Abstract
The obligate intracellular bacterium Chlamydia trachomatis rapidly induces its own entry into host cells. Initial attachment is mediated by electrostatic interactions to heparan sulfate moieties on the host cell, followed by irreversible binding to an unknown secondary receptor. This secondary binding leads to the recruitment of actin to the site of attachment, formation of an actin-rich, pedestallike structure, and finally internalization of the bacteria. How chlamydiae induce this process is unknown. We have identified a high-molecular-mass tyrosine-phosphorylated protein that is rapidly phosphorylated on attachment to the host cell. Immunoelectron microscopy studies revealed that this tyrosine-phosphorylated protein is localized to the cytoplasmic face of the plasma membrane at the site of attachment of surface-associated chlamydiae. The phosphoprotein was isolated by immunoprecipitation with the antiphosphotyrosine antibody 4G10 and identified as the chlamydial protein CT456, a hypothetical protein with unknown function. The chlamydial protein (Tarp) appears to be translocated into the host cell by type III secretion because it is exported in a Yersinia heterologous expression assay. Phosphotyrosine signaling across the plasma membrane preceded the recruitment of actin to the site of chlamydial attachment and may represent the initial signal transduced from pathogen to the host cell. These results suggest that C. trachomatis internalization is mediated by a chlamydial type III-secreted effector protein.
Chlamydia trachomatis is a Gram-negative obligate intracellular bacterium that is a leading cause of sexually transmitted diseases and blindness worldwide (1). Chlamydiae have a biphasic developmental cycle characterized by an infectious but metabolically inactive extracellular form, called the elementary body (EB), that initiates infection by attaching to and inducing uptake by the host cell. Once internalized, chlamydiae remain within a membrane-bound vacuole termed an inclusion, where the organism differentiates into the larger, metabolically active reticulate body. Reticulate bodies replicate and differentiate back to EBs before release at the end of the developmental cycle.
EBs attach to and enter cultured eukaryotic cells so efficiently that the process has been termed parasite-specified phagocytosis (2). Despite the importance of this event to chlamydial pathogenesis, little consensus exists regarding the identity of the chlamydial ligands and respective host receptors (for a review, see ref. 3). Considerable evidence suggests that electrostatic interactions mediate attachment with heparan sulfate-like proteoglycans involved in an initial, reversible interaction with the eukaryotic host cell for many, but not all, strains and species of chlamydiae (4–9). Recent studies using chemically mutagenized cell lines distinguished a subsequent, irreversible secondary binding step in the entry process, although the receptor was not identified (8, 10). Entry of C. trachomatis requires participation of the actin cytoskeleton and involves the formation of actin-rich pedestals at the site of entry (11). It is likely that chlamydiae initiate signaling cascades that recruit actin to promote internalization.
Tyrosine phosphorylation of host and/or bacterial proteins have been implicated in signaling pathways triggering the entry of many intracellular pathogens (12). Studies of tyrosine phosphorylation in response to chlamydial infection have described several proteins; a triplet of 64, 66, and 68 kDa, and proteins migrating at 97 and 140 kDa (13). Similar studies reported tyrosine phosphorylated proteins of 75–85 kDa and 100 kDa (14). In both cases, tyrosine-phosphorylated proteins were observed in association with EBs by immunofluorescence. Although definitive identification of the proteins was not achieved, they were presumed to be of host origin.
We have confirmed tyrosine phosphorylation in response to C. trachomatis infection but find only a single unique protein that is phosphorylated in a multiplicity-dependent fashion. This protein was partially purified and identified by matrix-assisted laser desorption ionization-time of flight MS as a chlamydial protein of unknown function that appears to function in signaling of the cytoskeletal rearrangements that lead to the endocytosis of EBs.
Materials and Methods
Organisms and Cell Culture. C. trachomatis serovars L2 (LGV 434) or D (UW3-Cx) were grown in HeLa 229 cells as described and EBs were purified by Renografin (Squibb) density gradient centrifugation (15). Fluorescent CMTMR-labeled EBs were prepared as described (11, 16). HeLa cells were infected at a multiplicity of infection (MOI) of 100 (unless otherwise noted) for both serovar L2 and D in Hanks' balanced salt solution (HBSS; Invitrogen) at 4°C. The inoculum was removed (time = 0), cells were rinsed with HBSS, and warm media were added for the additional times indicated.
Cloning and Sequencing. Full-length CT456 from L2 genomic DNA was PCR-amplified and cloned in pCR-Blunt (Invitrogen). Sequencing was performed by the DNA-sequencing facility at Iowa State University (Ames) by using M13 universal and reverse primers and progressive step primers. Sequences were compiled and analyzed with macvector and assemblylign. The L2 CT456 DNA sequence was deposited in GenBank (accession no. AY623902).
Antibodies. The C-terminal 580 aa of CT456 was cloned into pRSET-B (Invitrogen) and expressed as a His-tagged fusion. The fusion protein was purified on a nickel column (Invitrogen) and used as an immunogen for the production of specific polyclonal antibodies in female New Zealand White rabbits as described (17). Other antibodies used were anti-phosphotyrosine mAb clone 4G10 and 4G10 coupled to agarose (Upstate USA, Charlottesville, VA), anti-Rickettsia rickettsii ompB mAb 13–3 (18), anti-C. trachomatis major outer-membrane protein (MOMP) mAbs L2-I-10 and L2-I-45 (19), anti-Flag M2-alkaline phosphatase (Sigma-Aldrich), Texas red-X phalloidin (Molecular Probes), and anti-C. trachomatis LcrH (K.A.F. and T.H., unpublished data). Secondary antibodies for microscopy were AlexaFluor 594-conjugated anti-rabbit IgG (Molecular Probes), FITC-conjugated anti-rabbit IgG (Zymed), and tetramethylrhodamine B isothiocyanate-conjugated anti-mouse IgG (Zymed).
Analysis of Type III Secretion. Type III secretion of CT456 by heterologous expression in Yersinia pseudotuberculosis was performed by using type III competent YPIII pIB102 WT or type III null YPIII pIB68 (yscS) strains expressing CT456-Flag-tag (FT) fusions as described (20, 21).
Transfections. Full-length CT229 was PCR-amplified from L2 genomic DNA and ligated into pcDNA3.1/CT-GFP-TOPO (Invitrogen). HeLa cells were seeded on 12-mm glass coverslips in 24-well plates to obtain a monolayer of ≈50% confluency. Transfection of CT456-enhanced GFP (EGFP)-C3 (BD Biosciences, Palo Alto, CA) or CT229-GFP constructs were performed by using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Expression from the transfection vectors proceeded for 24 h at 37°C before fixation.
SDS/PAGE and Immunoblotting. Proteins were separated on SDS-7.5% polyacrylamide gels (22) and transferred to 0.45-μm Transblot nitrocellulose membrane (Bio-Rad). Immunoblots were developed by using Super Signal West Pico chemiluminescence reagent (Pierce). For immunoprecipitations, cultures were lysed in cold immunoprecipitation buffer [50 mM Tris, pH 8/150 mM NaCl/1% Nonidet P-40/1% sodium deoxycholate/0.1% SDS/1 mM Na3VO4/1 mM NaF/protease inhibitor mixture (Complete mini, EDTA-free, Roche Diagnostics)]. Supernatants were incubated overnight at 4°C with antibody and precipitated with protein A-Sepharose (Sigma-Aldrich). For 2D gel electrophoresis, cultures were scraped into buffer and acetoneprecipitated; the pellet was solubilized in rehydration buffer {7 M urea/2 M thiourea/2% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/5% (vol/vol) 2-mercaptoethanol/1 mM NaF/0.2 mM Na3VO4} and resolved in the first dimension by using the IPGphor isoelectric focusing system with pH 3–10 drystrips according to the manufacturer's instructions (Amersham Pharmacia Biosciences). The second dimension was separated and transferred to nitrocellulose as above.
MS. CT456 was enriched by immunoprecipitation from L2-infected HeLa cell lysates by using 4G10, separated by SDS/PAGE, and visualized by the SilverQuest silver stain kit (Invitrogen). Gel slices were processed as described in detail in the Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Matrix-assisted laser desorption ionization-time of flight spectra were collected by using a Voyager-DE STR mass spectrometer (PerSeptive Biosystems, Framington, MA).
Microscopy. Infected cells on 12-mm glass coverslips were processed as described (23), except that all buffers contained 1 mM Na3VO4/1 mM NaF. Confocal images were acquired on a Perkin–Elmer UltraView spinning disk connected to a Nikon Eclipse TE2000-S microscope with a ×60, 1.4 numerical aperture oil-immersion objective. All images were processed with imagej (written by Wayne Rasband at the National Institutes of Health and available by anonymous FTP from http://zippy.nimh.nih.gov) and/or photoshop software (Adobe Systems, Mountain View, CA). Images are representative of at least three separate experiments.
Live-Cell Confocal Microscopy. Cos-7 cells were grown on 25-mm glass coverslips and electroporated with 500 μg of actin-EGFP-C1 (24) per well with the BTX 35-mm Petri Pulser (Genetronics, San Diego) connected to an ECM 630 generator (Genetronics) set at 150 V/975 μF in serum-free Optimem (Invitrogen). Fresh serum-containing medium was added, and cells were allowed to express overnight. 4G10 was labeled by using the Alexa Fluor 660 protein-labeling kit (Molecular Probes) according to the manufacturer's instructions and was microinjected into ≈50 cells per coverslip as described (25), except that Oregon-green dextran was not used. GFP-expressing and microinjected cells were identified by spinning-disk confocal microscopy, and, after an initial image was taken, cells were infected with CMTMR-labeled L2 EBs. Images were acquired every 2 s for 10 min per coverslip. Results are representative of more than four experiments.
Immunoelectron Microscopy. Confluent HeLa cells on Thermanox coverslips were infected with L2 (MOI ≈ 300). Coverslips were washed two times with HBSS and then processed as described (17), except that cells were stained with 4G10 overnight at 4°C, and PBS washes, permeabilization, and primary antibody solutions included 1 mM Na3VO4 and 1 mM NaF. Sections were observed at 60 kV on a Philips CM-10 transmission electron microscope (FEI Co., Hillsboro, OR). Images were acquired with an AMT digital camera system (Advanced Microscopy Techniques, Chazy, NY) and processed with photoshop software.
Results
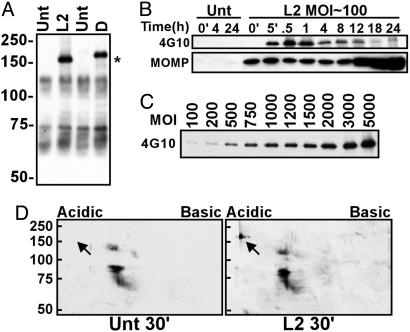
A Single Unique Tyrosine-Phosphorylated Protein Is Induced During Infection with C. trachomatis. Immunoblotting with the mAb 4G10 was used to identify tyrosine-phosphorylated proteins in response to C. trachomatis L2 infection. In contrast to previous studies that had identified two or more tyrosine-phosphorylated putative host proteins in C. trachomatis-infected cultures (13, 14), we observed only a single unique tyrosine-phosphorylated protein that increased in prominence in a multiplicity-dependent fashion during infection with C. trachomatis (Fig. 1 A and C). This phosphoprotein was not detected in lysates of purified EBs (data not shown) and was observed during infection with either C. trachomatis serovars L2 or D, although the apparent Mr of the D protein was slightly higher. Reactivity with 4G10 was abolished by treatment of the lysates with calf intestinal alkaline phosphatase (data not shown) and occurred even in the presence of the tyrosine kinase inhibitor, genistein, at 200 μM final concentration (Fig. 7, which is published as supporting information on the PNAS web site). EBs allowed to adhere at 4°C then shifted at 37°C to trigger entry resulted in rapid tyrosine phosphorylation of the protein (Fig. 1B). Signal was detected by 5-min after temperature shift and was maximal at 30–60 min after infection. Tyrosine phosphorylation was detectable for at least 24 h although the intensity diminished at later time points. Chlamydial transcription and translation were not required for this phosphorylation as incubation in the presence of rifampicin or chloramphenicol did not inhibit its occurrence (Fig. 7). Two-dimensional gel electrophoresis demonstrated that the phosphoprotein is highly acidic (Fig. 1D).
Fig. 1.
Detection of a unique tyrosine-phosphorylated protein in C. trachomatis-infected cells. (A) Mock-infected (Unt) and C. trachomatis serovar L2- or D-infected HeLa cells were incubated at 37°C for 30 min. Total cell lysates were separated by SDS/PAGE, and immunoblots were probed with mAb 4G10. The asterisk indicates the unique phosphoproteins. Molecular mass in kDa is indicated on the left. (B) HeLa cells were infected with L2 or mock-infected (Unt) for the indicated times (in hours, except where ′= minute) and subjected to SDS/PAGE and immunoblotting. The immunoblot was split, and upper blot was probed with 4G10 and lower blot was probed with anti-MOMP mAb I-45. (C) HeLa cells were infected with L2 with the indicated MOI for 30 min and immunoblotted against 4G10. (D) HeLa cells were infected (L2) or left uninfected (Unt) for 30 min, and total cell lysates were subjected to 2D gel electrophoresis and probed with 4G10. The arrow identifies the unique phosphoprotein. Molecular mass markers (kDa) are indicated.
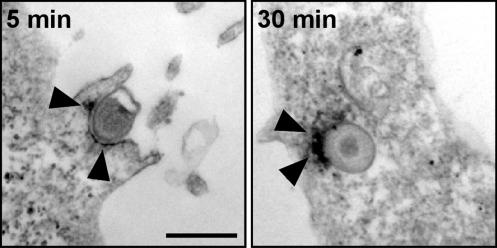
We confirmed tyrosine phosphorylation in association with internalized EBs by immunofluorescence microscopy with mAb 4G10. As shown (13, 14), immunoreactivity was observed soon after initiation of infection by warming L2-infected cultures to 37°C and appeared to stain the EBs in a polar fashion (Fig. 8, which is published as supporting information on the PNAS web site). By immunoelectron microscopy with a horseradish peroxidase-conjugated secondary antibody, reaction product could readily be visualized on the cytoplasmic face of the plasma membrane, even as EBs remained extracellular at the apex of the characteristic actin-rich pedestals formed during internalization (11). Immunoreactivity remained localized to one hemisphere of the nascent inclusion after endocytosis (Fig. 2).
Fig. 2.
Immunoelectron microscopy reveals polar localization of 4G10 with EBs. HeLa cells were infected with L2 EBs (MOI ≈ 300) for 5 or 30 min. Coverslips were stained with 4G10 and visualized with an immunoperoxidase-conjugated secondary antibody. Arrowheads indicate areas of positive 4G10 reactivity. (Bar = 500 nm.)
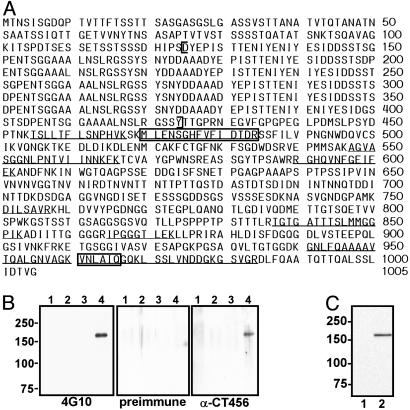
Identification of CT456. Immunoprecipitation with 4G10 was used to enrich for the tyrosine-phosphorylated protein from C. trachomatis L2-infected cultures. A single comigrating band corresponding to 4G10 reactivity was visualized on silver-stained SDS-polyacrylamide gels. No comigrating bands were observed from uninfected cultures immunoprecipitated with 4G10 or infected cultures precipitated with an irrelevant, isotype-matched mAb. The unique band was excised and identified by matrix-assisted laser desorption ionization-time of flight MS as the hypothetical protein CT456 from the C. trachomatis serovar D genome sequence (26). The corresponding gene from C. trachomatis L2 was cloned, sequenced, and found to exhibit 94% identity to the serovar D protein sequence (by bestfit alignment, http://molbio.info.nih.gov/molbio/gcglite/compare.html). The predicted molecular mass of the protein product is 103 kDa with a pI of 3.92. CT456 thus appears to migrate aberrantly on SDS/PAGE. Fig. 3A shows the serovar L2 CT456 protein sequence with peptides identified by MS demarcated. An interesting structural feature of CT456 is the presence of six near-identical tandem repeats of ≈50 aa in length (Fig. 9, which is published as supporting information on the PNAS web site). The purified C-terminal domain (downstream of the repeats) of serovar L2 CT456 was used as an immunogen to inoculate rabbits for antibody production. The anti-CT456 antiserum reacts with the protein immunoprecipitated by 4G10, thereby confirming the identity of the tyrosine-phosphorylated protein in C. trachomatis-infected cell lysates as CT456 (Fig. 3B). Conversely, immunoprecipitation of L2-infected cultures with anti-CT456 similarly precipitated CT456, which was reactive with 4G10, whereas immunoprecipitation of equivalent amounts of Tarp from L2-infected cultures maintained at 4°C confirmed that Tarp is present but not phosphorylated in EBs (Fig. 9). The anti-456 antiserum also reacted with a prominent band of the corresponding size from either L2 or D EBs (Fig. 3C and data not shown); thus, the nonphosphorylated precursor is preexisting in EBs.
Fig. 3.
Identification of the unique phosphoprotein as hypothetical protein CT456. (A) The complete protein sequence for L2 CT456. Peptide fragments that match the serovar D CT456 protein sequence are underlined and sequenced peptides are boxed. Brackets indicate the repeat region. (B) HeLa cells were mock-(lanes 1 and 2) or L2-infected (MOI ≈ 1,000; lanes 3 and 4) for 30 min, and cell lysates were immunoprecipitated with 4G10 (lanes 2 and 4) or the irrelevant isotype control mAb 13-3 (lanes 1 and 3) and separated by SDS/PAGE. Immunoblots were probed with 4G10, rabbit preimmune sera, or a rabbit polyclonal antiserum prepared against CT456. Molecular mass markers (kDa) are indicated. (C) An immunoblot of L2 EB total lysates was probed with anti-CT456 pAb (lane 2) or preimmune sera (lane 1).
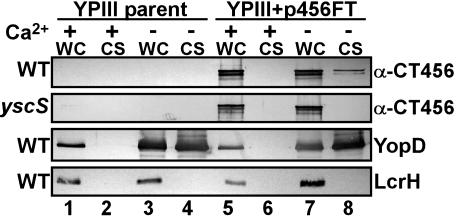
CT456 Is Secreted by a Type III System. Many Gram-negative pathogens deliver effector proteins to the host cell by a type III secretion system (27). Because tyrosine phosphorylation is observed on the cytoplasmic face of the host plasma membrane at the site of intimate attachment of EBs and on the nascent inclusion membrane of internalized EBs, it is likely that CT456 is translocated across the plasma membrane. To test this, we used a heterologous type III system used previously for identifying suspected chlamydial type III effectors (20, 21). Type III competent (WT) or null (yscS) Y. pseudotuberculosis alone or expressing full-length CT456-Flag-tagged (FT) fusions were grown in media inductive (-Ca2+) or repressive (+Ca2+) for type III secretion, and cultures were fractionated into cell-free culture supernatants and yersiniae-containing whole-cell pellets. Immunoblot analysis (Fig. 4) of parent and yscS cultures revealed essentially equal CT456-FT-specific signals in +Ca2+ (lane 5) and -Ca2+ (lane 7) whole-cell fractions but not in vector-only controls (lanes 1–4). CT456-FT was detected in the -Ca2+ supernatant (lane 8) of WT but not yscS Yersinia cultures. This outcome was not due to bacterial lysis, because the Yersinia cytoplasmic protein LcrH was detected only in whole-cell pellet samples and not released into supernatant fractions. Additionally, expression of CT456-FT had no adverse effects on the ability of Yersinia to properly regulate and secrete YopD, an endogenous type III substrate. Therefore, these data indicate that CT456-FT is specifically recognized and secreted as a type III substrate.
Fig. 4.
CT456 is secreted by a type III system. WT Y. pseudotuberculosis YPIII pIB102 (WT) or a type III secretion null strain YPIII pIB68 (yscS) expressing CT456-FT were grown in the presence (lanes 1, 2, 5, and 6) or absence (lanes 3, 4, 7, and 8) of 2.5 mM Ca2+. Cultures were induced at 37°C for 4 h and fractionated into cell-free supernatants (CS); whole cells (WC) were then subjected to SDS/PAGE. Immunoblots were probed with α-Flag M2 to detect CT456-FT, α-YopD, or α-LcrH.
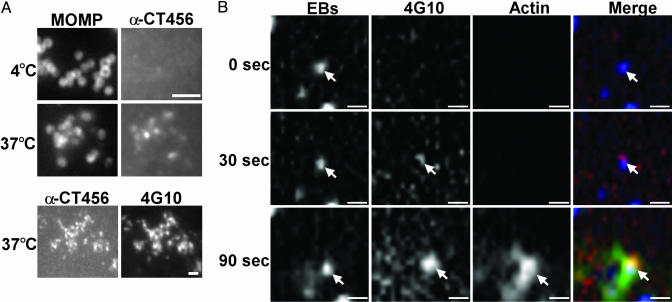
CT456 Translocation and Actin Recruitment. To characterize exposure of CT456 on the EB surface, HeLa cells were infected at 4°C, then shifted to 37°C for 30 min to trigger entry. Cultures were stained with anti-CT456 and counterstained with anti-MOMP mAb L2-I-10 to localize EBs. Surface-associated EBs show little or no reactivity with anti-CT456 at 4°C, supporting the concept that the protein is not exposed on the EB surface (Fig. 5A). After temperature shift, CT456 staining colocalized well with anti-MOMP staining. The staining pattern at 37°C with anti-CT456 also corresponded to 4G10 staining (Fig. 5A). Thus, CT456 does not appear to be exposed on the EB surface but is translocated such that it becomes antibody-accessible during the entry process.
Fig. 5.
CT456 is not exposed on the EB surface but is translocated and phosphorylated before actin recruitment. (A) HeLa cells were infected with L2 at 4°C, the medium was replaced, and the cultures were then held at 4°C or shifted to 37°C for 30 min. Cells were fixed and stained by indirect immunofluorescence with anti-CT456 and anti-MOMP mAb L2-I-10 or 4G10. (Bar = 1 μm.) (B) Cos7 cells were electroporated with actin-EGFP, microinjected with Alexa Fluor 660-labeled 4G10, and infected with CMTMR-labeled L2 EBs. Images of a GFP-expressing, microinjected, and infected cell were acquired every 2 s by spinning-disk confocal microscopy. The timeline begins with the first visualization of EB fluorescence in contact with the cell. The EB is indicated by the arrow. A representative experiment is shown. (Bar = 1 μm.)
Because CT456 phosphorylation occurs rapidly and is observed at the site of pedestal formation, we investigated whether it might be involved in signaling for actin recruitment. A three-color live-cell experiment was performed on Cos7 cells expressing actin C-terminally fused to enhanced GFP (actin-EGFP) and microinjected with fluorescently labeled 4G10 antibody. Fluorescent CMTMR-labeled L2 EBs were added and spinning-disk confocal microscopy was used to acquire images at 2-s intervals. With time 0 defined as the first sighting of an EB in the image, 4G10 staining was first observed by 30 s, adjacent to the EB, with robust recruitment of actin observed by 90 s (Fig. 5B). CT456 phosphorylation detected by the intracellular pool of 4G10 confirms that CT456 is translocated and exposed to the cytoplasm. Further, the sequence of events suggests that CT456 phosphorylation may play an important role in recruitment of actin and initiation of EB entry.
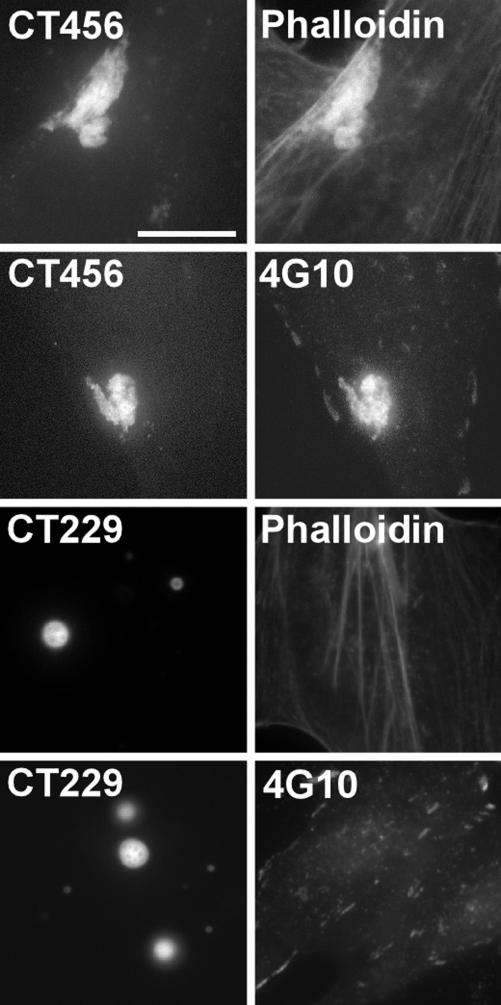
Transfected CT456 Is Tyrosine Phosphorylated and Recruits Actin. To further examine the role of CT456 in recruitment of actin, full-length CT456 from serovar L2 was expressed as a C-terminal EGFP-tagged fusion protein in HeLa cells. CT456-EGFP expressed in HeLa cells formed aggregates that stained with phalloidin to indicate recruitment or bundling of actin (Fig. 6). CT456-EGFP also colocalized with 4G10 staining to demon-strate phosphorylation of the transfected protein. HeLa cells expressing EGFP alone (data not shown) or a chlamydial inclusion membrane protein (CT229) fused to GFP showed no colocalization with phalloidin nor reactivity with mAb 4G10 (Fig. 6). Thus, it is likely that CT456 phosphorylation is important in the initiation of as-yet-unidentified signaling pathways that initiate the entry process by triggering recruitment of actin.
Fig. 6.
Cells expressing full-length CT456-EGFP colocalize with phalloidin and 4G10. HeLa cells expressing CT456-EGFP or CT229-EGFP were fixed, and the EGFP signal was viewed in conjunction with staining of actin with phalloidin or indirect immunofluorescence of 4G10. (Bar = 10 μm.)
Discussion
Although chlamydiae enter nonphagocytic cells very efficiently, the molecular mechanisms involved have remained elusive. An initial electrostatic interaction involving heparan sulfate-like glycosaminoglycans is believed to precede an irreversible engagement with unknown receptors that leads to internalization (5, 8, 10). The earliest known signaling of chlamydiae to the host cell is tyrosine phosphorylation of unknown proteins in immediate apposition to the internalized EBs (13, 14). We find that this signal is transduced to the cytoplasmic face of the plasma membrane when the chlamydiae are still extracellular and have identified a prominent tyrosine-phosphorylated protein as the C. trachomatis hypothetical protein CT456. CT456 is present in EBs but not exposed on the EB surface and appears to be translocated into the host cell by the chlamydial type III secretion system. Once exposed to the cytoplasm, CT456 is rapidly tyrosine-phosphorylated by unknown mechanisms. Tyrosine phosphorylation of CT456 is temporally and spatially associated with recruitment of actin to the site of chlamydial entry. Furthermore, CT456-EGFP expressed in HeLa cells is tyrosine-phosphorylated and recruits or bundles actin. CT456 thus appears to initiate or participate in signaling events that regulate the actin recruitment, which ultimately leads to internalization. We have therefore named CT456 “Tarp”, for translocated actin-recruiting phosphoprotein.
A striking feature of L2 Tarp is the presence of six tandem repeats of ≈50 aa. C. trachomatis serovar D is highly homologous to that of L2 but contains only three repeat units (Fig. 9). The repeat region is absent in other chlamydial strains or species. C. trachomatis strain MoPn (28), Chlamydia psittaci strain GPIC (29), and Chlamydia pneumoniae (28) all possess orthologs of CT456, but the greatest similarity occurs within the C-terminal domain downstream of the repeat region. The phosphorylated residues have not been defined but the repeat region of L2 is rich in tyrosine, containing 26 of the 31 residues present in the protein. The large number of potential tyrosine phosphorylation sites may explain the strong reactivity of Tarp with the 4G10 antibody even though the protein appears to be of low abundance in EBs. Despite the absence of repeats, C. pneumoniae entry involves the formation of identical short pedestal structures indistinguishable from those observed with C. trachomatis serovars L2 and D (K. Wolf, E.R.F., and T.H., unpublished observations). The function, if any, of the distinct domains of Tarp may provide unexpected insights into the different pathologies of chlamydial species and serovariants.
Phosphorylated Tarp appears to remain cytoplasmically exposed on the inclusion membrane at one side of internalized EBs for several hours after entry (data not shown). However, Tarp does not have any predicted transmembrane domains, thus it is unclear how the association with EBs through the inclusion membrane is maintained. An unusual protein structure may yield a domain that could span the membrane; however, Tarp does not become associated with membranes in the Triton X-100-soluble fraction after secretion and remains associated either with the cytoskeleton in cell debris or with the EBs themselves. Neither high salt (500 mM NaCl) nor reducing agents increased the Triton solubility of cell-associated Tarp (D.R.C. and T.H., unpublished observations). Identification of the means of Tarp attachment to EBs may shed light on Tarp function(s).
Although chlamydial EBs have been considered metabolically dormant, these findings suggest that they are primed for the translocation of preexisting effectors, such as CT456, by type III secretion. CT456 is transcribed from mid to late cycle in the chlamydial developmental cycle (30), and unphosphorylated Tarp is detected in purified, infectious EBs. In addition, inhibitors of chlamydial transcription or translation have no effect on Tarp phosphorylation. Tarp does not appear to be exposed on the surface of EBs because it is resistant to trypsinolysis on purified EBs (Fig. 10, which is published as supporting information on the PNAS web site) and is not reactive with an anti-Tarp antibody but becomes exposed after association of EBs with host cells at 37°C. Specific type III secretion from a heterologous bacterial host, Y. pseudotuberculosis, further suggests that Tarp is a chlamydial type III-secreted effector molecule. Whereas an active process on the part of extracellular EBs challenges some paradigms of this developmental stage, it should be noted that EBs contain an exceptionally high concentration of ATP (≈40 mM) (31) that is presumably retained for activities required before the developing reticulate bodies become fully metabolically active.
Several examples exist of bacteria that secrete type III effectors as a means of altering cytoskeletal processes. For example, enteropathogenic Escherichia coli (EPEC) translocates its own receptor, Tir, into the host cell where it inserts into the plasma membrane and binds to the EPEC outer-membrane protein intimin, which is required for intimate attachment to the host cell surface (32). Tyrosine phosphorylation of Tir recruits Nck and leads to the formation of a stable actin pedestal (33–35). In contrast, chlamydiae are internalized rapidly after recruitment of actin, whereas EPECs typically remain extracellular at the apex of the pedestals. EPEC also requires Nck for actin recruitment and pedestal formation, whereas chlamydial internalization functions independently of Nck (R.A.C., S.S.G., and T.H., unpublished observations). Salmonella and Shigella inject several type III effectors that interact with multiple host signaling pathways, including activating Rho/Rac/Cdc42 GTPases (36–40). This process leads to distinctive transient membrane ruffling that results in entry of the organism. Chlamydial entry depends on Rac activation (41), thus it is tempting to speculate that Tarp may trigger Rac signaling pathways that culminate in actin recruitment. However, the sequence similarity between chlamydial Tarp and other known bacterial secreted effectors is minimal, suggesting that any functional similarities arose independently.
The initial stage in the interaction of chlamydiae with host cells is a critical time in the infectious process. Tarp translocation and its tyrosine phosphorylation appear to be one of the first signals to the host cell to actively subvert host processes for parasite purposes. We believe Tarp plays a role in the recruitment of actin to initiate entry, but other functions are possible. Although we have no evidence that Tarp is involved in attachment, per se, the fact that it is rapidly translocated but remains associated with internalized EBs suggests it may be important in forming the irreversible attachment thought to follow initial electrostatic interactions (8). Additional, unidentified signaling pathways may also be activated that together initiate entry of the organism or promote events that favor intracellular survival after entry. For example, the nascent inclusion appears to be nonfusogenic and shows no demonstrable interaction with the host cell until de novo synthesized chlamydial proteins initiate interactions with the cytoskeleton and exocytic lipid-trafficking pathways (23, 25). Because Tarp is exposed to the cytoplasm and remains associated with EBs after entry, a role in controlling interactions of the early inclusion is possible.
Supplementary Material
Acknowledgments
We thank Janet Sager for excellent technical assistance; Drs. Robert Heinzen, Olivia Steele-Mortimer, and James Carroll for helpful discussions; Dr. Greg Plano for sharing the anti-YopD antibody; and the members of the Hackstadt laboratory for critical reading of the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EB, elementary body; MOI, multiplicity of infection; Tarp, translocated actin-recruiting phosphoprotein; MOMP, major outer-membrane protein; EGFP, enhanced GFP; FT, Flag-tag.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY623902).
See Commentary on page 9947.
References
- 1.Schachter, J. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity, ed. Stephens, R. S. (Am. Soc. Microbiol., Washington, DC), pp. 139-169.
- 2.Byrne, G. I. & Moulder, J. W. (1978) Infect. Immun. 19, 598-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackstadt, T. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity., ed. Stephens, R. S. (Am. Soc. Microbiol., Washington, DC), pp. 101-138.
- 4.Zhang, J. P. & Stephens, R. S. (1992) Cell 69, 861-869. [DOI] [PubMed] [Google Scholar]
- 5.Su, H., Raymond, L., Rockey, D. D., Fischer, E., Hackstadt, T. & Caldwell, H. D. (1996) Proc. Natl. Acad. Sci. USA 93, 11143-11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, C. H. & Wyrick, P. B. (1997) Infect. Immun. 65, 2914-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez-Martin, C. B., Ojcius, D. M., Hsia, R., Hellio, R., Bavoil, P. M. & Dautry-Varsat, A. (1997) Microb. Pathog. 22, 47-57. [DOI] [PubMed] [Google Scholar]
- 8.Carabeo, R. A. & Hackstadt, T. (2001) Infect. Immun. 69, 5899-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wuppermann, F. N., Hegemann, J. H. & Jantos, C. A. (2001) J. Infect. Dis. 184, 181-187. [DOI] [PubMed] [Google Scholar]
- 10.Fudyk, T., Olinger, L. & Stephens, R. S. (2002) Infect. Immun. 70, 6444-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carabeo, R. A., Grieshaber, S. S., Fischer, E. & Hackstadt, T. (2002) Infect. Immun. 70, 3793-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay, B. B. & Cossart, P. (1997) Science 276, 718-725. [DOI] [PubMed] [Google Scholar]
- 13.Birkelund, S., Johnsen, H. & Christiansen, G. (1994) Infect. Immun. 62, 4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fawaz, F. S., van Ooij, C., Homola, E., Mutka, S. C. & Engel, J. N. (1997) Infect. Immun. 65, 5301-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caldwell, H. D., Kromhout, J. & Schachter, J. (1981) Infect. Immun. 31, 1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boleti, H., Ojcius, D. M. & Dautry-Varsat, A. (2000) J. Microbiol. Methods 40, 265-274. [DOI] [PubMed] [Google Scholar]
- 17.Scidmore-Carlson, M. A., Shaw, E. I., Dooley, C. A., Fischer, E. R. & Hackstadt, T. (1999) Mol. Microbiol. 33, 753-765. [DOI] [PubMed] [Google Scholar]
- 18.Anacker, R. L., Mann, R. E. & Gonzales, C. (1987) J. Clin. Microbiol. 25, 167-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baehr, W., Zhang, Y. X., Joseph, T., Su, H., Nano, F. E., Everett, K. D. & Caldwell, H. D. (1988) Proc. Natl. Acad. Sci. USA 85, 4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields, K. A. & Hackstadt, T. (2000) Mol. Microbiol. 38, 1048-1060. [DOI] [PubMed] [Google Scholar]
- 21.Fields, K. A., Mead, D. J., Dooley, C. A. & Hackstadt, T. (2003) Mol. Microbiol. 48, 671-683. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 23.Hackstadt, T., Rockey, D. D., Heinzen, R. A. & Scidmore, M. A. (1996) EMBO J. 15, 964-977. [PMC free article] [PubMed] [Google Scholar]
- 24.Heinzen, R. A., Grieshaber, S. S., Van Kirk, L. S. & Devin, C. J. (1999) Infect. Immun. 67, 4201-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grieshaber, S. S., Grieshaber, N. A. & Hackstadt, T. (2003) J. Cell. Sci. 116, 3793-3802. [DOI] [PubMed] [Google Scholar]
- 26.Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R. L., Zhao, Q., et al. (1998) Science 282, 754-759. [DOI] [PubMed] [Google Scholar]
- 27.Cornelis, G. R. & Van Gijsegem, F. (2000) Annu. Rev. Microbiol. 54, 735-774. [DOI] [PubMed] [Google Scholar]
- 28.Read, T. D., Brunham, R. C., Shen, C., Gill, S. R., Heidelberg, J. F., White, O., Hickey, E. K., Peterson, J., Utterback, T., Berry, K., et al. (2000) Nucleic Acids Res. 28, 1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read, T. D., Myers, G. S., Brunham, R. C., Nelson, W. C., Paulsen, I. T., Heidelberg, J., Holtzapple, E., Khouri, H., Federova, N. B., Carty, H. A., et al. (2003) Nucleic Acids Res. 31, 2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., Beatty, W. L. & Caldwell, H. D. (2003) Proc. Natl. Acad. Sci. USA 100, 8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tipples, G. & McClarty, G. (1993) Mol. Microbiol. 8, 1105-1114. [DOI] [PubMed] [Google Scholar]
- 32.Rosenshine, I., Ruschkowski, S., Stein, M., Reinscheid, D. J., Mills, S. D. & Finlay, B. B. (1996) EMBO J. 15, 2613-2624. [PMC free article] [PubMed] [Google Scholar]
- 33.Kenny, B., DeVinney, R., Stein, M., Reinscheid, D. J., Frey, E. A. & Finlay, B. B. (1997) Cell 91, 511-520. [DOI] [PubMed] [Google Scholar]
- 34.Gruenheid, S., DeVinney, R., Bladt, F., Goosney, D., Gelkop, S., Gish, G. D., Pawson, T. & Finlay, B. B. (2001) Nat. Cell Biol. 3, 856-859. [DOI] [PubMed] [Google Scholar]
- 35.Campellone, K. G., Giese, A., Tipper, D. J. & Leong, J. M. (2002) Mol. Microbiol. 43, 1227-1241. [DOI] [PubMed] [Google Scholar]
- 36.Hardt, W. D., Chen, L. M., Schuebel, K. E., Bustelo, X. R. & Galan, J. E. (1998) Cell 93, 815-826. [DOI] [PubMed] [Google Scholar]
- 37.Stender, S., Friebel, A., Linder, S., Rohde, M., Mirold, S. & Hardt, W. D. (2000) Mol. Microbiol. 36, 1206-1221. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, D., Chen, L. M., Hernandez, L., Shears, S. B. & Galan, J. E. (2001) Mol. Microbiol. 39, 248-259. [DOI] [PubMed] [Google Scholar]
- 39.Mounier, J., Laurent, V., Hall, A., Fort, P., Carlier, M. F., Sansonetti, P. J. & Egile, C. (1999) J. Cell Sci. 112, 2069-2080. [DOI] [PubMed] [Google Scholar]
- 40.Tran Van Nhieu, G., Caron, E., Hall, A. & Sansonetti, P. J. (1999) EMBO J. 18, 3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carabeo, R. A., Grieshaber, S.S., Hasenkrug, A., Dooley, C. & Hackstadt, T. (2004) Traffic 5, 418-425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.