Highlights
-
•
Mycoplasmas may colonize tumor tissue in patients.
-
•
Mycoplasma-encoded cytidine deaminase deaminates cytosine-based anticancer drugs.
-
•
The activity of gemcitabine is compromised in mycoplasma-infected tumor cells.
-
•
Gemcitabine activity can be restored by nucleosides or a PNP inhibitor.
Abbreviations: 3TC, 2′,3′-dideoxy-3′-thiacytidine; ara-Cyd, cytosine arabinoside; CDA, cytidine deaminase; (d)Ado, (2′-deoxy)adenosine; (d)Guo, (2′-deoxy)guanosine; (d)Ino, (2′-deoxy)inosine; (d)Urd, (2′-deoxy)uridine; ddC, 2′,3′-dideoxycytidine; dThd, thymidine; dFdC, gemcitabine; dFdU, 2′,2′-difluoro-2′-deoxyuridine; Imm-H, Immucillin-H; NA, nucleoside analogue; PNP, purine nucleoside phosphorylase
Keywords: Mycoplasma, Cytidine deaminase, Cancer, Nucleoside analogue, Purine nucleoside phosphorylase, Gemcitabine
Abstract
Mycoplasmas may colonize tumor tissue in patients. The cytostatic activity of gemcitabine was dramatically decreased in Mycoplasma hyorhinis-infected tumor cell cultures compared with non-infected tumor cell cultures. This mycoplasma-driven drug deamination could be prevented by exogenous administration of the cytidine deaminase (CDA) inhibitor tetrahydrouridine, but also by the natural nucleosides or by a purine nucleoside phosphorylase inhibitor. The M. hyorhinis-encoded CDAHyor gene was cloned, expressed as a recombinant protein and purified. CDAHyor was found to be more catalytically active than its human equivalent and efficiently deaminates (inactivates) cytosine-based anticancer drugs. CDAHyor expression at the tumor site may result in selective drug inactivation and suboptimal therapeutic efficiency.
1. Introduction
Mycoplasmas are considered to be the smallest self-replicating organisms, both in dimension and genome size [1]. They often lack genes that are crucial for different synthetic pathways, including the de novo synthesis of purine and pyrimidine bases [2,3]. Therefore mycoplasmas rely on their host tissue from which they scavenge and recycle DNA/RNA precursors using various nucleo(s)(t)ide transporters and salvage enzymes [2,4,5]. Recently, we and others showed that certain catabolic mycoplasma enzymes (i.e. pyrimidine nucleoside phosphorylase, purine nucleoside phosphorylase and cytidine deaminase) interfere with the biological (i.e. cytostatic and antiviral) activity of different therapeutic nucleoside analogues (NAs) by producing less active or inactive drug metabolites. This was demonstrated for both pyrimidine- and purine-derived antimetabolites including gemcitabine, floxuridine, trifluridine, cladribine, and others [6–10]. There have been several reports that mycoplasmas have been shown to preferentially colonize tumor tissue in patients [11–20]. If this phenomenon can be broadly confirmed and since nucleoside-derived drugs are established cornerstones in the chemotherapy of several cancers [21], the presence of such prokaryotes in the tumor microenvironment may be a confounding factor for the efficiency of anticancer nucleoside analogues and of importance for optimization of nucleoside-based cancer treatment [22,23].
Recently, we reported efficient CDA-catalyzed deamination of gemcitabine (2′,2′-difluoro-2′-deoxycytidine; dFdC) resulting in a dramatically decreased cytostatic activity (up to 60-fold) of this drug in different Mycoplasma hyorhinis-infected tumor cell cultures [10]. Similarly, the response of M. hyorhinis-infected tumor xenografts in mice to gemcitabine treatment was significantly lower compared with uninfected control tumors [10]. The biological function of CDA is to catalyze the irreversible deamination of the natural pyrimidine nucleosides cytidine (Cyd) and 2′-deoxycytidine (dCyd) to uridine (Urd) and 2′-deoxyuridine (dUrd), respectively [24]. However, several clinical anticancer (d)Cyd analogues, including gemcitabine and cytarabine (cytosine arabinoside; ara-Cyd) (Fig. 1), can be catabolized by (cellular) drug deamination producing the corresponding, less active, (2′-deoxy)uridine metabolites. These molecules therefore show a decreased cytostatic activity in CDA-overexpressing tumor cells [25,26]. In the present study we biochemically and kinetically characterized M. hyorhinis-encoded CDA and report on a surprising interaction between mycoplasma CDA and purine nucleoside phosphorylase (PNP) activity in mycoplasma-infected tumor cells.
Fig. 1.
Molecular structure of the (2′-deoxy)cytidine analogues gemcitabine (A) and cytarabine (B).
2. Materials and methods
2.1. Chemicals
Nucleosides, nucleoside analogues and inorganic agents were purchased from Sigma–Aldrich (St-Louis, MO) unless stated differently. Gemcitabine (2′,2′-difluoro-2′-deoxycytidine; dFdC) was purchased from Carbosynth (Berkshire, UK). Radioactive [5-3H]-gemcitabine ([5-3H]-dFdC) (radiospecificity: 12 Ci/mmol) was obtained from Moravek Biochemicals Inc. (Brea, CA). Immucillin-H (Imm-H) was kindly provided by Dr. V. Schramm (Albert Einstein College of Medicine, Bronx, NY).
2.2. Cell cultures
Human breast carcinoma MDA-MB-231 cells and M. hyorhinis were obtained from the American Tissue Culture Collection (Rockville, MD). Human breast carcinoma MCF-7 cells were kindly provided by Prof. G.J. Peters (Amsterdam, The Netherlands). Cells were infected with M. hyorhinis and after two or more passages (to avoid bias by the initial inoculum) successful infection was confirmed using the MycoAlert™ mycoplasma detection kit (Lonza, Basel, Switzerland). Although this assay is only semi-quantitative, a maximal infection was observed three to four days after subculturing the mycoplasma-exposed cells. Chronically M. hyorhinis-infected tumor cells are further referred to as MDA-MB-231.Hyor and MCF-7.Hyor. All cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% foetal bovine serum (Integro, Dieren, The Netherlands), 10 mM HEPES and 1 mM sodium pyruvate (Invitrogen) and grown at 37 °C in a humidified CO2-controlled incubator.
2.3. Biological assays
The cytostatic activity of dFdC (gemcitabine) was compared in mycoplasma-infected and uninfected tumor cells. MDA-MB-231 and MDA-MB-231.Hyor cells were seeded in 48-well plates (Nunc™, Roskilde, Denmark) at 10,000 cells/well. After 24 h, an equal volume of fresh medium containing gemcitabine [in the presence or absence of natural purine nucleosides (100 μM) or the PNP inhibitor Imm-H (10 μM)] was added. Three days later (to ensure sufficient cell-proliferation and mycoplasma growth), cells were trypsinized and counted in a Coulter counter (Analis, Suarlée, Belgium). The 50% inhibitory concentration (IC50) was defined as the compound concentration required to reduce tumor cell proliferation by 50%.
2.4. Gemcitabine stability in the supernatant of mycoplasma-infected and uninfected cell cultures
The stability of gemcitabine in spent cell-free but mycoplasma-containing culture medium of confluent MDA-MB-231, MDA-MB-231.Hyor, MCF-7 and MCF-7.Hyor tumor cells was evaluated. Tumor cells were seeded in 75 cm2 culture flasks (TTP, Trasadingen, Switzerland). After five days, supernatant was withdrawn and cleared by centrifugation at 300g for 6 min to remove (debris of) the tumor cells. Reactions were performed in a final volume of 300 μL containing dFdC (5 μM), [5-3H]dFdC (1 μCi), different concentrations of thymidine (dThd), uridine (Urd), adenosine (Ado) or inosine (Ino) and 240 μL spent culture medium. Samples were incubated at 37 °C and after 60 min incubation, 100 μL was withdrawn and ice-cold MeOH was added to a final concentration of 66% MeOH to terminate the enzymatic reactions and to precipitate (remove) macromolecules such as DNA, RNA and proteins. Samples were kept on ice for 10 min and cleared by centrifugation at 16,000g for 15 min. The supernatants were withdrawn and analyzed on a reverse phase RP-8 column (Merck, Darmstadt, Germany) using HPLC (Alliance 2690, Waters, Milford, MA). The following gradient (further referred to as gradient A) was used: 10 min linear gradient of 100% buffer A [50 mM NaH2PO4 (Acros Organics, Geel, Belgium); 5 mM heptane sulfonic acid; pH 3.2] to 98% buffer A + 2% acetonitrile (BioSolve BV, Valkenswaard, the Netherlands); 10 min linear gradient to 90% buffer A + 10% acetonitrile; 5 min linear gradient to 75% buffer A + 25% acetonitrile; 5 min linear gradient to 100% buffer A followed by 10 min equilibration at 100% buffer A. Fractions of 1 mL were collected, transferred to 9 mL OptiPhase HiSafe 3 and radioactivity was counted in a liquid scintillation analyzer.
2.5. Purification of M. hyorhinis CDA (CDAHyor)
A codon-optimized DNA sequence encoding the M. hyorhinis cytidine deaminase (CDAHyor) was synthetically assembled between the EcoRI and NotI restriction sites of a pIDTsmart vector (Integrated DNA technologies, Coralville, IO). The fragment was subsequently subcloned between the EcoRI and NotI sites of the pGEX-5X-1 bacterial expression vector (Amersham Pharmacia Biotech, Uppsala, Sweden) and CDAHyor was expressed in Escherichia coli as a GST-fusion protein (hereafter referred to as CDAHyor) according to a procedure previously described by Liekens et al. [27]. SDS–PAGE revealed that the protein was of expected size (∼38–40 kDa) and purity (⩾95%) (Fig. 2). Since E. coli CDA consists of 294 amino acids, it would be characterized by a molecular weight of around 31 kDa [28]. Therefore, the contaminating protein bands shown in Fig. 2 are not likely related to E. coli-encoded CDA.
Fig. 2.
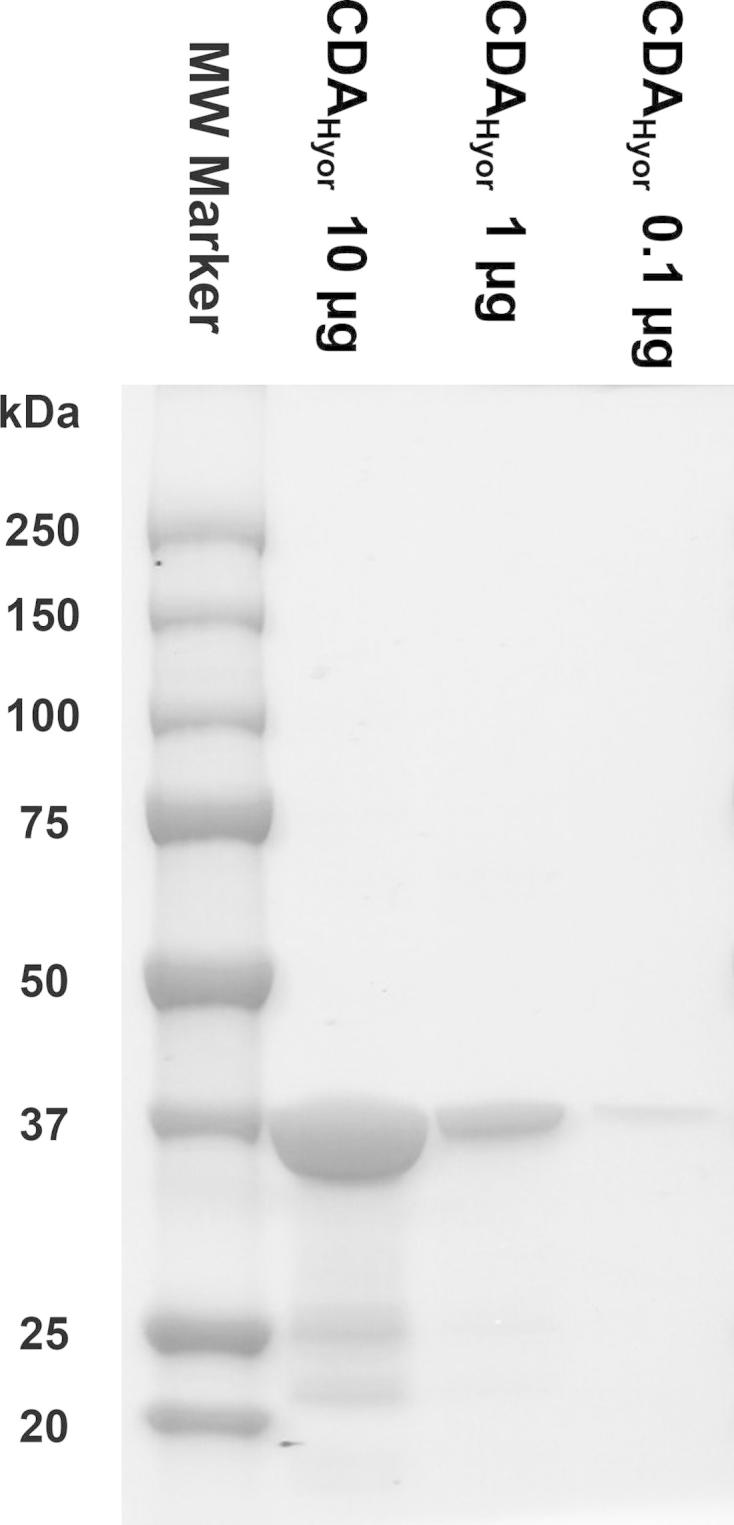
Purity evaluation of the CDAHyor-GST fusion protein. Three different concentrations (i.e. 10, 1 and 0.1 μg) of the purified enzyme preparation were analyzed using SDS–PAGE. Proteins were stained using Bio-Safe™ Coomassie G-250 Stain (Bio-Rad Laboratories, CA, USA).
2.6. Enzyme assays
2.6.1. Determination of the substrate specificity of CDAHyor and CDAHuman
To study the deamination of different nucleosides and nucleoside analogues by CDAHyor and CDAHuman (ProSpec, Rehovot, Israel) different potential substrates (100 μM) were exposed to both enzymes (80 nM CDAHyor or 27 nM CDAHuman) and incubated at 37 °C in PBS in a total volume of 300 μL. At different time points, 100 μL-fractions were withdrawn and the reaction was terminated by heat-inactivation of the enzyme at 95 °C for 3 min. Next, the samples were rapidly cooled on ice for 15 min and cleared by centrifugation at 16,000g for 15 min. Nucleosides were separated on a reverse phase RP-8 column (Merck) and quantified by HPLC analysis. For each product UV-based detection was performed at the specific wavelength of optimal absorption.
The separation of (2′-deoxy)cytidine [(d)Cyd] from (2′-deoxy)uridine [(d)Urd] was performed by HPLC using linear gradient B [from 100% buffer A and 2% acetonitrile to 80% buffer A and 20% acetonitrile] as follows: 5 min 100% buffer A; 5 min linear gradient to 80% buffer A + 20% acetonitrile; 5 min linear gradient to 100% buffer A followed by 5 min equilibration at 100% buffer A. Samples containing dFdC, ara-Cyd, 5-aza-2′-deoxycytidine, 5-aza-cytidine, 2′,3′-dideoxycytidine (ddC) or 2′,3′-dideoxy-3′-thiacytidine (3TC) were analyzed by HPLC using linear gradient A as described above.
2.6.2. Kinetic assays
Kinetic studies were performed for the deamination of different substrates [Cyd, dCyd, dFdC and ara-Cyd] by CDAHyor and CDAHuman. Deamination was studied at varying substrate concentrations ranging from 100 μM to 45 mM in a reaction containing 4 nM CDAHyor or 11 nM CDAHuman incubated in PBS at 37 °C for 10 min. Samples were processed and analyzed by HPLC as described above. Kinetic parameters (KM and kcat) were determined by means of non-linear regression analysis (using GraphPad Prism5) and the ratio kcat/KM (catalytic efficiency) was calculated.
3. Results
3.1. Deamination of gemcitabine by mycoplasma CDA can be prevented in the presence of exogenous natural purine and pyrimidine nucleosides
The cytostatic activity of gemcitabine (dFdC) was decreased by ∼36 fold in M. hyorhinis-infected MDA-MB-231 breast cancer cell cultures compared with uninfected control tumor cells (Table 1). This could be prevented by co-administration of 250 μM of the cytidine deaminase inhibitor tetrahydrouridine (THU), but also by natural purine nucleosides (i.e. Ado, Ino or Guo) as well as by Immucillin-H (Imm-H), a potent inhibitor of mycoplasma PNP (Table 1).
Table 1.
Cytostatic activity of gemcitabine (dFdC) in MDA-MB-231 and MDA-MB-231. Hyor cells in the absence/presence of the selective CDA inhibitor tetrahydrouridine, natural purine nucleosides or the selective purine nucleoside phosphorylase inhibitor Immucillin-H (Imm-H). Results are the mean ± S.D. of at least two independent experiments.
| IC50a value of dFdC (μM) |
||||||
|---|---|---|---|---|---|---|
| As such | +THU (250 μM) | +Ado (100 μM) | +Ino (100 μM) | +Guo (100 μM) | +Imm-H (10 μM) | |
| MDA-MB-231 | 0.0042 ± 0.00041 | 0.004 ± 0.0003 | 0.0031 ± 0.0014 | 0.0037 ± 0.00076 | 0.0042 ± 0.0012 | 0.0037 ± 0.00076 |
| MDA-MB-231.Hyor | 0.15 ± 0.016 | 0.004 ± 0.0001 | 0.0096 ± 0.0049 | 0.0099 ± 0.0042 | 0.025 ± 0.0089 | 0.0051 ± 0.0021 |
| Fold difference | ∼36 | 1 | ∼3 | ∼3 | ∼6 | ∼1.5 |
50% Inhibitory concentration or compound concentration required to inhibit tumor cell proliferation by 50%.
Since THU could efficiently restore the cytostatic activity of dFdC in the presence of M. hyorhinis in the tumor cell culture medium, the stability of radiolabeled dFdC ([5-3H]dFdC) was studied in the tumor cell-free (but mycoplasma-containing) culture medium of M. hyorhinis-infected MDA-MB-231 (Fig. 3A) and MCF-7 (Fig. 3B) breast cancer cells. A pronounced inhibition of [5-3H]-dFdC deamination (i.e. decreased formation of the inactive metabolite [5-3H]-dFdU) by 1 mM THU could be observed [10], but also, a dose-dependent inhibition of [5-3H]dFdC deamination by the exogenous supply of natural pyrimidine (i.e. dThd and Urd) or purine (i.e. Ado and Ino) nucleosides was observed in the spent tumor cell culture medium (Fig. 3). Also, [5-3H]-dFdU formation could be inhibited by exogenous administration of other natural nucleosides such as Guo, dAdo, dIno or dGuo (data not shown).
Fig. 3.
Inhibition of mycoplasma-associated [5-3H]dFdU formation by natural nucleosides. Formation of [5-3H]dFdU from [5-3H]dFdC in the tumor cell-free supernatant of mycoplasma-infected and control MDA.MB.231 (A) and MCF-7 (B) breast cancer cell cultures in the presence/absence of different concentrations of pyrimidine (dThd and Urd) and purine (Ado and Ino) nucleosides. The data are the mean of at least two independent experiments (±S.E.M.).
3.2. Substrate specificity of human- and mycoplasma-encoded CDA
The substrate specificity of recombinant M. hyorhinis CDA (CDAHyor) was studied and compared with CDAHuman. Both enzymes catalyzed the deamination of the natural pyrimidine nucleosides Cyd and dCyd, and the well-known anticancer drugs dFdC and ara-Cyd. Deamination of 5-aza-2′-deoxycytidine (decitabine) and 5-aza-cytidine (vidaza), both used for the treatment of myelodysplastic syndromes [29], was also observed in the presence of CDAHyor but could not be demonstrated for CDAHuman. The antiviral (i.e. HIV) drugs 2′,3′-dideoxycytidine (ddC; zalcitabine) and 2′,3′-dideoxy-3′-thiacytidine (3TC; lamivudine) were found to be insensitive to deamination by both human and mycoplasma CDA.
3.3. Kinetics of human- and mycoplasma-encoded CDA
The kinetic parameters (KM and kcat) for CDAHyor- and CDAHuman-catalyzed deamination of Cyd, dCyd, dFdC and ara-Cyd were determined. Relatively high KM values (ranging from high micromolar to low millimolar concentrations) were observed for both enzymes. CDAHyor typically displayed higher KM values compared with CDAHuman (Tables 2 and 3 and Fig. 4). However, the catalytic efficiency (calculated as kcat/KM) of CDAHyor-catalyzed reactions was ∼2–4 fold higher compared with CDAHuman (Tables 2 and 3).
Table 2.
Kinetic parameters of CDAHyor. The KM and kcat values ± S.E.M. for the natural substrates of CDAHyor and for gemcitabine and cytarabine were determined using nonlinear regression analysis (using GraphPad Prism 5) from data obtained in at least two independent experiments.
| KM (μM) | kcat (s−1) | kcat/KM (s μM)−1 | |
|---|---|---|---|
| Cytidine | 1898 ± 163 | 171 ± 5 | 0.090 |
| 2′-Deoxycytidine | 2586 ± 114 | 127 ± 2 | 0.049 |
| dFdC | 9064 ± 1487 | 105 ± 7 | 0.012 |
| ara-Cyd | 6172 ± 2336 | 119 ± 13 | 0.019 |
Table 3.
Kinetic parameters of CDAHuman. The KM and kcat values ± S.E.M. for the natural substrates of CDAHuman and for gemcitabine and cytarabine were determined using nonlinear regression analysis (using GraphPad Prism 5) from data obtained in at least two independent experiments.
| KM (μM) | kcat (s−1) | kcat/KM (s μM)−1 | |
|---|---|---|---|
| Cytidine | 811 ± 181 | 17 ± 1 | 0.021 |
| 2′-Deoxycytidine | 373 ± 138 | 7.8 ± 0.64 | 0.021 |
| dFdC | 3080 ± 598 | 14 ± 0.91 | 0.004 |
| ara-Cyd | 2853 ± 741 | 15 ± 1.3 | 0.005 |
Fig. 4.
Kinetic analysis of CDAHyor- and CDAHuman-catalyzed deamination of natural nucleosides and nucleoside analogues Deamination of different concentrations of Cyd (A and B), dCyd (C and D), dFdC (E and F) and ara-Cyd (G and H) by CDAHyor (A, C, E and G) or CDAHuman (B, D, F and H). The data are the mean of at least two independent experiments (±S.E.M.).
Interestingly, deamination of both dFdC and ara-Cyd by CDAHyor, but not by CDAHuman, was characterized by a second KM at very high (and presumably biologically irrelevant) concentrations. However, precise values could not be determined due to insolubility of the highest drug concentrations (∼45 mM in the reaction mixture) tested. The calculated KM for dFdC and ara-Cyd as represented in Table 2 was therefore based on the measurements obtained when using up to 30 mM substrate. Measurements obtained using higher concentrations were excluded from the non-linear regression analysis but are still displayed in Fig. 4E (for dFdC) and Fig. 4G (for ara-Cyd). For these above-mentioned reasons, we preferred not to draw a curve line fitting the experimental data points in Fig. 4E and G.
4. Discussion
Recent studies have focused on the role of commensal prokaryotes in the efficiency of chemotherapeutic cancer treatment. For example, an intact intestinal microbiome seems to be essential for optimal response to immune therapy and platinum-, doxorubicin- or cyclophosphamide-based cancer chemotherapy [30,31]. Conversely, the presence of some prokaryotes (e.g. mycoplasmas) in the tumor microenvironment may negatively influence the outcome of nucleoside-based treatment of cancer [23]. A thorough understanding of the implications of the (tumor) microbiome on cancer chemotherapy may ultimately lead to a more optimal treatment strategy (e.g. by combination therapy with antibiotics or specific prokaryotic enzyme inhibitors) and may also attenuate adverse side-effects.
In this report, we investigated the catabolic action of mycoplasma-encoded cytidine deaminase against the anticancer nucleoside analogues gemcitabine and cytarabine. To the best of our knowledge, this study is the first to describe the biochemical properties of the CDA encoded by M. hyorhinis, a commensal that has been repeatedly reported to preferentially associate with tumor tissue in patients. We found that CDAHyor does not only catalyze the deamination of natural cytidine and 2′-deoxycytidine but also of several clinical nucleoside antimetabolites, including gemcitabine and cytarabine. The efficiency of substrate conversion catalyzed by CDAHyor was consistently higher compared with the human equivalent enzyme. In addition, we observed deamination of 5-aza-2′-deoxycytidine and 5-aza-cytidine by CDAHyor but not by CDAHuman under similar experimental conditions. Based on these observations it could be expected that pronounced inactivation of different anticancer (2′deoxy)cytidine analogues may occur in mycoplasma-infected tumors leading to reduced efficacy.
Indeed, expression of CDAHyor explains the predominantly high levels of (the poorly cytostatic) [5-3H]dFdU derived from radiolabeled gemcitabine in the culture medium of mycoplasma-infected tumor cell cultures and, as a result, the consequently decreased cytostatic activity of this drug in these tumor cell cultures (Table 1) and xenografts in mice [10]. Somewhat surprisingly, we found that the stability and biological (cytostatic) activity of gemcitabine in mycoplasma-infected tumor cell cultures could be restored by (i) the co-administration of natural pyrimidine (i.e. dThd and Urd) and purine (i.e. Ado, Ino and Guo) nucleosides and (ii) administration of a specific PNP inhibitor. Interestingly, inhibition of Escherichia coli and human CDA by different nucleosides [i.e. dThd, (d)Urd, (d)Ado and (d)Guo] has been reported earlier [32–34]. Previously, we have shown that M. hyorhinis PNP is responsible for the catabolism of different purine nucleoside analogues, including cladribine and fludarabine [9]. It can be hypothesized that M. hyorhinis-related PNP activity may also indirectly contribute to the deamination of cytidine analogues by depletion of those intracellular purine nucleosides that seem to act as natural inhibitors of CDAHyor. Exogenous administration of these natural nucleosides may then restore the depleted purine nucleoside pools and, as a result, also the cytostatic potential of gemcitabine. This hypothesis would then also explain the rescuing cytostatic activity of gemcitabine by Immucillin-H, a potent and selective PNP inhibitor. Earlier, we reported a similar concerted action between CDAHyor and the M. hyorhinis-encoded pyrimidine nucleoside phosphorylase which catabolizes dThd, Urd and dUrd [10]. However, when exposing purified CDAHyor to different substrates (Cyd, dCyd or dFdC) in the presence of natural purine nucleosides or dThd, we could not observe decreased deamination (data not shown). It is therefore unlikely that the observed rescue of gemcitabine is due to a direct interaction of CDAHyor with purine nucleosides or dThd. Although inhibition of CDA by (d)Urd may be explained by end-product feedback inhibition, the mechanism of CDAHyor-inhibition by dThd and the purine nucleosides remains unclear. In this respect, it cannot be excluded that interaction (i.e. inhibition) of CDA can be attributed to the mono-, di- or triphosphorylated derivatives of the natural nucleosides that were found inhibitory to CDAHyor-catalyzed deamination of gemcitabine. Alternatively, the natural nucleosides may compete for uptake with dFdC by M. hyorhinis and therefore lower the amount of drug to be deaminated intracellularly by the mycoplasma.
In conclusion, we have kinetically characterized the M. hyorhinis-encoded CDA and found it more catalytically efficient than human CDA. It deaminates (inactivates) anticancer drugs such as gemcitabine and cytarabine, but also 5-aza-(2′-deoxy)cytidine. Its deaminating action may negatively affect the cytostatic activity of anti-cancer drugs such as gemcitabine, but could be annihilated by co-administration of natural nucleosides or a specific PNP inhibitor.
Acknowledgments
We thank Christiane Callebaut, Eef Meyen, Lizette van Berckelaer, and Ria Van Berwaer for their excellent technical assistance. We are also grateful to Dr. Vern Schramm (Albert Einstein College of Medicine, Bronx, NY) for providing Imm-H. This work was supported in part by KU Leuven Grant GOA/15/019/TBA. JVV and PV performed the experiments; JVV, PV, SL and JB analyzed and interpreted the data; JVV, SL and JB designed and wrote the manuscript.
References
- 1.Razin S., Yogev D., Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finch L.R., Mitchell A. Sources of nucleotides. In: Maniloff J., McElhaney R.N., Finch L.R., Baseman J.B., editors. American Society for Microbiology; Washington: 1992. pp. 211–230. (Mycoplasmas, Molecular Biology and Pathogenesis). [Google Scholar]
- 3.Pollack J.D., Williams M.V., McElhaney R.N. The comparative metabolism of the mollicutes (Mycoplasmas): the utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit. Rev. Microbiol. 1997;23:269–354. doi: 10.3109/10408419709115140. [DOI] [PubMed] [Google Scholar]
- 4.Neale G.A., Mitchell A., Finch L.R. Uptake and utilization of deoxynucleoside 5′-monophosphates by Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 1984;158:943–947. doi: 10.1128/jb.158.3.943-947.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tham T.N., Ferris S., Kovacic R., Montagnier L., Blanchard A. Identification of Mycoplasma pirum genes involved in the salvage pathways for nucleosides. J. Bacteriol. 1993;175:5281–5285. doi: 10.1128/jb.175.16.5281-5285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronckaers A., Balzarini J., Liekens S. The cytostatic activity of pyrimidine nucleosides is strongly modulated by Mycoplasma hyorhinis infection: implications for cancer therapy. Biochem. Pharmacol. 2008;76:188–197. doi: 10.1016/j.bcp.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 7.Jetté L., Bissoon-Haqqani S., Le Francois B., Maroun J.A., Birnboim H.C. Resistance of colorectal cancer cells to 5-FUdR and 5-FU caused by Mycoplasma infection. Anticancer Res. 2008;28:2175–2180. [PubMed] [Google Scholar]
- 8.Vande Voorde J., Gago F., Vrancken K., Liekens S., Balzarini J. Characterization of pyrimidine nucleoside phosphorylase of Mycoplasma hyorhinis: implications for the clinical efficacy of nucleoside analogues. Biochem. J. 2012;445:113–123. doi: 10.1042/BJ20112225. [DOI] [PubMed] [Google Scholar]
- 9.Vande Voorde J., Liekens S., Balzarini J. Mycoplasma hyorhinis-encoded purine nucleoside phosphorylase: kinetic properties and its effect on the cytostatic potential of purine-based anticancer drugs. Mol. Pharmacol. 2013;84:865–875. doi: 10.1124/mol.113.088625. [DOI] [PubMed] [Google Scholar]
- 10.Vande Voorde J., Sabuncuoglu S., Noppen S., Hofer A., Ranjbarian F., Fieuws S., Balzarini J., Liekens S. Nucleoside-catabolizing enzymes in mycoplasma-infected tumour cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J. Biol. Chem. 2014;289:13054–13065. doi: 10.1074/jbc.M114.558924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan P.J., Seraj I.M., Kalugdan T.H., King A. Prevalence of mycoplasma conserved DNA in malignant ovarian cancer detected using sensitive PCR-ELISA. Gynecol. Oncol. 1996;63:258–260. doi: 10.1006/gyno.1996.0316. [DOI] [PubMed] [Google Scholar]
- 12.Kidder M., Chan P.J., Seraj I.M., Patton W.C., King A. Assessment of archived paraffin-embedded cervical condyloma tissues for mycoplasma-conserved DNA using sensitive PCR-ELISA. Gynecol. Oncol. 1998;71:254–257. doi: 10.1006/gyno.1998.5177. [DOI] [PubMed] [Google Scholar]
- 13.Huang S., Li J.Y., Wu J., Meng L., Shou C.C. Mycoplasma infections and different human carcinomas. World J. Gastroenterol. 2001;7:266–269. doi: 10.3748/wjg.v7.i2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pehlivan M., Itirli G., Onay H., Bulut H., Koyuncuoglu M., Pehlivan S. Does Mycoplasma sp. play role in small cell lung cancer? Lung Cancer. 2004;45:129–130. doi: 10.1016/j.lungcan.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Pehlivan M., Pehlivan S., Onay H., Koyuncuoglu M., Kirkali Z. Can mycoplasma-mediated oncogenesis be responsible for formation of conventional renal cell carcinoma? Urology. 2005;65:411–414. doi: 10.1016/j.urology.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Yang H., Qu L., Ma H., Chen L., Liu W., Liu C., Meng L., Wu J., Shou C. Mycoplasma hyorhinis infection in gastric carcinoma and its effects on the malignant phenotypes of gastric cancer cells. BMC Gastroenterol. 2010;10:132. doi: 10.1186/1471-230X-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apostolou P., Tsantsaridou A., Papasotiriou I., Toloudi M., Chatziioannou M., Giamouzi G. Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg. 2011;6:137. doi: 10.1186/1749-8090-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbanek C., Goodison S., Chang M., Porvasnik S., Sakamoto N., Li C.Z., Boehlein S.K., Rosser C.J. Detection of antibodies directed at M. hyorhinis p37 in the serum of men with newly diagnosed prostate cancer. BMC Cancer. 2011;11:233. doi: 10.1186/1471-2407-11-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barykova Y.A., Logunov D.Y., Shmarov M.M., Vinarov A.Z., Fiev D.N., Vinarova N.A., Rakovskaya I.V., Baker P.S., Shyshynova I., Stephenson A.J., Klein E.A., Naroditsky B.S., Gintsburg A.L., Gudkov A.V. Association of Mycoplasma hominis infection with prostate cancer. Oncotarget. 2011;2:289–297. doi: 10.18632/oncotarget.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erturhan S.M., Bayrak O., Pehlivan S., Ozgul H., Seckiner I., Sever T., Karakok M. Can mycoplasma contribute to formation of prostate cancer? Int. Urol. Nephrol. 2013;45:33–38. doi: 10.1007/s11255-012-0299-5. [DOI] [PubMed] [Google Scholar]
- 21.Jordheim L.P., Durantel D., Zoulim F., Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- 22.Liekens S., Bronckaers A., Balzarini J. Improvement of purine and pyrimidine antimetabolite-based anticancer treatment by selective suppression of mycoplasma-encoded catabolic enzymes. Lancet Oncol. 2009;10:628–635. doi: 10.1016/S1470-2045(09)70037-3. [DOI] [PubMed] [Google Scholar]
- 23.Vande Voorde J., Balzarini J., Liekens S. An emerging understanding of the Janus face of the human microbiome: enhancement versus impairment of cancer therapy. J. Antimicrob. Chemother. 2014;69:2878–2880. doi: 10.1093/jac/dku201. [DOI] [PubMed] [Google Scholar]
- 24.Vincenzetti S., Cambi A., Neuhard J., Garattini E., Vita A. Recombinant human cytidine deaminase: expression, purification, and characterization. Protein Expr. Purif. 1996;8:247–253. doi: 10.1006/prep.1996.0097. [DOI] [PubMed] [Google Scholar]
- 25.Neff T., Blau C.A. Forced expression of cytidine deaminase confers resistance to cytosine arabinoside and gemcitabine. Exp. Hematol. 1996;24:1340–1346. [PubMed] [Google Scholar]
- 26.Yoshida T., Endo Y., Obata T., Kosugi Y., Sakamoto K., Sasaki T. Influence of cytidine deaminase on antitumour activity of 2′-deoxycytidine analogs in vitro and in vivo. Drug Metab. Dispos. 2010;38:1814–1819. doi: 10.1124/dmd.110.034397. [DOI] [PubMed] [Google Scholar]
- 27.Liekens S., Bilsen F., De Clercq E., Priego E.M., Camarasa M.J., Perez-Perez M.J., Balzarini J. Anti-angiogenic activity of a novel multi-substrate analogue inhibitor of thymidine phosphorylase. FEBS Lett. 2002;510:83–88. doi: 10.1016/s0014-5793(01)03233-1. [DOI] [PubMed] [Google Scholar]
- 28.Carlow D.C., Carter C.W., Jr, Mejlhede N., Neuhard J., Wolfenden R. Cytidine deaminases from B. subtilis and E. coli: compensating effects of changing zinc coordination and quaternary structure. Biochemistry. 1999;38:12258–12265. doi: 10.1021/bi990819t. [DOI] [PubMed] [Google Scholar]
- 29.Parker W.B. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem. Rev. 2009;109:2880–2893. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viaud S., Saccheri F., Mignot G., Yamazaki T., Daillere R., Hannani D., Enot D.P., Pfirschke C., Engblom C., Pittet M.J., Schlitzer A., Ginhoux F., Apetoh L., Chachaty E., Woerther P.L., Eberl G., Berard M., Ecobichon C., Clermont D., Bizet C., Gaboriau-Routhiau V., Cerf-Bensussan N., Opolon P., Yessaad N., Vivier E., Ryffel B., Elson C.O., Dore J., Kroemer G., Lepage P., Boneca I.G., Ghiringhelli F., Zitvogel L. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., Dai R.M., Kiu H., Cardone M., Naik S., Patri A.K., Wang E., Marincola F.M., Frank K.M., Belkaid Y., Trinchieri G., Goldszmid R.S. Commensal bacteria control cancer response to therapy by modulating the tumour microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosono H., Kuno S. The purification and properties of cytidine deaminase from Escherichia coli. J. Biochem. 1973;74:797–803. doi: 10.1093/oxfordjournals.jbchem.a130305. [DOI] [PubMed] [Google Scholar]
- 33.Vita A., Amici A., Cacciamani T., Lanciotti M., Magni G. Cytidine deaminase from Escherichia coli B. Purification and enzymatic and molecular properties. Biochemistry. 1985;24:6020–6024. doi: 10.1021/bi00342a049. [DOI] [PubMed] [Google Scholar]
- 34.Cacciamani T., Vita A., Cristalli G., Vincenzetti S., Natalini P., Ruggieri S., Amici A., Magni G. Purification of human cytidine deaminase: molecular and enzymatic characterization and inhibition by synthetic pyrimidine analogs. Arch. Biochem. Biophys. 1991;290:285–292. doi: 10.1016/0003-9861(91)90543-r. [DOI] [PubMed] [Google Scholar]





