 3). Post hoc analysis revealed that concentrations of these AA's were lower in the EPS male PVN relative to EPS female PVN (P < .05) (Figure 4B and C). In addition, PVN glycine concentration was lower in EPS males relative to control males (P < .05). After applying Bonferroni correction for multiple comparisons, there was no effect of sex, treatment, or its interaction on AA concentration in the PvThal (Supplemental Figure 4). In the hippocampus, there was a main effect of EPS on concentrations of glycine (F1,20 = 17.48, P = .0004), threonine (F1,20 = 13.18, P = .0018), and arginine (F1,20 = 13.46, P = .0015) (Supplemental Figure 5). No other comparisons were significant.
Discussion
Maternal stress has been identified as a significant contributor in the etiology of offspring stress dysregulation, a pervasive symptom across neuropsychiatric disorders (64, 65). Vertical transmission of maternal microbiota and the subsequent bacterial assembly of the neonate gut overlap with a critical period of neurodevelopment (8, 66). Therefore, the ability of maternal stress to alter the composition of colonizing microbiota may ultimately have negative downstream effects on immunologic, nutritional, and metabolic function in the neonate. Because the developing brain exhibits a substantially high metabolic demand during this critical window, colonization of a dysbiotic microbial community may influence efficiency of nutrient metabolism and the availability of circulating energy substrates necessary for normal development (6–8, 37, 39).
To determine whether stress during early pregnancy disrupts the vaginal mucosal ecosystem, vaginal proteomic profiles were compared at E7.5 of pregnancy and again at PN2 postpartum. As would be expected, the overall vaginal proteomic composition dramatically shifted throughout pregnancy. At E7.5, we found an increased abundance of proteins that function as structural constituents of the cytoskeleton involved in the regulation of endometrium development and maintenance of placental implantation during pregnancy (67). Although maternal stress did not broadly disrupt vaginal proteomic composition, the antimicrobial protein lactoferrin was dramatically reduced after stress and remained low throughout pregnancy. A reduction in lactoferrin has been associated with a global reduction in the bactericidal efficiency of neutrophils, suggesting that lactoferrin depletion in the vaginal mucosa in females exposed to stress may serve as a catalyst of dysregulation of vaginal immune function (68). Consistent with this, the impact of stress was most striking at PN2 with disruption in vaginal proteins involved in mobilizing defense and immune responses. Along with lactoferrin, these proteins are constituents of neutrophil granules with potent antimicrobial capacity that contribute to the protection of epithelial cells, and are involved in the maintenance of commensal bacteria in the vagina, including for Lactobacillus (69, 70). Further, as predicted, stress during pregnancy suppressed the abundance of Lactobacillus, an effect that was independent of total microbial abundance. Taken together, these results are consistent with previous studies in pregnant women showing that stress experience suppresses the vaginal immune response and induce microbial dysbiosis (71).
To evaluate the impact of maternal stress on microbiota transmission, we similarly assessed offspring gut Lactobacillus at PN2. Indeed, Lactobacillus abundance was reduced in the gut of offspring exposed to EPS relative to control offspring. These results are consistent with the previously reported reduction of Lactobacillus abundance in infant rhesus macaques exposed to prenatal stress, although the contribution of stress-induced changes to maternal Lactobacillus was not considered (31). Our results now demonstrate that stress-induced suppression of maternal Lactobacillus is correlated with altered Lactobacillus abundance in the neonate gut. Moreover, this depletion in EPS offspring was consistent with broad microbial compositional differences. Comparison of microbiota composition at the genus level revealed sex-specific changes, where EPS males exhibited a profile more similar to control females, and EPS females showed a profile more similar to control males. Depletion of the facultative anaerobe Lactobacillus corresponded to a sex-specific enrichment of strict anaerobes such as Clostridium and Bacteroides in EPS males, but not in EPS females. These results are intriguing in light of recent studies demonstrating sex differences in the gut microbiome modulating immune function and disease risk, and warrant further consideration as to the importance of sex differences in microbiota composition early in life on host metabolism and neurodevelopment (72, 73).
In order to confirm that total bacteria colonizing the offspring gut is directly influenced by vertical transmission of maternal vaginal microbiota in mice, we compared the microbial census from offspring delivered vaginally with offspring delivered by cesarean that were either seeded within minutes of delivery with maternal vaginal fluid or left unmanipulated. Cesarean delivery was associated with a reduction in total bacterial counts, consistent with previous mode-of-delivery studies in rodents and humans (23, 74). Remarkably, seeding cesarean delivered offspring with maternal vaginal fluid within minutes of delivery rescued the microbial abundance in the offspring gut. These findings confirm that the correlation between maternal vaginal and pup gut microbiota in EPS exposed mice is driven in part by microbial exposure during parturition.
To determine the potential metabolic consequences of an altered gut microbiome after EPS exposure, we examined metabolite composition in the neonatal colon and plasma using metabolomic profiling. Because the community structure of the colonizing microbiota plays a key role in the host metabolome, we hypothesized that EPS changes to the gut microbe composition, such as enrichment of strict anaerobes in EPS offspring, would be associated with altered metabolite profiles in EPS neonates (75). Indeed, combined multivariate modeling and metabolite set enrichment analyses revealed that EPS exposure altered both gut microbial and host metabolism. Specifically, EPS altered levels of indolelactic acid, a microbial cometabolite that is produced as a result of AA and tryptophan metabolism by a variety of gut bacterial species such as Candida and Bifidobacterium, although the mechanistic relevance of increased indolelactic acid levels on brain development and stress dysregulation is currently unknown (76, 77). Further, EPS altered hippuric acid levels, consistent with previous studies demonstrating an association between hippuric acid dysregulation and neuropsychiatric disorders including autism spectrum disorders and depression (78, 79). The metabolite sets enriched in the distal colon and plasma of EPS offspring suggest significant reprogramming of pathways related to energy homeostasis, most notably metabolites involved in gluconeogenesis and glycolysis. The PN2 plasma exhibited distinct metabolic profiles in EPS exposed offspring, including alterations in metabolites involved in nutrient metabolism, oxidative stress regulation, and mitochondrial function, metabolic processes that have been previously implicated with increased neuropsychiatric disease risk (80–83). Together, these results suggest that the composition of the early colonizing microbiota may exert a significant impact on metabolism and provide a novel mode of transmission by which EPS may influence developmental outcomes.
To determine whether these peripheral metabolic changes related to energy balance and AA metabolism influence the metabolic programming of the developing brain, we measured free AA profiles of the developing PVN, hippocampus, and PvThal, brain regions important in central stress axis regulation and, and where we have previously demonstrated changes in gene expression in our EPS male mice (40–46). The postpartum window of development is a time of increased capillary transport of AAs into the brain where they provide the necessary substrate for rapid cerebral protein formation (84–88). Consistent with this, we found by proteomics assessment that EPS produced brain region and sex-specific changes in AA profiles. Specifically, levels of serine, histidine, asparagine, glutamine, glutamate, and glycine were significantly reduced in EPS males, but not EPS females. This appeared to be specific to the PVN, as similar sex-specific effects of EPS were not detected in either the hippocampus or PvThal. Because the developing hypothalamus is relatively accessible to circulating factors and readily responds to peripheral metabolic signals, this may provide a mechanism whereby changes in gut microbiota composition and peripheral metabolism would have a region-specific effect on central stress axis regulation (84, 85). Sex-specific changes in AA availability may also exert downstream effects on a variety of processes in the developing brain, including metabolism, neurotransmission, and synaptic plasticity (89). In addition, these results support our previous studies demonstrating an effect of EPS in the hypothalamus that correlate with long-term and sex-specific effects on growth, metabolism, and stress dysregulation (42, 43, 45, 46).
Vertical transmission of maternal microbiota to offspring is an important factor in mediating disease risk and resilience. Although the impact of maternal stress on neurodevelopment likely involves complex mechanisms at multiple levels, the current study examined the correlation of the maternal vaginal microbiota and subsequent colonization of the neonate gut as an important contributing factor in prenatal stress programming (47–49). As a first step in defining this model, the current studies identified complex associations between the maternal vaginal and offspring gut microbiome, and the altered peripheral and central metabolite profiles in EPS offspring, many of which were sex-specific. Future studies will now need to focus on further defining the mechanisms of these associations, and producing studies in which causality can be demonstrated between the neonate microbiome and the developing brain.
Acknowledgments
We thank Dan Beiting and Ana Misic for technical assistance with microbiome analysis, Aaalim Weljie and Arjun Sengupta for assistance with multivariate analysis, Illana Nissim at the University of Pennsylvania CHOP Metabolomics Core for HPLC analysis, and the Perelman School of Medicine Proteomics and Systems Biology Core for proteomics assessment. We also thank Stefanie Bronson, Jen Chen, Katie Morrison, Chris Morgan, Bridget Nugent, and Ali Rodgers for insightful discussion. We thank the University of Pennsylvania Diabetes Research Center (DRC) for the use of the Metabolomics Core (P30-DK19525).
This work was supported by a pilot award from the PennVet Center for Host-Microbial Interactions at the University of Pennsylvania and by National Institutes of Health Grants MH104184, MH091258, and MH087597.
Disclosure Summary: The authors have nothing to disclose.
For News & Views see page 3066
- AA
- amino acid
- ADP
- adenonine diphosphate
- DAVID
- Database for Visualization and Integrative Discovery
- E
- embryonic day
- EPS
- early prenatal stress
- GTP
- Guanosine-5′-triphosphate
- MSEA
- metabolite set enrichment analysis
- NAD+
- Nicotinamide adenine dinucleotide
- OPLS-DA
- orthogonal partial least squares discriminant analysis
- PN
- postnatal day
- PVN
- paraventricular nucleus of the hypothalamus
- PvThal
- paraventricular nucleus of the thalamus
- UDP
- uridine diphosphate.
References
- 1. Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(suppl 1):4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. [DOI] [PubMed] [Google Scholar]
- 4. Cilieborg MS, Boye M, Sangild PT. Bacterial colonization and gut development in preterm neonates. Early Hum Dev. 2012;88(suppl 1):S41–S49. [DOI] [PubMed] [Google Scholar]
- 5. Siggers RH, Siggers J, Thymann T, Boye M, Sangild PT. Nutritional modulation of the gut microbiota and immune system in preterm neonates susceptible to necrotizing enterocolitis. J Nutr Biochem. 2011;22:511–521. [DOI] [PubMed] [Google Scholar]
- 6. Borre YE, O'Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med. 2014;20:509–518. [DOI] [PubMed] [Google Scholar]
- 7. Gur TL, Worly BL, Bailey MT. Stress and the commensal microbiota: importance in parturition and infant neurodevelopment. Front Psychiatry. 2015;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jašarevic E, Rodgers AB, Bale TL. A novel role for maternal stress and microbial transmission in early life programming and neurodevelopment. Neurobiol Stress. 2015;1:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. [DOI] [PubMed] [Google Scholar]
- 10. Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108:3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37:1369–1378. [DOI] [PubMed] [Google Scholar]
- 12. Finegold SM, Molitoris D, Song Y, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35:S6–S16. [DOI] [PubMed] [Google Scholar]
- 13. Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Williams BL, Hornig M, Buie T, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Braniste V, Al-Asmakh M, Kowal C, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sudo N, Chida Y, Aiba Y, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ashcraft KA, Bonneau RH. Psychological stress exacerbates primary vaginal herpes simplex virus type 1 (HSV-1) infection by impairing both innate and adaptive immune responses. Brain Behav Immun. 2008;22:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Culhane JF, Rauh V, McCollum KF, Elo IT, Hogan V. Exposure to chronic stress and ethnic differences in rates of bacterial vaginosis among pregnant women. Am J Obstet Gynecol. 2002;187:1272–1276. [DOI] [PubMed] [Google Scholar]
- 21. Culhane JF, Rauh V, McCollum KF, Hogan VK, Agnew K, Wadhwa PD. Maternal stress is associated with bacterial vaginosis in human pregnancy. Matern Child Health J. 2001;5:127–134. [DOI] [PubMed] [Google Scholar]
- 22. Wadhwa PD, Culhane JF, Rauh V, et al. Stress, infection and preterm birth: a biobehavioural perspective. Paediatr Perinat Ep. 2001;15:17–29. [DOI] [PubMed] [Google Scholar]
- 23. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(suppl 1):4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Mahony SM, Felice VD, Nally K, et al. Disturbance of the gut microbiota in early-life selectively affects visceral pain in adulthood without impacting cognitive or anxiety-related behaviors in male rats. Neuroscience. 2014;277:885–901. [DOI] [PubMed] [Google Scholar]
- 26. O'Mahony SM, Hyland NP, Dinan TG, Cryan JF. Maternal separation as a model of brain-gut axis dysfunction. Psychopharmacology. 2011;214:71–88. [DOI] [PubMed] [Google Scholar]
- 27. Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(suppl 1):13–15. [DOI] [PubMed] [Google Scholar]
- 28. Bouhanick B, Ehlinger V, Delpierre C, Chamontin B, Lang T, Kelly-Irving M. Mode of delivery at birth and the metabolic syndrome in midlife: the role of the birth environment in a prospective birth cohort study. BMJ Open. 2014;4:e005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hansen CH, Andersen LS, Krych L, et al. Mode of delivery shapes gut colonization pattern and modulates regulatory immunity in mice. J Immunol. 2014;193:1213–1222. [DOI] [PubMed] [Google Scholar]
- 30. Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol. 1999;35:146–155. [PubMed] [Google Scholar]
- 31. Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J Pediatr Gastroenterol Nutr. 2004;38:414–421. [DOI] [PubMed] [Google Scholar]
- 32. Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keski-Nisula L, Kyynäräinen HR, Kärkkäinen U, Karhukorpi J, Heinonen S, Pekkanen J. Maternal intrapartum antibiotics and decreased vertical transmission of Lactobacillus to neonates during birth. Acta Paediatr. 2013;102:480–485. [DOI] [PubMed] [Google Scholar]
- 34. Matsumiya Y, Kato N, Watanabe K, Kato H. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J Infect Chemother. 2002;8:43–49. [DOI] [PubMed] [Google Scholar]
- 35. Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. [DOI] [PubMed] [Google Scholar]
- 37. Georgieff MK. Nutrition and the developing brain: nutrient priorities and measurement. Am J Clin Nutr. 2007;85:614S–620S [DOI] [PubMed] [Google Scholar]
- 38. Manco M. Gut microbiota and developmental programming of the brain: from evidence in behavioral endophenotypes to novel perspective in obesity. Front Cell Infect Microbiol. 2012;2:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keunen K, van Elburg RM, van Bel F, Benders MJ. Impact of nutrition on brain development and its neuroprotective implications following preterm birth. Pediatr Res. 2015;77:148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bronson SL, Bale TL. Prenatal stress-induced increases in placental inflammation and offspring hyperactivity are male-specific and ameliorated by maternal antiinflammatory treatment. Endocrinology. 2014;155:2635–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Howerton CL, Bale TL. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc Natl Acad Sci USA. 2014;111:9639–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci USA. 2013;110:5169–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mueller BR, Bale TL. Impact of prenatal stress on long term body weight is dependent on timing and maternal sensitivity. Physiol Behav. 2006;88:605–614. [DOI] [PubMed] [Google Scholar]
- 44. Mueller BR, Bale TL. Early prenatal stress impact on coping strategies and learning performance is sex dependent. Physiol Behav. 2007;91:55–65. [DOI] [PubMed] [Google Scholar]
- 45. Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pankevich DE, Mueller BR, Brockel B, Bale TL. Prenatal stress programming of offspring feeding behavior and energy balance begins early in pregnancy. Physiol Behav. 2009;98:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Champagne FA, Meaney MJ. Stress during gestation alters postpartum maternal care and the development of the offspring in a rodent model. Biol Psychiatry. 2006;59:1227–1235. [DOI] [PubMed] [Google Scholar]
- 48. Purcell RH, Sun B, Pass LL, Power ML, Moran TH, Tamashiro KL. Maternal stress and high-fat diet effect on maternal behavior, milk composition, and pup ingestive behavior. Physiol Behav. 2011;104:474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Welberg LA, Seckl JR. Prenatal stress, glucocorticoids and the programming of the brain. J Neuroendocrinol. 2001;13:113–128. [DOI] [PubMed] [Google Scholar]
- 50. Kamphorst JJ, Fan J, Lu W, White E, Rabinowitz JD. Liquid chromatography-high resolution mass spectrometry analysis of fatty acid metabolism. Anal Chem. 2011;83:9114–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones BN, Gilligan JP. Ortho-phthaldialdehyde precolumn derivatization and reversed-phase high-performance liquid-chromatography of polypeptide hydrolysates and physiological fluids. J Chromatogr A. 1983;266:471–482. [DOI] [PubMed] [Google Scholar]
- 53. Nagy A, Gertsenstein M, Vintersten K, Behringer R. Caesarean section and fostering. CSH Protoc. 2006;2006. [DOI] [PubMed] [Google Scholar]
- 54. Murphy D. Caesarean section and fostering. Methods Mol Biol. 1993;18:177–178. [DOI] [PubMed] [Google Scholar]
- 55. Castillo M, Martín-Orúe SM, Manzanilla EG, Badiola I, Martín M, Gasa J. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol. 2006;114:165–170. [DOI] [PubMed] [Google Scholar]
- 56. Eriksson L, Antti H, Gottfries J, et al. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal Bioanal Chem. 2004;380:419–429. [DOI] [PubMed] [Google Scholar]
- 57. Eriksson L, Trygg J, Wold S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemometr. 2008;22:594–600. [Google Scholar]
- 58. Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. [DOI] [PubMed] [Google Scholar]
- 59. Trygg J, Wold S. Orthogonal projections to latent structures (OPLS). J Chemom. 2002;16:119–128. [Google Scholar]
- 60. Wheelock ÅM, Wheelock CE. Trials and tribulations of 'omics data analysis: assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol Biosyst. 2013;9:2589–2596. [DOI] [PubMed] [Google Scholar]
- 61. Sherman BT, Huang da W, Tan Q, et al. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–W133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Howerton CL, Bale TL. Prenatal programing: at the intersection of maternal stress and immune activation. Horm Behav. 2012;62:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bale TL, Baram TZ, Brown AS, et al. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68:314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11:e1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chae JI, Kim J, Lee SG, et al. Proteomic analysis of pregnancy-related proteins from pig uterus endometrium during pregnancy. Proteome Sci. 2011;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moreno-Navarrete JM, Ortega FJ, Bassols J, Ricart W, Fernández-Real JM. Decreased circulating lactoferrin in insulin resistance and altered glucose tolerance as a possible marker of neutrophil dysfunction in type 2 diabetes. J Clin Endocrinol Metab. 2009;94:4036–4044. [DOI] [PubMed] [Google Scholar]
- 69. Lönnerdal B, Iyer S. Lactoferrin: molecular structure and biological function. Annu Rev Nutr. 1995;15:93–110. [DOI] [PubMed] [Google Scholar]
- 70. Sandrini SM, Shergill R, Woodward J, et al. Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J Bacteriol. 2010;192:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ehrström SM, Kornfeld D, Thuresson J, Rylander E. Signs of chronic stress in women with recurrent candida vulvovaginitis. Am J Obstet Gynecol. 2005;193:1376–1381. [DOI] [PubMed] [Google Scholar]
- 72. Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. [DOI] [PubMed] [Google Scholar]
- 73. Yurkovetskiy L, Burrows M, Khan AA, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–240. [DOI] [PubMed] [Google Scholar]
- 75. Ursell LK, Haiser HJ, Van Treuren W, et al. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Aragozzini F, Ferrari A, Pacini N, Gualandris R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol.. 1979;38:544–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Narayanan TK, Rao GR. β-Indoleethanol and β-indolelactic acid production by Candida species: their antibacterial and autoantibiotic action. Antimicrob Agents Chemother. 1976;9:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yap IK, Angley M, Veselkov KA, Holmes E, Lindon JC, Nicholson JK. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J Proteome Res. 2010;9:2996–3004. [DOI] [PubMed] [Google Scholar]
- 79. Zheng P, Chen JJ, Huang T, et al. A novel urinary metabolite signature for diagnosing major depressive disorder. J Proteome Res. 2013;12:5904–5911. [DOI] [PubMed] [Google Scholar]
- 80. Giulivi C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304:2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Melnyk S, Fuchs GJ, Schulz E, et al. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012;42:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Frye RE, Delatorre R, Taylor H, et al. Redox metabolism abnormalities in autistic children associated with mitochondrial disease. Transl Psychiatry. 2013;3:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Frye RE, Rossignol DA. Mitochondrial dysfunction can connect the diverse medical symptoms associated with autism spectrum disorders. Pediatr Res. 2011;69:41R–47R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Levin BE. Metabolic sensing neurons and the control of energy homeostasis. Physiol Behav. 2006;89:486–489. [DOI] [PubMed] [Google Scholar]
- 85. Horvath TL, Diano S, Sotonyi P, Heiman M, Tschöp M. Minireview: ghrelin and the regulation of energy balance–a hypothalamic perspective. Endocrinology. 2001;142:4163–4169. [DOI] [PubMed] [Google Scholar]
- 86. Pascucci T, Andolina D, Ventura R, Puglisi-Allegra S, Cabib S. Reduced availability of brain amines during critical phases of postnatal development in a genetic mouse model of cognitive delay. Brain Res. 2008;1217:232–238. [DOI] [PubMed] [Google Scholar]
- 87. Sershen H, Lajtha A. Capillar transport of amino acids in the developing brain. Exp Neurol. 1976;53:465–474. [DOI] [PubMed] [Google Scholar]
- 88. Baños G, Daniel PM, Pratt OE. The effect of age upon the entry of some amino acids into the brain, and their incorporation into cerebral protein. Dev Med Child Neurol. 1978;20:335–346. [DOI] [PubMed] [Google Scholar]
- 89. McDonald JW, Johnston MV. Physiological and pathophysiological roles of excitatory amino acids during central nervous system development. Brain Res Brain Res Rev. 1990;15:41–70. [DOI] [PubMed] [Google Scholar]