Abstract
Background
Sudden Cardiac Death (SCD) follows a diurnal variation. Data suggest the timing of SCD is influenced by circadian (~24 hour) changes in neurohumoral and cardiomyocyte-specific regulation of the heart’s electrical properties.
Objective
The basic helix-loop-helix transcription factors BMAL1 and CLOCK coordinate the circadian expression of select genes. We tested whether Bmal1 expression in cardiomyocytes contributes to K+ channel expression and diurnal changes in ventricular repolarization.
Methods
We utilized transgenic mice that allow for the inducible cardiomyocyte-specific deletion of Bmal1 (iCSΔBmal1−/−). We used quantitative PCR, voltage-clamping, promoter-reporter bioluminescence assays, and electrocardiographic (ECG) telemetry.
Results
Although several K+ channel gene transcripts were downregulated in iCSΔBmal1−/− mouse hearts, only Kcnh2 exhibited a robust circadian pattern of expression that was disrupted in iCSΔBmal1−/− hearts. Kcnh2 underlies the rapidly activating delayed-rectifier K+ current (IKr), and IKr recorded from iCSΔBmal1−/− ventricular cardiomyocytes was ~50% compared to control myocytes. Promoter-reporter assays demonstrated that the human Kcnh2 promoter is transactivated by the co-expression of BMAL1 and CLOCK. ECG analysis showed iCSΔBmal1−/− mice developed a prolongation in the heart rate corrected QT (QTc) interval during the light (resting)-phase. This was secondary to an augmented circadian rhythm in the uncorrected QT interval without a corresponding change in the RR interval.
Conclusion
The molecular clock in the heart regulates the circadian expression of Kcnh2, modifies K+ channel gene expression and is important for normal ventricular repolarization. Disruption of the cardiomyocyte circadian clock mechanism likely unmasks diurnal changes in ventricular repolarization that could contribute to an increased risk of cardiac arrhythmias/SCD.
Keywords: circadian rhythm, repolarization, ECG, QTc, Bmal1, Kcnh2
INTRODUCTION
The discovery that the incidence of sudden cardiac death (SCD) follows a time-of-day dependence suggests that circadian factors participate in the initiation of lethal arrhythmias.1, 2 Biological circadian rhythms are evolutionarily conserved cycles that repeat every ~24 hours, and synchronize behavior and physiology with the daily environment.3, 4 The cardiovascular system shows robust circadian rhythms in blood pressure, heart rate, electrocardiographic (ECG) properties, and gene expression.5 Historically, the early morning rise in SCD has been linked to the circadian variations in myocardial ischemic events and autonomic signaling, however, more recent studies also support a cardiomyocyte-specific circadian or molecular clock mechanism in the heart.6–8
The cellular mechanism responsible for generating biological circadian rhythms is a conserved gene regulatory network composed of a transcriptional-translational feedback loop called the molecular clock, which is expressed in almost all cells including cardiomyocytes.9, 10 The positive limb of the molecular clock is formed by the transcription factors BMAL1 (brain muscle arnt-like1) and CLOCK (circadian locomotor output control kaput), and the negative limb is regulated by PERs (Period 1, 2, and 3) and CRYs (Cryptochrome 1 and 2). Circadian changes in gene expression and cardiac function are linked to the cardiomyocyte molecular clock mechanism.11, 12 Bray and colleagues (2008) showed transgenic mice that selectively overexpress a dominant-negative CLOCK mutation in cardiomyocytes alters gene expression in both atrial and ventricular myocytes, heart rate, contractile function, and metabolism.13 We recently found that the inducible cardiomyocyte-specific deletion of Bmal1 (iCSΔBmal1−/−) in adult mice disrupts molecular clock signaling in the heart, causes a loss in the circadian expression of the Na+ channel gene Scn5a (Nav1.5), decreases macroscopic Na+ current (INa) recorded from isolated ventricular cardiomyocytes, slows the heart rate, and increases the frequency of cardiac arrhythmias.8 Emerging evidence suggests that the cardiomyocyte molecular clock might regulate the expression of several cardiac K+ channels and ventricular repolarization as well.6, 7 However, these studies do not distinguish the relative contribution of the cardiomyocyte molecular clock from circadian clock signaling in other tissues. The purpose of this study is to determine how Bmal1 expression in cardiomyocytes contributes to changes in K+ channel expression and ventricular repolarization.
METHODS
Inducible deletion of Bmal1 in adult cardiomyocytes
All animal procedures were conducted in compliance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care and were approved by the Institutional Animal Care and Use Committee at University of Kentucky. The inducible cardiac specific ΔBmal1 (iCSΔBmal1) mouse model used for these studies was described previously.8 Cre-recombination was activated once the mice reached 12 weeks of age by intraperitoneal injections of tamoxifen (2 mg/day) for 5 consecutive days. This injection protocol causes effective recombination without any obvious long-term tamoxifen toxicity. Mice were housed in a 12-hour light/dark (L/D) cycle for all experiments except the time course collection, which is detailed below.
Circadian Collections
Circadian collections were done as described previously.8 Briefly, 64 iCSΔBmal1 mice (mixed gender, 14–16 weeks of age) were housed in individual cages in light boxes and entrained to a 12-hour L/D cycle for 2 weeks. Mice had ad libitum access to food and water. Following the entrainment period, half of the mice were injected with vehicle and the other half with tamoxifen, generating 32 control iCSΔBmal1+/+ and 32 iCSΔBmal1−/− mice, respectively. Two-weeks after the final injection, the mice were then released into constant darkness (D/D), and after 30 hours in D/D, we collected the ventricular apex every 4 hours from 3–4 animals in each group for a total of 8 time points. Circadian collections from control WKY (Wistar Kyoto) rats were done similarly. RNA was prepared for quantitative PCR (qtPCR) using TaqMan (Applied Biosystems) assays to examine the gene expression of Kcnd2, Kcnh2, Kcnip2, Kcna5, Kcnb1, Kcnj2, and Kcnq1 mRNA. The ΔΔCT method was used for the quantification of qtPCR data in the circadian collections. Gene expression in each sample was shown as the relative value compared with the mean vehicle value.
Adult cardiomyocyte isolation and electrophysiology
Adult ventricular myocytes were isolated for voltage-clamp experiments as described previously.8 Isolations were performed at 6–8 weeks following vehicle or tamoxifen injections. Voltage-clamp was performed with an Axopatch 200B patch-clamp amplifier (Axon Instruments, Foster City, CA) and pClamp10 software (Axon Instruments, Foster City, CA). Because IKr recorded using conventional approaches in mouse ventricular myocytes is small and contaminated by other currents, we isolated IKr using Cs+ as the charge carrier similar to that described previously.14–16 Unlike other K+ channels, IKr channels readily permeate Cs+ in the absence of K+ and using Cs+ as the charge carrier allows us to measure IKr directly (without current subtraction using IKr blockers, which improves the signal to noise ratio). The extracellular solution contained (in mM): NaCl 5, CsCl 90, CaCl 1, MgCl 1.2, glucose 11, TEA-Cl 10, HEPES 5 (pH 7.3 set with CsOH), and the pipette (intracellular) solution contained CsF 120, CsCl 20, EGTA 10, TEA-Cl 10, Na2ATP 1, HEPES 5 (pH 7.3 set with CsOH). Heterologously expressed Kcnh2 channels (Kv11.1) in HEK293 cells generated large currents with similar gating properties as native IKr (data not shown). The holding potential was −140 mV. Cells were depolarized from −80 to 40 mV in 10 mV increments for 1 sec, followed by a test-pulse to −80 mV. The peak current measured during the test-pulse to −80 mV was plotted as a function of the pre-pulse potential and the individual current-voltage (I–V) relations were described using the following Boltzmann equation:
IMIN is the minimally activated current, IMAX is the maximally activated current, V½ is the midpoint potential for half maximal current activation, and k is the slope factor of activation (mV/e-fold change). All voltage-clamp experiments from isolated iCSΔBmal1+/+ or iCSΔBmal1−/− ventricular myocytes were performed at 22–23°C within 4 hours of isolation.
Promoter-reporter bioluminescence assays
Heterologous expression of promoter-reporter constructs was performed in C2C12 myoblasts similar to that described previously.8 For control studies, we utilized the Per1 promoter-reporter construct 6.8Per1-Luc. We cloned the 734 bp human Kcnh2 5′-promoter sequence into the pGL3 basic vector (Promega) using human genomic DNA (hKcnh2-luc). The primers used for amplification of the 5′ promoter sequence were 5′-CACGGTACC TCTTAGTCGCTAATCTGGGGTGG -3′ (forward) and 5′-CACGCTAGC ACCGGCATCCTGAGCCCAT -3′ (reverse). The sequence of the hKcnh2 promoter-reporter construct (hKcnh2-luc) was verified by DNA sequencing at the Advanced Genetic Testing Center, University of Kentucky. Lipofectamine2000 was used at a 3:1 ratio. To control for the total amount of in each transfection, transfected DNA was adjusted to 390 ng with the empty pcDNA3.1 plasmid. Forty-eight hours after transfection, luminescence of the lysate (20 μl) was measured using the Dual-Luciferase Reporter Assay System (Promega) in a Lumat LB 9507 (EG&G Berthold). Similar to what we have shown previously in NIH/3T3 expression of the mouse Bmal1 and Clock cDNA cloned in pcDNA3.1 (mBmal1- and mClock-pcDNA3.1, respectively) enhanced Per1 promoter activity several fold in C2C12 cells (data not shown).8 We assessed hKcnh2-luc promoter-reporter expression in four conditions: 1) promoter-reporter only, 2) co-transfected with mBmal1- and mClock-pcDNA3.1; 3) co-transfected with mClock-pcDNA3.1 and a missense mutation in Bmal1 cDNA that changes an arginine to an alanine at amino acid 91 (mBmal1R91A-pcDNA3.1) and disrupts BMAL1 function; and 4) co-transfected with mBmal1- and a deletion mutant in Clock cDNA at amino acid 19 (mClockΔ19) that disrupts CLOCK function. 17 These latter two experiments with the mutant constructs served as negative controls. The pRL null vector was co-transfected in each experiment to provide a control for variations in transfection efficiency.
ECG telemetry
In vivo ECG telemetry was performed as described previously.8, 18 Briefly, mice were anesthetized with isoflurane and telemetry transmitter units (PhysioTel ETA-F10; Data Sciences International) were implanted in the peritoneal cavity. Two ECG leads were secured near the apex of the heart and the right acromion. Mice were allowed to recover for 2 weeks. We used two complementary methods to quantify the QT interval.18 The first method was to manually measure and calculate the QTc interval using a modified version of the Bazett’s formula adjusted for mice and described in detail by Mitchell and colleagues (1998).19 For the second approach, we used an automated program written in Matlab (Mathworks) to compute hourly uncorrected QT intervals similar as described previously.18 Briefly, ECG traces recorded for each hour were aligned to the peak of the R wave to generate an hourly averaged trace and the QT calculation was done using the threshold method. The Q was defined as the base of the QRS complex (where the slope of the profile changed from negative or isoelectric to positive) and the T was defined as the point where the ECG returned 95% of the way from the T-wave minima to the isoelectric level. The hourly RR and QT intervals were plotted for ~ 2–3 consecutive days and were fit with the following non-linear sinusoidal model:
This model allowed calculation of the period (T) - the time between the peak amplitudes; phase (τ) - time of the peak rhythm in reference to the onset of the lights on (Zeitgeber Time or ZT= 0); the circadian amplitude (A) - ½ of the peak-to-trough amplitude; and the circadian mean (m) - a rhythm-adjusted mean halfway between the peak and trough amplitude.
Statistical analysis
Results are reported as mean ± SEM where P < 0.05 is considered significant. For gene expression studies, the statistical JTK_CYCLE package was used to identify circadian rhythms in the qtPCR expression profiles. JTK_CYCLE reliably distinguishes between rhythmic and non-rhythmic transcripts and exhibits an increased resistance to outliers in the data, giving it considerably greater sensitivity and specificity.20 Additionally, JTK_CYCLE accurately measures the period, phase, and amplitude of cycling transcripts, facilitating comparative analyses. Unpaired t-tests were performed to determine significance for electrophysiological measures, and statistical analysis of the ECG telemetry data was done using a one-way ANOVA with the Bonferroni post-hoc analysis (Prism, GraphPad Software, Inc.).
RESULTS
Kcnh2 is a candidate clock controlled gene
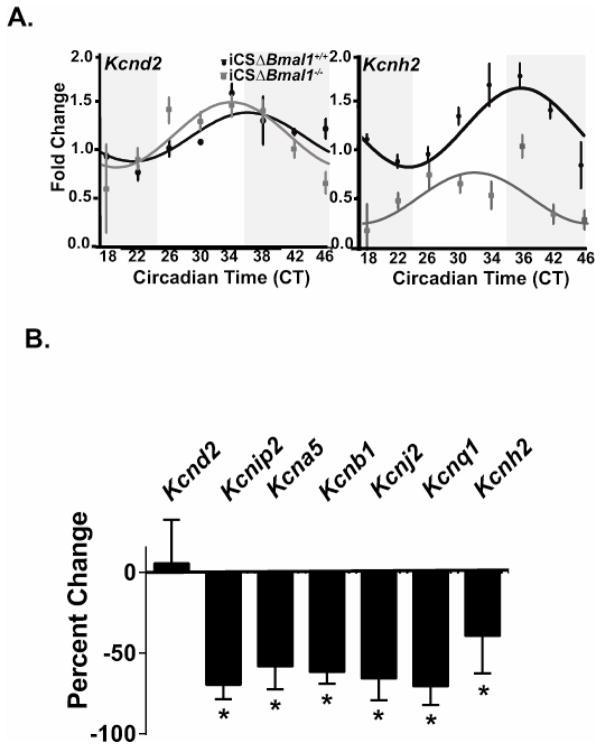
We investigated the role of cardiomyocyte molecular clock in the regulation of cardiac K+ channel gene expression important in mouse and human ventricular repolarization (Supplemental Table 1). To identify K+ channel gene candidates that are expressed in a circadian pattern, we utilized the high resolution CircaDB microarray dataset for the heart (http://circadb), which analyzed gene expression every 2 hours for 48 hours in combination JTK_CYCLE analysis.21 We then confirmed these results in control (iCSΔBmal1+/+) hearts by measuring the mRNA expression levels every 4 hours for 28 hours using qtPCR and JTK_CYCLE statistical analysis (Figure 1A). If an mRNA is determined to be circadian following JTK-CYCLE analysis in both datasets, then we concluded that the mRNA transcripts are circadian. The results of the two datasets suggest that only Kcnd2 and Kcnh2 transcripts exhibit a robust circadian oscillation in control hearts. We next tested whether cardiac Bmal1 expression contributed to the circadian expression pattern of Kcnd2 and Kcnh2 by determining whether the circadian oscillation in transcripts was lost in iCSΔBmal1−/− hearts. The circadian pattern of the Kcnh2 transcript, but not the Kcnd2 transcript, was lost in iCSΔBmal1−/− hearts (Figure 1A). This suggested that Kcnh2 is controlled by the cardiomyocyte molecular clock but the central clock likely regulates the circadian expression of Kcnd2. Surprisingly, we found that the average 24-hour expression levels of several other cardiac K+ channel transcripts (which did not follow a circadian pattern) were lower in iCSΔBmal1−/− hearts (Figure 1B). These data suggested that the cardiomyocyte molecular clock signaling might indirectly contribute to the expression of non-circadian K+ channel genes.
Figure 1. The cardiomyocyte molecular clock regulates the circadian expression of Kcnh2 transcripts as well as the expression of several K+ channel gene transcripts that are not circadian.
A) Shown are the JTK_CYCLE best fit data for the mRNA expression profiles for the K+ channel genes Kcnd2 and Kcnh2 from iCSΔBmal1+/+ (solid circles) and iCSΔBmal1−/− (grey squares) mouse hearts (n=3–4 animals per time point). The dark and light bars on the graph represent extrapolated subjective day and night as defined by Circadian Time (CT) according to the prior L:D cycle before release into DD. JTK_CYCLE statistics were used to determine if the expression pattern was circadian. The JTK_CYCLE calculated p value for Kcnd2 and Kcnh2 in the iCSΔBmal1+/+ mice is 0.003 and 7.02E-05 respectively, and the JTK_CYCLE calculate p value for Kcnd2 and Kcnh2 in the iCSΔBmal1−/− mice is 0.0095 and 0.060 respectively. Channels with JTK_CYCLE p values less than 0.05 were considered circadian in expression. B) Shown is the percent change in the mRNA transcript over a 24-hour period for Kcnd2, Kcnip2, Kcna5, Kncb1, Kcnj2, Kcnq1, and Kcnh2 in the iCSΔBmal1−/− mice compared to iCSΔBmal1+/+ mice.
Disruption of the cardiomyocyte molecular clock decreases IKr
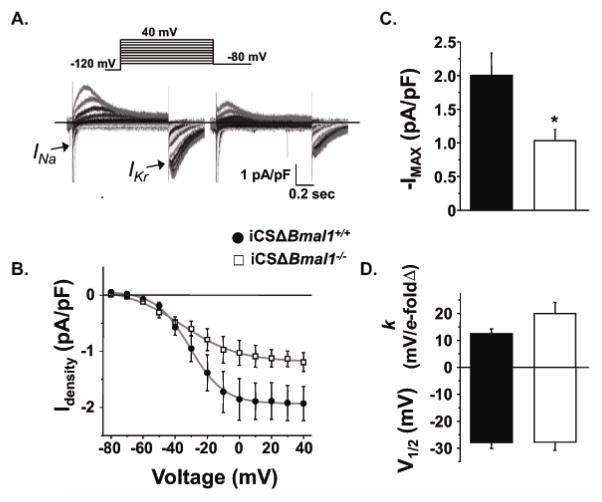
Since Kcnh2 is a candidate clock controlled gene, we focused our mechanistic investigation on the consequences that deleting Bmal1 has on the functional expression of Kcnh2. Kcnh2 encodes the pore-forming α-subunit that conducts the rapidly activating delayed-rectifier K+ current (IKr,).22 Loss-of-function mutations in Kcnh2 are a common cause of congenital long QT syndrome (LQTS), and IKr channels are prolific drug targets that contribute to drug-induced LQTS.23 IKr recorded from iCSΔBmal1−/− ventricular myocytes hearts was ~50% smaller than control ventricular myocytes (Figure 2A). The reduction in IKr was consistent with a decrease in gene expression, because there were no obvious differences in the gating properties of the IKr (Figure 2B).
Figure 2. Disruption of BMAL1 signaling reduces IKr.
A) Shown are representative families of currents recorded from iCSΔBmal1+/+ or iCSΔBmal1−/− ventricular cardiomyocytes (isolated from mice after 58 hours in constant darkness). The currents were recorded by pre-pulsing cells from −80 to 40 mV in 10 mV increments, followed by a test-pulse to −80 mV. The peak INa measured during the initial pre-pulse is cropped for presentation purposes. B) The peak inward IKr measured during the test-pulse is plotted as a function of the pre-pulse potential and the data are described using a Boltzmann equation (grey line). C–D) The mean Boltzmann data from the fits to the individual cells for maximally activated IKr (IMAX, pA/pF), the slope factor (k, mV/e-fold change), and the midpoint potential for half-maximal activation (V1/2, mV) are shown for iCSΔBmal1+/+ (solid bars) and iCSΔBmal1−/− (open bars) mice (n=11–13 cells, *P<0.05).
The circadian pattern of Kcnh2 expression is conserved and co-expression of BMAL1 and CLOCK transactivates the human Kcnh2 promoter
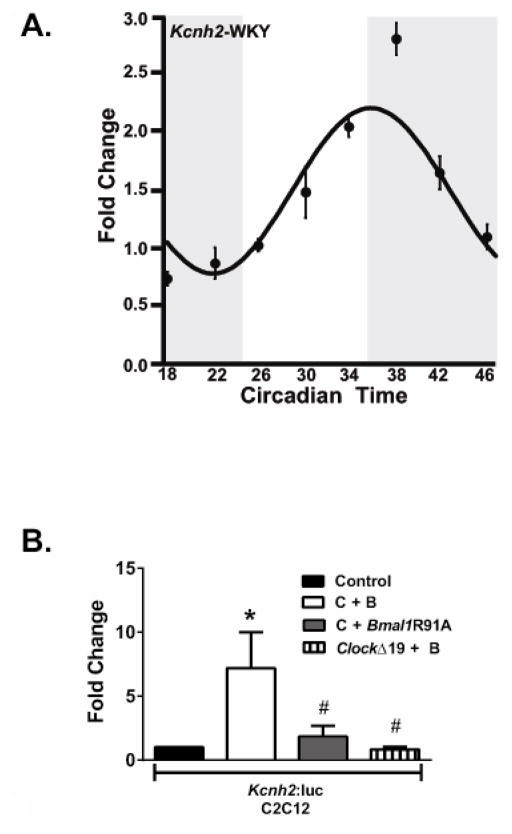
We next determined whether Kcnh2 transcripts follow a circadian pattern of expression in another species. We measured Kcnh2 expression every 4 hours for 28 hours from rat ventricle, similar to what was done for the iCSΔBmal1 mice. JTK_CYLCE analysis of the Kcnh2 mRNA confirmed that the transcripts followed a robust circadian pattern (Figure 3A). Furthermore, we investigated whether heterologous expression of BMAL1 and CLOCK could enhance the transcriptional activity of the cloned human Kcnh2 promoter. Heterologous expression of hKCNH2-Luc in C2C12 cells showed that, compared to cells expressing hKCNH2-Luc alone, co-expressing mBmal1 and mClock DNA increased luciferase activity several fold (Figure 3B). Importantly, negative control experiments that co-expressed cDNAs with mutations (Bmal1R91A or ClockΔ19) that disrupt BMAL1 or CLOCK function did not enhance hKCNH2-Luc luciferase activity (Figure 3B). Together the data suggest that the circadian expression of Kcnh2 is conserved, and BMAL1 and CLOCK can enhance the transcriptional activity of the human KCNH2 promoter.
Figure 3. Kcnh2 transcripts follow a circadian pattern of expression in rat hearts and the human Kcnh2 promoter is transactivated by the co-expression of BMAL1 and CLOCK.
A) The circadian expression profile for Kcnh2 in the WKY rat heart is shown (n=4 animals per time point). Shaded and light regions represent subjective dark and light cycle. B) BMAL1 and CLOCK transactivate the human Kcnh2 promoter reporter. Luciferase assay results of transfection experiments using the hKcnh2 reporter gene in C2C12 myoblasts (n = 6–10/condition). CLOCK (C) and BMAL1 (B) significantly transactivate the hKcnh2 reporter gene (solid bars) relative to control transfections (open bars). Activation of the hKcnh2 reporter is significantly diminished when Bmal1R91A was overexpressed with CLOCK (gray bar) or ClockΔ19 was overexpressed with BMAL1 (vertical striped bar). Values are expressed as means ± SE. *P < 0.05 compared with the hKcnh2 reporter gene; #P < 0.05 compared with hKcnh2 reporter gene + CLOCK + BMAL.
Disruption of the cardiomyocyte molecular clock unmasks a circadian rhythm in ventricular repolarization
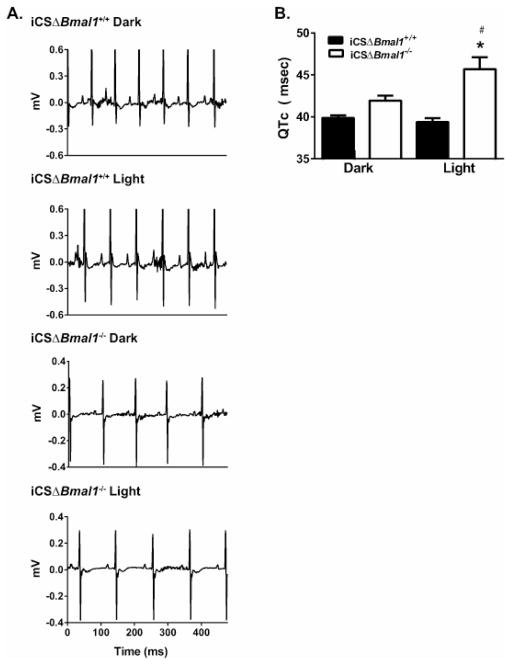
Since disrupting the cardiomyocyte molecular clock in the mouse heart decreased the expression of several K+ channel transcripts important for mouse ventricular repolarization, we tested whether the QTc intervals measured from iCSΔBmal1−/− mice were different compared to iCSΔBmal1+/+ mice. Similar to what was previously shown with in vivo ECG telemetry of wild type mice, the QTc interval was constant in the iCSΔBmal1+/+ mice throughout the day.19 However, the QTc intervals measured from the iCSΔBmal1−/− showed a time-of-day prolongation during the light-phase (Figure 4). These data are surprising because they suggest that disruption of Bmal1 in cardiomyocytes unmasked a diurnal prolongation in the QTc interval that is not normally seen in control mice.
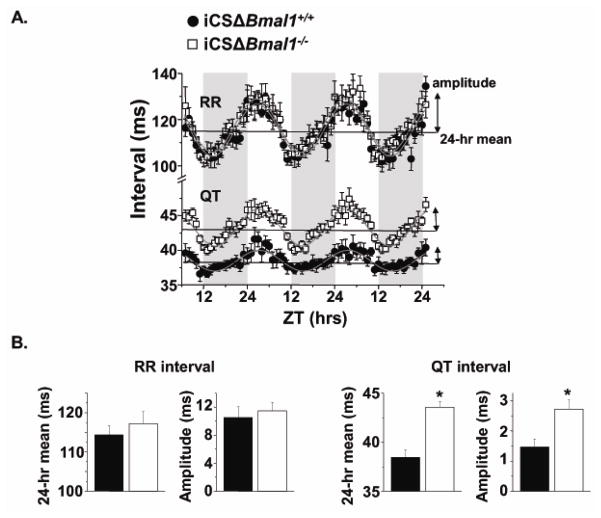
Figure 4. iCSΔBmal1−/− mice have a prolonged QTc intervals during the light-phase.
A) Shown are representative ECG traces measured during the dark and light from iCSΔBmal1+/+ or iCSΔBmal1−/− mice. B) QTc intervals measured at peak and trough RR interval hours during the light- and dark-phases from iCSΔBmal1−/− (open bars) or iCSΔBmal1+/+ mice (solid bars) (n=6–7 animals, *P<0.05 compared to dark values; #<0.05 compared to iCSΔBmal1+/+).
In order to understand how the cardiomyocyte-specific deletion of Bmal1 prolongs the QTc interval during the light-phase, we quantified the circadian rhythms in the hourly RR and uncorrected QT intervals for control and iCSΔBmal1−/− mice over several days. The data were fit with a non-linear sinusoidal function to calculate the period, phase, circadian mean, and circadian amplitude of the rhythms (Figure 5; Table 1). The rhythms in the RR and QT intervals were all circadian (~24 hrs) and peaked 2–5 hours after the beginning of the light phase (ZT 2–5). The mean and amplitude of the circadian rhythm in the RR intervals were not different between the iCSΔBmal1+/+ and iCSΔBmal1−/− mice, but the mean and the amplitude of the circadian rhythm in the QT intervals were larger for the iCSΔBmal1−/− mice (Figure 5; Table 1). Importantly, these changes were only seen after inducing the deletion of Bmal1 with tamoxifen treatment (Table 1), and they did not occur in parental Mer-Cre-Mer mice treated with tamoxifen (n=4–5 mice each, p>0.05, data not shown). Together, these data show the cardiomyocyte deletion of Bmal1 increased the QTc interval during the light-phase because it augmented the circadian rhythm in the uncorrected QT interval without a corresponding change in the RR interval.
Figure 5. iCSΔBmal1−/− mice have a larger circadian rhythm mean and amplitude in their uncorrected QT intervals.
A) ECGs were continuously measured using telemetry for ~3 days and the hourly RR or uncorrected QT intervals were plotted as a function of ZT for iCSΔBmal1+/+ (solid circles) and iCSΔBmal1−/− (open squares) mice (n=6–7). The shaded and light regions represent the dark and light, respectively. The grey line is a non-linear sinusoidal fit to the data. B) Shown are the average circadian rhythm mean and the amplitude for the RR interval and QT interval calculated from the sinusoidal fits to individual iCSΔBmal1+/+ (solid bars) or iCSΔBmal1−/− (open bars) mouse data (n=6–7 each, *P<0.05)
Table 1. Disruption of the cardiomyocyte molecular clock selectively increases the mean and amplitude of the circadian rhythm in the QT interval.
The table shows the circadian RR or QT interval parameters from iCSΔBmal1+/+ and iCSΔBmal1−/− mice before (Pre-) and after (Post-) vehicle or tamoxifen treatment.
| RR or QT interval | Pre-iCSΔBmal1+/+ vehicle | Post-iCSΔBmal1+/+ vehicle | Pre-iCSΔBmal1−/− tamoxifen | Post-iCSΔBmal1−/− tamoxifen |
|---|---|---|---|---|
| RR midline (ms) | 111.7 ± 1.8 | 114.3 ± 2.4 | 108.3 ± 3.1 | 117.3 ± 3.1 |
| RR amplitude (ms) | 25.5 ± 0.9 | 10.5 ± 1.5 | 8.4 ± 1.2 | 11.5 ± 1.2 |
| RR phase (hrs) | 2.3 ± 0.7 | 2.6 ± 0.5 | 3.0 ± 0.6 | 2.4 ± 0.3 |
| RR period (hrs) | 25.5 ± 0.9 | 24.9 ± 0.6 | 24.3 ± 0.4 | 24.2 ± 0.1 |
| QT midline (ms) | 38.8 ± 0.9 | 38.5 ± 0.7 | 38.8 ± 1.1 | 43.6 ± 0.6* |
| QT amplitude (ms) | 1.2 ± 0.1 | 1.4 ± 0.3 | 1.6 ± 0.4 | 2.7 ± 0.3* |
| QT phase (hrs) | 3.9 ± 0.9 | 2.9 ± 0.9 | 4.9 ± 0.4 | 3.2 ± 0.5 |
| QT period (hrs) | 23.9 ± 0.3 | 24.4 ± 0.3 | 23.7 ± 0.2 | 24.0 ± 0.1 |
| N | 7 | 7 | 6 | 6 |
p<0.05 compared to pre-iCSΔBmal1+/+, post-iCSΔBmal1+/+ and pre-iCSΔBmal1−/−
DISCUSSION
The circadian molecular clock is a highly conserved, cell-autonomous, transcriptionally mediated mechanism that provides an evolutionary advantage by optimizing an organism’s physiology to anticipate the daily variation in the environment.24 To date there is only tangential and circumstantial evidence linking the cardiomyocyte molecular clock to circadian changes in K+ channel expression and cardiac electrophysiology. In 2003, Yamashita and colleagues hypothesized that the expression of certain cardiac K+ channel genes might be circadian, and they showed that Kcnd2 and Kcna5 transcripts followed a circadian pattern of expression in the female rat heart.6 Jeyaraj and colleagues (2012) recently published that Kcnd2 and Kchip2 are also expressed in a circadian pattern in the heart.7 However, neither study directly addressed the role that the cardiomyocyte molecular clock has on the circadian expression of these channels.
There are several similarities and differences between the results and conclusions from Yamashita and company (2003), Jeyaraj and colleagues (2012), and our study.6, 7 All three studies agree that Kcnd2 transcripts are expressed in a circadian pattern and Kcnj2 transcripts are not. However in contrast to Yamashita and colleagues, we conclude that Kcnh2 transcripts are expressed in a circadian pattern but Kcna5 transcripts are not. Moreover, unlike Jeyaraj and colleagues, we did not find that Kchip2 transcripts follow a circadian pattern of expression. The reasons for these discrepancies likely include the statistical approaches used to determine which transcripts exhibit a circadian pattern of expression and the sampling frequency of the transcripts tested (typically every 3–5 hours over a single day). Yamashita and colleagues relied on an ANOVA around a single time point whereas Jeyaraj and colleagues used consiner analysis. We used JTK_CYCLE analysis, which is a statistical approach that is resistant to outliers in the data and results in considerably greater sensitivity and specificity.20 The identification of circadian transcripts was done using data from a high resolution data set (CircaDB) in which expression is tracked every 2 hours for 48 hours coupled with our own JTK_CYCLE analyses.21 Importantly, we also found that Kcnh2 transcripts undergo a robust circadian pattern of expression in rat hearts (Figure 3A). Together, these datasets allow us to confidently conclude that Kcnh2 transcripts follow a circadian pattern of expression.
To directly test the role that the cardiomyocyte molecular clock has on the circadian expression pattern of Kcnd2 and Kcnh2 transcripts, we utilized a transgenic mouse model that disrupts the core circadian clock gene Bmal1 in adult cardiomyocytes. We found that the circadian expression of Kcnh2 (but not Kcnd2) transcripts was disrupted in iCSΔBmal1−/− hearts (Figure 1A), the corresponding IKr was smaller in these iCSΔBmal1−/− ventricular myocytes (Figure 2), and co-expressing Bmal1 and Clock cDNA caused the transactivation of the cloned human Kcnh2 promoter in C2C12 cells (Figures 3B). Together, these data strongly suggest that Kcnh2 is a direct target of the cardiomyocyte molecular clock mechanism. Our finding that the circadian expression of Kcnd2 is not altered in iCSΔBmal1−/− hearts suggests its expression is likely influenced by clock mechanisms originating outside the heart (i.e. neurohumoral signaling). Consistent with this, germline Bmal1-null mice do not show a circadian pattern of Kcnd2 expression in the heart.7 Perhaps more surprising, is the finding that disruption of the molecular clock also caused the downregulation of a number of different K+ channel transcripts not expressed in a circadian pattern (Figure 1B). These data argue that several downstream mechanisms related to cardiomyocyte molecular clock signaling (i.e. regulation of transcription factors, metabolism, etc) can also influence K+ channel expression. Identification of the mechanism(s) which drives the circadian expression of Kcnd2 and the downregulation of non-circadian K+ channel transcripts warrants further investigation.
Surprisingly, we found that the deletion of Bmal1 from cardiomyocytes unmasked a time-of-day prolongation in the QTc interval during the light-phase (Figure 4). This finding is counterintuitive because it suggests that the cardiomyocyte molecular clock functions to limit the circadian rhythm in the QT interval. These data are exciting because they highlight a new role for the molecular clock mechanism: not only does the molecular clock mechanism initiate anticipatory circadian rhythms to adjust to daily changes in the environment, but it can also help to offset or blunt circadian rhythms that might prove detrimental.
Based on the ECG data, our working model for how the cardiomyocyte molecular clock contributes to ventricular repolarization is quite different than Jeyaraj and colleagues.7 Jeyaraj and colleagues suggested that the transcription factor, Krüppel-like factor 15 gene (Klf15) is a direct target of BMAL1 in the heart, and they used several different transgenic Klf15 mouse models to argue KLF15 underlies the circadian expression of Kchip2 in the heart. They speculate that, since Kchip2 is an obligatory subunit for the transient outward K+ current (Ito), the KLF15-mediated circadian expression of Kchip2 produces a daily rise and fall in Ito to generate a circadian rhythm in the QTc interval of WT mice. We and several other studies have not been able to resolve a circadian rhythm in the QTc interval in WT or control mice.18, 19 More importantly, our work suggests that disruption of the cardiomyocyte molecular clock likely downregulates the expression for a number of different K+ genes. A loss in overall K+ channel expression would be expected to prolong the QT interval. Interestingly, our data suggest that disruption of the cardiomyocyte molecular clock contributes to a disproportionate prolongation of the QT interval during the light-phase when the heart rate is slowest in mice (Figure 5; Table 1). These findings are analogous to reports investigating the circadian relation between the RR interval and QT interval in a transgenic mouse model of long QT syndrome (LQTS), as well as clinical data investigating the relation between the RR and QT intervals in long QT syndrome patients.18, 25
There are several limitations to this study. Since this work is done in mice, the question as to whether a link exists between cardiac ion channel expression, diurnal alterations in ventricular repolarization, and susceptibility to lethal arrhythmias underlie the diurnal variation in SCD in humans requires further study. There are undoubtedly multiple factors that contribute to the heightened early morning susceptibility to SCD, including changes in autonomic signaling.
CONCLUSION
This is the first work to clearly demonstrate that intrinsic circadian mediators, including the cardiomyocyte molecular clock, are modulators of the electrical properties in the heart, which possibly contributes to the daily variation in SCD.
Supplementary Material
CLINICAL PERSPECTIVES.
Is there a diurnal rhythm in the QTc interval? For over 25 years clinician scientists have been working to answer this question. It is an important question because if there is a circadian rhythm in the QTc interval, then the time-of-day it is measured could influence its utility in determining of whether patients can safely tolerate drugs known to affect the QTc interval or in the diagnosis of the congenital long QT syndrome (LQTS). The general consensus is that, if there is a diurnal variation in the QTc interval, it is very small. This observation has also been confirmed in mice. However, data in our manuscript now suggests that an internal time-keeping mechanism intrinsic to cardiomyocytes directly contributes to buffering against daily changes in the QTc interval. If the intrinsic timing-keeping mechanism in the heart is disrupted, then the diurnal change in the QTc interval is amplified by as much as 10%. In humans, this would translate into about a 40 ms difference in the QTc measurement! In other words, although most patients might not normally show a large circadian flux in their QTc interval, they might if the internal clock mechanism in the heart is disrupted. Instead of simply taking single snaps shots of ventricular repolarization, taking diurnal temporal changes into consideration may better delineate at-risk patients for arrhythmias. Our findings may ultimately help physicians to increase their clinical suspicion for prolonged QT syndrome or predict the QT prolongation while initiating a QT prolonging drug.
Acknowledgments
Acknowledgement of Financial Support: Funding Sources
This work was supported by the following NIH grants RC1ES018636 and AR55246 (KAE), and R01 HL087039 (BPD).
Abbreviations
- SCD
Sudden Cardiac Death
- ECG
Electrocardiographic
- qtPCR
quantitative PCR
- BMAL1
Brain muscle arnt-like1
- Clock
Circadian locomotor output control kaput
- PER
Period
- CRY
cryptochrome
- iCSΔBmal1
inducible cardiomyocyte specific deletion of Bmal1
- ZT
Zeitgeber time
- QTc
heart rate corrected QT
- INa
Na+ current
- IKr
rapidly activating delayed-rectifier K+ current
- LQTS
long QT syndrome
- Klf15
krüppel-like factor 15
Footnotes
Conflicts of Interests: Brian Delisle has a research contract with Gilead Scientific.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography & References Cited
- 1.Muller JE, Ludmer PL, Willich SN, Tofler GH, Aylmer G, Klangos I, Stone PH. Circadian variation in the frequency of sudden cardiac death. Circulation. 1987 Jan;75:131–138. doi: 10.1161/01.cir.75.1.131. [DOI] [PubMed] [Google Scholar]
- 2.Willich SN, Levy D, Rocco MB, Tofler GH, Stone PH, Muller JE. Circadian variation in the incidence of sudden cardiac death in the Framingham Heart Study population. The American journal of cardiology. 1987 Oct 1;60:801–806. doi: 10.1016/0002-9149(87)91027-7. [DOI] [PubMed] [Google Scholar]
- 3.Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep. 2005 Jul;6(S9-13) doi: 10.1038/sj.embor.7400424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Naef F. Cellular oscillators: rhythmic gene expression and metabolism. Current opinion in cell biology. 2005 Apr;17:223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Martino TA, Sole MJ. Molecular Time: An Often Overlooked Dimension to Cardiovascular Disease. Circulation Research. 2009 Nov 20;105:1047–1061. doi: 10.1161/CIRCRESAHA.109.206201. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita T, Sekiguchi A, Iwasaki YK, Sagara K, Iinuma H, Hatano S, Fu LT, Watanabe H. Circadian variation of cardiac K+ channel gene expression. Circulation. 2003 Apr 15;107:1917–1922. doi: 10.1161/01.CIR.0000058752.79734.F0. [DOI] [PubMed] [Google Scholar]
- 7.Jeyaraj D, Haldar SM, Wan X, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012 Mar 1;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP. The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol. 2013 May 15;304:C954–965. doi: 10.1152/ajpcell.00383.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohawk JA, Green CB, Takahashi JS. Central and Peripheral Circadian Clocks in Mammals. Annual Review of Neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young ME, Razeghi P, Taegtmeyer H. Clock genes in the heart: characterization and attenuation with hypertrophy. Circ Res. 2001 Jun 8;88:1142–1150. doi: 10.1161/hh1101.091190. [DOI] [PubMed] [Google Scholar]
- 11.Durgan DJ, Hotze MA, Tomlin TM, Egbejimi O, Graveleau C, Abel ED, Shaw CA, Bray MS, Hardin PE, Young ME. The intrinsic circadian clock within the cardiomyocyte. American journal of physiology Heart and circulatory physiology. 2005 Oct;289:H1530–1541. doi: 10.1152/ajpheart.00406.2005. [DOI] [PubMed] [Google Scholar]
- 12.Young ME, Brewer RA, Peliciari-Garcia RA, et al. Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. Journal of biological rhythms. 2014 Aug;29:257–276. doi: 10.1177/0748730414543141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray MS, Shaw CA, Moore MW, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008 Feb;294:H1036–1047. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S. Isolation and characterization of I(Kr) in cardiac myocytes by Cs+ permeation. American journal of physiology Heart and circulatory physiology. 2006 Mar;290:H1038–1049. doi: 10.1152/ajpheart.00679.2005. [DOI] [PubMed] [Google Scholar]
- 15.Guo J, Massaeli H, Li W, Xu J, Luo T, Shaw J, Kirshenbaum LA, Zhang S. Identification of IKr and its trafficking disruption induced by probucol in cultured neonatal rat cardiomyocytes. The Journal of pharmacology and experimental therapeutics. 2007 Jun;321:911–920. doi: 10.1124/jpet.107.120931. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Massaeli H, Xu J, Jia Z, Wigle JT, Mesaeli N, Zhang S. Extracellular K+ concentration controls cell surface density of IKr in rabbit hearts and of the HERG channel in human cell lines. The Journal of clinical investigation. 2009 Sep;119:2745–2757. doi: 10.1172/JCI39027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Patel SP, McCarthy JJ, Rabchevsky AG, Goldhamer DJ, Esser KA. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Research. 2012 Apr 1;40:3419–3430. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder EA, Burgess DE, Manning CL, Zhao Y, Moss AJ, Patwardhan A, Elayi CS, Esser KA, Delisle BP. Light phase-restricted feeding slows basal heart rate to exaggerate the type-3 long QT syndrome phenotype in mice. 3072014 doi: 10.1152/ajpheart.00341.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell GF, Jeron A, Koren G. Measurement of heart rate and Q-T interval in the conscious mouse. Am J Physiol. 1998 Mar;274:H747–751. doi: 10.1152/ajpheart.1998.274.3.H747. [DOI] [PubMed] [Google Scholar]
- 20.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An Efficient Nonparametric Algorithm for Detecting Rhythmic Components in Genome-Scale Data Sets. Journal of Biological Rhythms. 2010 Oct 1;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Research. 2013 Jan 1;41:D1009–D1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995 Apr 21;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 23.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. 03/23/print. [DOI] [PubMed] [Google Scholar]
- 24.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. 11//print. [DOI] [PubMed] [Google Scholar]
- 25.Merri M, Moss AJ, Benhorin J, Locati EH, Alberti M, Badilini F. Relation between ventricular repolarization duration and cardiac cycle length during 24-hour Holter recordings. Findings in normal patients and patients with long QT syndrome. Circulation. 1992 May 1;85:1816–1821. doi: 10.1161/01.cir.85.5.1816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.