Abstract
The lectin complement pathway in innate immunity is closely related to the classical complement pathway in adaptive immunity, with respect to the structures and functions of their components. Both pathways are initiated by complexes consisting of collagenous proteins and serine proteases of the mannose-binding lectin (MBL)-associated serine protease (MASP)/C1r/C1s family. It has been speculated that the classical pathway emerged after the lectin pathway, and that the activation mechanism of the latter was partially conserved. The classical and lectin pathways can be traced back to at least cartilaginous fish and ascidian (urochordata), respectively. To elucidate the evolution of the complement system, we isolated and characterized a GlcNAc-binding lectin from sera of lamprey (agnathans), the most primitive vertebrate that lacks the classical pathway. Lamprey GlcNAc-binding lectin was an oligomer consisting of 24-kDa subunits. cDNA and phylogenetic analyses revealed that the lamprey GlcNAc-binding lectin is an orthologue of mammalian C1q, a collagenous subcomponent of the first component involved in binding to immunoglobulins in the classical pathway. Lamprey C1q copurified with MASP-A, a serine protease of the MASP/C1r/C1s family, which exhibited proteolytic activity against lamprey C3. Surface plasmon resonance analysis showed that lamprey C1q specifically bound to GlcNAc, but not various other carbohydrates tested. These results suggest that C1q may have emerged as a lectin and may have functioned as an initial recognition molecule of the complement system in innate immunity before the establishment of adaptive immunity such as immunoglobulins in the cartilaginous fish.
The complement system mediates a chain reaction of proteolysis and assembly of protein complexes that results in the elimination of invading microorganisms (1, 2). Three activation pathways (the classical, lectin, and alternative pathways) and a lytic pathway regulate these events. From an evolutionary perspective, the classical and lytic pathways seem to have emerged at the cartilaginous fish stage, when adaptive immunity was established (3, 4). The classical pathway is found in jawed vertebrates. In this pathway, C1q, a collagenous subcomponent of the first component (C1), binds to immunoglobulins within immune complexes, and its associated serine proteases, C1r and C1s, become activated. The complement cascade is initiated by the subsequent cleavage of C4 and C2, followed by C3 activation. The resulting C3b fragment not only acts as an opsonin but also leads to the membrane attack complex formation in the lytic pathway. In innate immunity, a complex composed of a recognition molecule (lectin) and serine proteases, termed the mannose-binding lectin (MBL)-associated serine protease (MASP), activates C4 and C2 upon binding to carbohydrates on the surface of microorganisms via the lectin pathway. This binding occurs in the absence of immunoglobulins (4). The recognition molecules of the lectin pathway found in jawed vertebrates are MBLs and ficolins, both of which are characterized by the presence of a collagen-like domain like C1q and a carbohydrate-binding domain having a common binding specificity for GlcNAc (5–8). MASPs and C1r/C1s share the same domain organization and form a subfamily of serine proteases.
In invertebrates, which lack immunoglobulins, the lectin pathway may play a crucial role in innate immunity, as revealed by the presence of MBL-like lectin (9), ficolins (10), MASP (11), and C3 (12) in ascidians (Urochordata). Activation of the lectin pathway of the ascidian complement system leads the generation of a C3 fragment, which facilitates phagocytosis through C3 receptors on phagocytes (13, 14). The fact that the lectin–MASP complex structurally and functionally resembles the C1 complex, together with the presence of an ancient lectin-based complement system, suggest that the lectin pathway evolved into the classical pathway (3, 4). To more clearly delineate the evolution of the complement system, we focused on lamprey (agnathans), the most primitive vertebrate lacking the classical pathway, in which MASP/C1r/C1s sequences (15, 16) and C3 protein (17) have been identified. Here, we show an orthologue of mammalian C1q that acts as a GlcNAc-specific lectin is expressed in lamprey. This lamprey C1q (LC1q) is associated with a serine protease of the MASP/C1r/C1s family that is capable of activating C3.
Materials and Methods
Purification of LC1q, MASP-A, and C3 from Lamprey Serum. Serum from Lampetra japonicus was applied to GlcNAc-agarose (Sigma) equilibrated with Tris buffer (50 mM Tris·HCl/200 mM NaCl/20 mM CaCl2, pH 7.8). Elution was carried out with 0.3 M mannose-containing buffer and then with 0.3 M GlcNAc-containing buffer. Esterolytic activity was monitored by hydrolysis of Boc-Leu-Ser-Thr-Arg-methylcoumarylamide (Peptide Institute, Osaka) to generate methylcoumarylamide. Samples were incubated with 20 μM Boc-Leu-Ser-Thr-Arg-methylcoumarylamide (final concentration) in 500 μl of 50 mM Tris·HCl/10 mM CaCl2, pH 8.0, at 20°C for 60 min. The samples were excited at 380 nm, and emission was at 460 nm. Eluates obtained with GlcNAc were pooled and dialyzed against 20 mM Tris·HCl/50 mM NaCl/10 mM CaCl2, pH 8.0, and were then chromatographed on a Mono Q column (Amersham Biosciences, Piscataway, NJ) that had been equilibrated with the above buffer.
Elution was carried out by applying a linear gradient to 0.5 M. Fractions having both GlcNAc-binding lectin (LC1q) and esterolytic activity (MASP-A) were dialyzed against 20 mM Tri·HCl/50 mM NaCl/10 mM EDTA, pH 8.0, and were then reapplied to a Mono Q column. LC1q passed through, whereas MASP-A was retained on the column. Fractions from the first Mono Q chromatographic separation were also analyzed by gel filtration on Superose 6 (Amersham Biosciences) equilibrated with 50 mM Tris·HCl/150 mM NaCl/10 mM CaCl2, pH 7.5, or 50 mM Tris·HCl/150 mM NaCl/10 mM EDTA, pH 7.5. Incorporation of tritium-labeled diisopropylfluorophosphate (New England Nuclear) into purified MASP-A was carried out as described (18). Collagenase digestion of LC1q was carried out by incubating 10 μg of purified LC1q with collagenase (50 units, Sigma) from Clostridium histolyticum in 50 mM Tris·HCl/150 mM NaCl/10 mM CaCl2, pH 7.5, at 37°C for 18 h. Lamprey C3 was purified according to published methods (17) with a modification. Briefly, lamprey serum was precipitated by polyethylene glycol and subjected to ion-exchange chromatography, followed by gel filtration. To examine C3 activation, 8 μg of lamprey C3 was incubated with 0.2 μg of LC1q-MASP-A or MASP-A in 50 mM Tris·HCl/150 mM NaCl/10 mM CaCl2, pH 7.5 at 37°C for 30 min, and the reaction mixture was then subjected to SDS/PAGE. SDS/PAGE was performed according to the Laemmli method and proteins were stained with Coomassie brilliant blue R-250.
Amino Acid Sequence Analysis. The N-terminal amino acid of LC1q was blocked. Therefore, its internal amino acid sequence was obtained. After LC1q was digested with collagenase as described above, digested products were subjected to SDS/PAGE under reducing conditions and were then electroblotted onto poly(vinylidene difluoride) membranes (Millipore). The bands were protein stained, excised, and then analyzed by using a protein sequencer (model 476A, Applied Biosystems). LC1q was also digested with Staphylococcus aureus V8 protease (Sigma) according to Cleveland's method (19). Digested peptides were electroblotted, by following the same method as described above. The N-terminal amino acid sequence of the 36-kDa polypeptide of MASP-A was also determined after electroblotting.
cDNA Cloning of LC1q. Three degenerated primers were synthesized, based on the amino acid sequences that had been determined for the products of LC1q digested with V8 protease or collagenase: CAGF/INGF/IPG (5′-TGYGCIGGIWTHAAYGGIW T HCCIGGICA-3′), PGQPGA PGQ (5′-CCIGGICARCCIGGIGCIGCICCIGGICARCA-3′), and GERGESDVG (5′-GGICCIACRTCISWYICICCICKYTCICC-3′). A nested PCR was carried out on lamprey liver cDNA by using two primer sets (5′-TGYGCIGGIWTHAAYGGIWTHCCIGGICA-3′ and 5′-GGICCIACRTCISWYICICCICKYTCICC-3′ for the first, and 5′-CCIGGICARCCIGGIGCIGCICCIGGICARCA-3′ and 5′-CCIGGICARCCIGGIGCIGCICCIGGICARCA-3′ for the second). A 248-bp PCR product was cloned into pGEM-T easy vector (Promega) and sequenced. 5′ and 3′ RACE were then conducted to complete the cDNA sequence by using a kit (Marathon, Clontech). LC1q was found to contain a 720-bp ORF encoding 240 aa.
Surface Plasmon Resonance Analysis. Neoglycoproteins GlcNAc-BSA, GalNAc–BSA, glucose-BSA, mannose-BSA, and galactose-BSA (Dextra Laboratories) or BSA were immobilized (≈10,000 units) by conventional amine coupling onto CM5 sensor chips with a BIAcore X instrument (Biacore). All sensorgrams were recorded at a flow rate of 20 μl/min at 25°C in running buffer (50 mM Hepes/150 mM NaCl, pH 7.5). Binding was measured as the response at 105 s after injection, relative to a baseline determined immediately before injection. In inhibition studies, LC1q (12.5 μg/ml) was preincubated with the various carbohydrates shown in Table 1 at concentrations ranging from 0 to 100 mM for 60 min at 4°C. Samples were then applied to a sensor chip containing immobilized GlcNAc-BSA. The concentrations of carbohydrates resulting in a 50% reduction in response (IC50) were determined graphically from the inhibition curve.
Table 1. Carbohydrate-binding characteristics of LC1q.
| Carbohydrates or carbohydrate residues composing neoglycoproteins | LC1q binding to neoglycoproteins,* RU | IC50,† mM |
|---|---|---|
| GlcNAc | 99.7 | 15 |
| N-acetylmannosamine | ND | 52 |
| GalNAc | 0 | ND |
| Glucose | 0 | >100 |
| Mannose | 0 | >100 |
| Galactose | 0 | >100 |
ND, not determined.
Binding of LC1q to each neoglycoprotein was evaluated in terms of resonance units (RU) obtained with surface plasmon resonance analysis
The IC50 value of each carbohydrate was determined as the concentration resulting in 50% inhibition of LC1q binding to GlcNAc-BSA
Results and Discussion
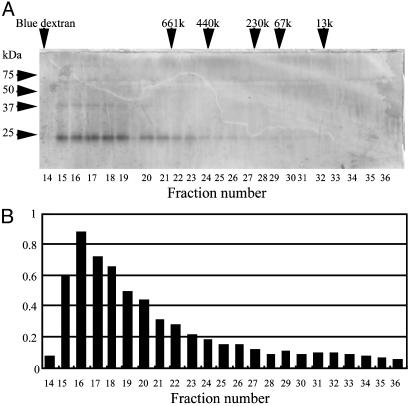
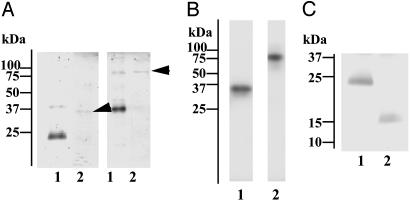
Serum from the L. japonicus lamprey was subjected to GlcNAc-agarose column chromatography in the presence of Ca2+. The column was sequentially eluted with mannose and with GlcNAc, as described in Materials and Methods. Human MBL can be eluted with mannose, and l-ficolin, one of the serum ficolins, can be subsequently eluted with GlcNAc (20). Both of these lectins are found to complex with MASPs (18, 20–22). MASP-like protease activity was monitored by esterolytic activity against Boc-Leu-Ser-Thr-Arg-methylcoumarylamide. Lamprey lectin eluted with mannose from GlcNAc-agarose was identified as MBL that was associated with MASP-A (4). Eluates obtained with GlcNAc were further purified by Mono Q chromatography. Fractions containing protease activity were then subjected to molecular sieve chromatography by using Superose 6 in the presence of Ca2+. As shown in Fig. 1, a GlcNAc-binding lectin with a molecular mass of 24 kDa under reducing conditions cofractionated with protease activity. On the other hand, in the presence of EDTA, the 24-kDa lectin and protease activity were separately recovered in the fractions ≈480 and 100 kDa on Superose 6, respectively (data not shown), suggesting a complex formation in the presence of Ca2+. Based on these results, to isolate the two components, fractions from the Mono Q column were dialyzed against EDTA-containing buffer and again applied to the column. The 24-kDa lectin passed through the column, whereas a 36-kDa protein associated with the protease activity was retained. An SDS/PAGE analysis of these proteins is shown in Fig. 2A. Under nonreducing conditions, the 24-kDa lectin showed an apparent molecular mass of ≈48 kDa. The 36-kDa protease migrates at ≈80 kDa under nonreducing conditions. It was determined to be a serine protease, because it had incorporated diisopropylfluorophosphate (Fig. 2B), and the N-terminal 18-aa sequence of the 36-kDa polypeptide was identical to a sequence starting at Ile-457 of lamprey MASP-A, a serine protease of the MASP/C1r/C1s family, with Ile-457 being the putative N-terminal amino acid of its light chain in the active form (15). Furthermore, digestion of the 24-kDa lectin with collagenase reduced it to a molecular mass of ≈15 kDa (Fig. 2C). Therefore, we concluded that the 24-kDa GlcNAc-binding lectin has a collagen-like sequence and is associated with lamprey MASP-A.
Fig. 1.
Gel filtration and SDS/PAGE analysis of lamprey GlcNAc-binding lectin. Lamprey serum was subjected to Mono Q column chromatography, followed by gel filtration on Superose 6 in the presence of Ca2+, as described in Materials and Methods. (A) SDS/PAGE profile under reducing conditions. Arrows indicate fractions in which standard substances were eluted. (B) Protease activity of each fraction.
Fig. 2.
Characterization of lamprey GlcNAc-binding lectin and its associated protease. (A) SDS/PAGE of purified GlcNAc-binding lectin (lane 1) and its associated protease (lane 2, arrows). Samples were run under reducing (Left) or nonreducing (Right) conditions and stained for proteins with Coomassie brilliant blue R-250. (B) Incorporation of tritium-labeled diisopropylfluorophosphate into GlcNAc-binding lectin-associated protease. The purified protease was treated with tritium-labeled diisopropylfluorophosphate and subjected to SDS/PAGE under reducing (lane 1) or nonreducing (lane 2) conditions, followed by autoradiography. (C) Collagenase digestion of GlcNAc-binding lectin. Lamprey GlcNAc-binding lectin was incubated in the absence (lane 1) or presence (lane 2) of collagenase and subjected to SDS/PAGE, followed by protein staining.
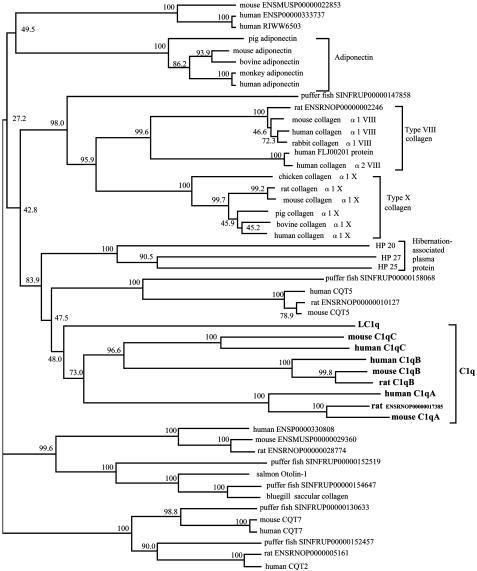
Based on the amino acid sequences of the peptides derived from V8 protease or collagenase digestion of the lamprey 24-kDa GlcNAc-binding lectin, we isolated the cDNA from lamprey liver by PCR and deduced its entire amino acid sequence. The lamprey lectin consisted of 240 aa with a putative signal peptide. Surprisingly, analysis of the peptide sequence indicated that the lamprey lectin was neither a ficolin nor any known lectin homologue. Instead, it was a homologue of mammalian C1q, because it contained a collagen-like sequence with 28 Gly-Xaa-Yaa repeats and a C-terminal globular C1q (gC1q) domain (Fig. 3). LC1q shared ≈35% identity at the amino acid level with mammalian C1q, which includes a hexamer of trimers, each consisting of three chains (A, B, and C), each of which contains a collagen-like domain and a gC1q domain with ≈135 aa. The gC1q domain of LC1q shared 25-33% identity with that of each chain of mammalian C1q. Similar motifs are found in other proteins, such as type X collagen and adiponectin (23). By using the neighbor-joining method, we constructed a phylogenetic tree of the gC1q domains of proteins that have both collagenous and gC1q domains. This analysis showed that LC1q is an orthologue of mammalian C1q protein (Fig. 4).
Fig. 3.
Alignment of amino acid sequences of LC1q and mammalian C1q. Sequences were aligned by using clustalw software. Residues that were identical in all proteins are indicated by asterisks. LC1q contains a collagen-like domain (residues 29–110) and a gC1q domain (residues 111–240).
Fig. 4.
Phylogenetic tree of gC1q domains of proteins that have both collagen-like and gC1q domains. The tree was constructed by the neighbor-joining method, based on the alignment of amino acid sequences from 50 proteins by using clustalw software. Numbers on branches are bootstrap percentages supporting a given partitioning. C1q, adiponectin, type VIII collagen, type X collagen, and chipmunk hibernation-associated plasma protein are grouped as indicated, but other proteins are not defined.
As described above, LC1q exhibited a molecular mass of ≈480 kDa on Superose 6, 48 kDa under nonreducing conditions, and 24 kDa under reducing conditions in SDS/PAGE, respectively. We obtained only one sequence for LC1q by both protein sequencing and cDNA cloning. Unlike mammalian C1q, therefore, LC1q consists of identical 24-kDa subunits that form a dimeric structural unit linked by disulfide bond(s). The estimated molecular size of LC1q on Superose 6 suggests that the nine dimeric structural units (18 chains) might be held noncovalently to give a hexameric structure like mammalian C1q (23).
To determine the specificity of LC1q binding to various carbohydrates, we used surface plasmon resonance. LC1q was injected over sensor chips with various immobilized neoglycoproteins, as shown in Table 1. Of these, only GlcNAc-BSA showed appreciable binding to LC1q (Table 1). LC1q bound to GlcNAc-BSA in a concentration-dependent manner (0–50 μg/ml; data not shown). In contrast, human C1q did not bind to any of the neoglycoproteins tested (data not shown). The binding of LC1q to GlcNAc-BSA was inhibited by GlcNAc and N-acetylmannosamine, but not by the other carbohydrates used.
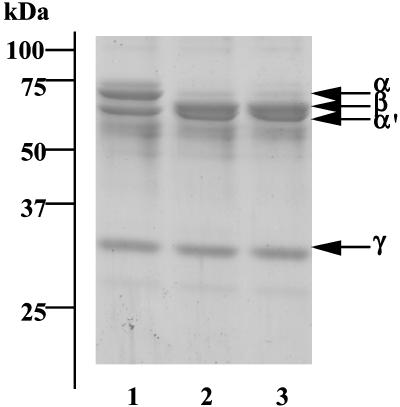
Next, we examined whether the LC1q–MASP-A complex activated lamprey C3, because ascidian MBL-like protein (glucose-binding lectin), upon association with MASPs, activates C3, the key component of the complement system (9). As shown in Fig. 5, the α′ chain appeared after lamprey C3 was exposed to LC1q-MASP-A or to MASP-A. This finding suggests that binding of LC1q-MASP-A to carbohydrates on pathogens results in the activation of C3 concomitant with the generation of C3b, which is deposited on the surfaces of pathogens and acts as an opsonin (17). MASPs are classified into three types (MASP-1, -2, and -3), based on the codon encoding the serine residue of the active center of the serine protease domain and the gene organization (3). In human and mouse, MASP-2 is involved the activation of C4 and C2, similar to what is observed with C1s, whereas we showed that MASP-1 directly cleaves C3 (3). The functions of MASP-3 have not yet been clarified. Lamprey express several types of MASPs, but their precise functions are not known (16). Thus, it appears that MASP-A plays a critical role in complement activation in lower vertebrates that lack MASP-2 and C1s, suggesting that MASP-A is the prototype of MASP-2 and C1s (4).
Fig. 5.
Cleavage of lamprey C3 by MASP-A. Lamprey C3 (lane 1) was incubated with LC1q–MASP-A complex (lane 2) or MASP-A (lane 3) and subjected to SDS/PAGE under reducing conditions, followed by protein staining.
The fact that ficolins are present as GlcNAc-binding lectin in the body fluids of ascidians prompted us to search for GlcNAc-binding lectins in lamprey serum (10). However, no ficolins were identified in lamprey serum in the present study. It is possible that lamprey ficolins, even if they are present in serum, might have a weak affinity for GlcNAc, as does human H-ficolin (Hakata antigen; ref. 24).
Adaptive immunity evolved at some point in the divergence of jawless (agnathan) and jawed (chondrichthian) vertebrates (25). However, recent reports (26) have shown that agnathans have cells that morphologically resemble lymphocytes and a gene homologous to PU.1, which controls lymphocyte development in jawed vertebrates (27–29). Our findings imply that although agnathans lack immunoglobulins, they have LC1q, a C1q homologue that is a GlcNAc-binding lectin, which plays a pivotal role in innate immunity by activating C3 in association with a serine protease of the MASP/C1r/C1s family. It is of particular interest that similar to MBL and ficolins, LC1q specifically binds to GlcNAc, which is widely distributed in lipopolysaccharides and in the cell walls of bacteria, whereas human C1q has lost this specificity. Therefore, it is likely that LC1q functionally resembles lectin pathway-activating lectins. Mammalian C1q binds to immunoglobulins through its gC1q domain, and the binding sites for gC1q on IgG and IgM are located in peptide sequences in the CH2 domain of IgG and in the CH3 domain of IgM (30). It is known that mammalian C1q binds not only to immunoglobulins but also to other substances, such as C-reactive protein and endotoxins through its collagen-like domain, resulting in complement activation (31). The binding region for GlcNAc on LC1q is not yet known. If the gC1q domain of LC1q is involved in GlcNAc binding, it is possible that the gC1q domain with lectin activity have acquired the capacity to bind to proteins (immunoglobulins) during evolution, as is proposed for the molecular evolution of fibrinogen-like domains (32). As described above, many C1q-related proteins have been identified, but their functions are not fully understood. Our finding that LC1q is a lectin raises the possibility that some of these proteins have lectin activity.
In conclusion, we have identified a protein homologous to mammalian C1q in lamprey that has no components of adaptive immunity such as immunoglobulins, T cell receptor, or MHC. LC1q acts as a recognition molecule in the lamprey complement system and seems to have developed into the more sophisticated, multifunctional system of the classical complement pathway.
Acknowledgments
This work was supported by grants from the Japan Society for the Promotion of Sciences and from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.M. and T.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LC1q, lamprey C1q; gC1q, globular C1q; MBL, mannose-binding lectin; MASP, MBL-associated serine protease.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB074155).
References
- 1.Walport, M. J. (2001) N. Engl. J. Med. 344, 1058-1066. [DOI] [PubMed] [Google Scholar]
- 2.Walport, M. J. (2001) N. Engl. J. Med. 344, 1140-1144. [DOI] [PubMed] [Google Scholar]
- 3.Fujita, T. (2002) Nat. Rev. Immunol. 2, 346-353. [DOI] [PubMed] [Google Scholar]
- 4.Fujita, T., Matsushita, M. & Endo, Y. (2004) Immunol. Rev. 198, 185-202. [DOI] [PubMed] [Google Scholar]
- 5.Turner, M. W. (1996) Immunol. Today 17, 532-540. [DOI] [PubMed] [Google Scholar]
- 6.Matsushita, M. & Fujita, T. (2001) Immunol. Rev. 180, 78-85. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita, M. & Fujita, T. (2002) Immunobiology 205, 490-497. [DOI] [PubMed] [Google Scholar]
- 8.Holmskov, U., Thiel, S. & Jensenius, J. C. (2003) Annu. Rev. Immunol. 21, 547-578. [DOI] [PubMed] [Google Scholar]
- 9.Sekine, H., Kenjo, A., Azumi, K., Ohi, G., Takahashi, M., Kasukawa, R., Ichikawa, N., Nakata, M, Mizuochi, T., Matsushita, M., et al. (2001) J. Immunol. 167, 4504-4510. [DOI] [PubMed] [Google Scholar]
- 10.Kenjo, A., Takahashi, M., Matsushita, M., Endo, Y., Nakata, M., Mizuochi, T. & Fujita, T. (2001) J. Biol. Chem. 276, 19959-19965. [DOI] [PubMed] [Google Scholar]
- 11.Ji, X., Azumi, K., Sasaki, M. & Nonaka, M. (1997) Proc. Natl. Acad. Sci. USA 94, 6340-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nonaka, M., Azumi, K., Ji, X., Namikawa-Yamada, C., Sasaki, M., Saiga, H., Dodds, A. W., Sekine, H., Homma, M. K., Matsushita, M., et al. (1999) J. Immunol. 162, 387-391. [PubMed] [Google Scholar]
- 13.Miyazawa, S., Azumi, K. & Nonaka, M. (2001) J. Immunol. 166, 1710-1715. [DOI] [PubMed] [Google Scholar]
- 14.Miyazawa, S. & Nonaka, M. (2004) Immunogenetics 55, 836-844. [DOI] [PubMed] [Google Scholar]
- 15.Endo, Y., Takahashi, M., Nakao, M., Saiga, H., Sekine, H., Matsushita, M., Nonaka, M. & Fujita, T. (1998) J. Immunol. 161, 4924-4930. [PubMed] [Google Scholar]
- 16.Endo, Y., Nonaka, M., Saiga, H., Kakinuma, Y., Matsushita, A., Takahashi, M., Matsushita, M. & Fujita, T. (2003) J. Immunol. 170, 4701-4707. [DOI] [PubMed] [Google Scholar]
- 17.Nonaka, M., Fujii, T., Kaidoh, T., Natsuume-Sakai, S., Nonaka, M., Yamaguchi, N. & Takahashi, M. (1984) J. Immunol. 133, 3242-3249. [PubMed] [Google Scholar]
- 18.Matsushita, M. & Fujita, T. (1992) J. Exp. Med. 176, 1497-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleveland, D. W. (1983) Methods Enzymol. 96, 222-229. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita, M., Endo, Y. & Fujita, T. (2000) J. Immunol. 164, 2281-2284. [DOI] [PubMed] [Google Scholar]
- 21.Thiel, S., Vorup-Jensen, T., Stover, C. M., Schwaeble, W., Laursen, S. B., Poulsen, K., Willis, A. C., Eggleton, P., Hansen, S., Holmskov, U., et al. (1997) Nature 386, 506-510. [DOI] [PubMed] [Google Scholar]
- 22.Dahl, M. R., Thiel, S., Matsushita, M., Fujita, T., Willis, A. C., Christensen, T., Vorup-Jensen, T. & Jensenius, J. C. (2001) Immunity 15, 127-135. [DOI] [PubMed] [Google Scholar]
- 23.Kishore, U. & Reid, K. B. M. (2000) Immunopharmacology 49, 159-170. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita, M., Kuraya, M., Hamasaki, N., Tsujimura, M., Shiraki, H. & Fujita, T. (2002) J. Immunol. 168, 3502-3506. [DOI] [PubMed] [Google Scholar]
- 25.Laird, D. J., De Tomaso, A. W., Cooper, M. D. & Weissman, I. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6924-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer, W. E., Uinuk-Ool, T., Tichy, H., Gartland, L. A., Klein, J. & Cooper, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 14350-14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shintani, S., Terzic, J., Sato, A., Saraga-Babic, M., O'hUigin, C., Tichy, H. & Klein, J. (2000) Proc. Natl. Acad. Sci. USA 97, 7417-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson, M. K., Sun, X., Miracle, A. L., Litman, G. W. & Rothenberg, E.V. (2001) Proc. Natl. Acad. Sci. USA 98, 553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uinuk-ool, T., Mayer, W. E., Sato, A., Dongak, R., Cooper, M. D. & Klein, J. (2002) Proc. Natl. Acad. Sci. USA 99, 14356-14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan, A. R. & Winter, G. (1988) Nature 332, 738-740. [DOI] [PubMed] [Google Scholar]
- 31.Gewurz, H., Ying, S.-C., Jiang, H. & Lint, T. F. (1993) Behring Inst. Mitt. 93, 138-147. [PubMed] [Google Scholar]
- 32.Kairies, N., Beisel, H. G., Fuentes-Prior, P., Tsuda, R., Muta, T., Iwanaga, S., Bode, W., Huber, R. & Kawabata, S. (2001) Proc. Natl. Acad. Sci. USA 98, 13519-13524. [DOI] [PMC free article] [PubMed] [Google Scholar]