Chlamydia species have been known to cause human disease since antiquity. Chlamydia trachomatis is the leading cause of preventable blindness (Trachoma) in developing nations and sexually transmitted diseases and noncongenital infertility in the Western world (1). Chlamydia psittaci causes illnesses in psatticine birds and occasionally humans by accident (Psittacosis). More recently, Chlamydia pneumoniae has been recognized as a significant cause of upper and lower respiratory infections. An intriguing association between seropositivity to C. pneumoniae and chronic human diseases, particularly atherosclerosis, has been observed, although causal effect has yet to be definitively demonstrated (2).
The rapid progress in understanding microbial pathogenesis afforded by the exploitation of bacteria genetics has, until recently, left chlamydiae behind due to the inability to stably introduce DNA into this obligate intracellular pathogen and the inability to grow this bacteria ex vivo. However, through recent advances in host cell biology, some of the intimate secrets and tricks of this enigmatic but fascinating pathogen are being revealed. In this issue of PNAS, Hackstadt and coworkers (3) now add another chapter into the fascinating story of C. trachomatis.
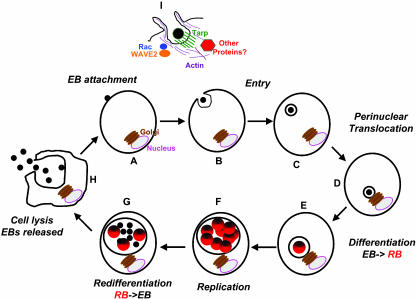
Chlamydia species undergo a biphasic developmental cycle (ref. 4 and Fig. 1). The infectious hardy, spore-like, and metabolically inert extracellular form [elementary body (EB)] induces its own uptake by a diverse range of both nonphagocytic and phagocytic cultured cells, including insect cells, epithelial cells, endothelial cells, and macrophage and monocyte-derived cell lines (Fig. 1. A–D). Once internalized in a membrane-bound compartment (the chlamydial inclusion), the EB transforms into a larger and more conventional bacterial form [the reticulate body (RB); Fig. 1E] that translocates through a dynein-dependent mechanism to the peri-Golgi region (5) and replicates by binary fission (Fig. 1F). Through unknown signals, the RBs reconvert to EBs after 24–72 h (Fig. 1G) and are released from the host cell, poised to infect adjacent cells or be spread to new hosts (Fig. 1H).
Fig. 1.
The Chlamydia life cycle. See text for details.
Getting in and Surviving
Like other intracellular pathogens, chlamydiae must survive within the hostile intracellular environment (6). It appears to do so by remaining sequestered within the chlamydial inclusion, separated from canonical intracellular compartments, such as early, late, or recycling endosomes. In a process that requires ongoing and active bacterial protein synthesis, the inclusion avoids phagolysosomal fusion and does not acquire lysosomal markers or contents (7). Rather, the inclusion contains some molecules characteristic of the host Golgi apparatus, including sphingolipids and cholesterol intercepted en route from the Golgi to the host cell plasma membrane (8, 9).
Although the mechanism by which chlamydiae accomplishes this redirection of host cell components is still under intense investigation, some clues come from the discovery of a unique and large family of chlamydial inclusion-associated (Inc) proteins. These proteins are translocated to the cytoplasmic face of the bacteria-containing inclusion by the chlamydial type III secretion system (10). A few host proteins have also been identified on the inclusion, including 14-3-3 family members and a subset of Rabs (including Rab 1, 4, and 11; ref. 11). New approaches to studying Chlamydia–host cell interactions, including examining the effects of expressing Incs in yeast on host cell trafficking and the use of RNA interference to identify host proteins required for chlamydial intracellular survival may yield new answers and surprises.
The chlamydial entry process has been quite mysterious. The observation that it can enter almost all cells examined suggests a ubiquitous receptor or a chlamydiae-encoded receptor, pathogen-directed entry, and/or multiple modes of entry. Initial attachment is mediated by electrostatic interactions with heparan sulfate moieties on the host cell (10). Characterization of chemically mutagenized host cells for mutants defective in chlamydiae entry suggests a two-step process; reversible binding, followed by irreversible attachment, but the subsequent identification of the site of mutation in the host cells has been difficult (12, 13).
In elegant applications of imaging and cell biology, Hackstadt and coworkers (14, 15) recently elucidated some early events downstream of entry (summarized in Fig. 1I). They have shown that EB entry is facilitated by an active actin remodeling process that is induced by the attachment of the pathogen, resulting in distinct microvillar reorganization throughout the cell surface and the formation of a pedestal-like structure at the site of attachment and entry. These protrusions are reminiscent of pedestal formation by enteropathogenic (EPEC) and enterohemorrhagic Escherichia coli, pathogens that regulate their entry in a type III secretion-dependent manner (16). Actin and the Rho GTPase family member Rac, but not Rho or Cdc42, colocalize with the EBs at the site of entry. Rac is specifically activated, and overexpression of dominant-negative Rac prevents EB internalization and recruitment of actin to the site of entry). By using live-cell confocal microscopy, components of the Arp2/3 complex, WAVE2, and various actin cytoskeleton-associated proteins also colocalize with entering EBs in a rapid and ordered fashion.† Whether Rac is necessary for all entry events is unclear, because the effect of expressing DN Rac when assaying both early and later events (such as inclusion formation or production of progeny) has not been reported.
Hackstadt and coworkers (3) now report that on entry, C. trachomatis translocates an actin-recruiting protein (Tarp) into the host cell. Previous investigators had observed the increased tyrosine phosphorylation of a number of proteins in response to chlamydial infection, including a complex of ≈70 kDa (17, 18). These proteins were presumed to be of host origin, because their tyrosine phosphorylated forms were not detectable in EBs or RBs, and tyrosine phosphorylation was independent of bacterial RNA or protein synthesis. Hackstadt and coworkers revisited these experiments and chose to concentrate on a high-molecular-weight protein (≈150 kDa), whose tyrosine phosphorylation could be detected within 5 min of infection and increased with higher levels of infection. Despite its low abundance, sufficient quantities of the tyrosine-phosphorylated protein were obtained by immunoprecipitation of infected host cell lysates with the antiphosphotyrosine antibody 4G10, likely made possible by its abundant phosphorylation. MS revealed the surprising finding that this protein corresponded to a chlamydial ORF, CT456, which encodes a 103-kDa protein (Tarp). The protein is present in EBs in an unphosphorylated state. By careful confocal immunofluoresence and electron microscopy, Hackstadt and coworkers find that a tyrosine-phosphorylated protein (presumably Tarp) is localized to the cytoplasmic face of the host plasma membrane at the site of attachment of Chlamydia and remains associated with the inclusion for hours. Its preexistence in EBs, its polar translocation across both bacterial and host cell membranes, and the lack of a canonical secretion signal suggest that Tarp is a type III-secreted effector. Indeed, by using Yersinia as a heterologous but genetically tractably host, Hackstadt and coworkers show that CT456 can be secreted from Yersinia in a type III secretion-dependent manner. After phosphorylation of CT456, actin is recruited to the site of EB attachment. Remarkably, transfected CT456 is tyrosine-phosphorylated in the host cell (suggesting that an additional chlamydial protein is not necessary for tyrosine phosphorylation) and is sufficient to induce actin aggregation, although simultaneous colocalization of this tyrosine phosphorylated protein and actin was not demonstrated.
New Answers, More Questions
These intriguing observations suggest several hypotheses and raise many new questions. Reminiscent of EPEC, chlamydiae “induces” its own uptake by translocation of an actin-recruiting bacterial protein. In the case of EPEC, the bacteria supplies both its own receptor (TIR) and its own ligand (intimin), although other molecules may also be involved (16). For chlamydiae, there may be a separate host cell receptor. It is possible that the observed involvement of heparan sulfate is related to its requirement for attachment of the type III secretion apparatus, as has been observed for Yersinia (19). Is the second “irreversible” step in chlamydiae entry the translocation of Tarp? Is Tarp translocation required for Rac recruitment and is tyrosine phosphorylation required for recruitment of actin and other associated proteins? Relevant to this query is the finding that Tarp is present in all of the sequenced chlamydial genomes, but the repeats containing the tyrosine residues, found in the central domain, are present in variable numbers. Indeed, C. pneumoniae lacks the repeats altogether, even though filopoidal extensions at the site of EB attachment and entry have been observed. Preliminary results from the Hackstadt laboratory suggest that tyrosine phosphorylation can be shown on a different domain from that required for the recruitment or bundling of actin. The latter C-terminal domain is present on all chlamydial strains and servovars thus far examined, and regions of conservation can be identified.† The Tarp phosphotyrosine residues may allow recruitment of additional cytoskeletal proteins through binding to Src homology 2 domains. The identity of the kinase(s) and the exact sites of phosphorylation will be of great interest. Again, similarity to EPEC and enterohemorrhagic is noted: the Tir protein from EPEC is tyrosine-phosphorylated and requires Nck, whereas the closely related Tir protein from enterohemorrhagic lacks the critical tyrosine phosphorylation residues and does not recruit Nck, despite inducing otherwise indistinguishable pedestals (16). Finally, what other bacterial proteins are translocated into the host by chlamydiae and how do they function in pathogenesis? How is Rac activated? We eagerly await the answers to some of these questions, as the tools of modern cell biology allow us to unravel the mysteries of this clever human pathogen.
See companion article on page 10166.
Footnotes
Carabeo, R. A., Grieshaber, S. S., Dooley, C. A. & Hackstadt, T., 104th General Meeting of the American Society for Microbiology, May 23–27, 2004, New Orleans, abstr. D-222.
References
- 1.Stamm, W. E. & Holmes, K. K. (1990) in Sexually Transmitted Diseases, eds. Holmes, K., Mardh, P.-A., Sparling, P. F., Weisner, P. J., Cates, W., Lemon, S. M. & Stamm, W. E. (McGraw–Hill, New York), pp. 181-194.2264006
- 2.Campbell, L. A. & Kuo, C. C. (2003) Semin. Respir. Infect. 18, 48-54. [DOI] [PubMed] [Google Scholar]
- 3.Clifton, D. R., Fields, K. A., Grieshaber, S. S., Dooley, C. A., Fischer, E. R., Mead, D. J., Carabeo, R. A. & Hackstadt, T. (2004) Proc. Natl. Acad. Sci. USA 101, 10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moulder, J. W. (1991) Microbiol. Rev. 55, 143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grieshaber, S. S., Grieshaber, N. A. & Hackstadt, T. (2003) J. Cell Sci. 116, 3793-3802. [DOI] [PubMed] [Google Scholar]
- 6.Fields, K. A. & Hackstadt, T. (2002) Annu. Rev. Cell Dev. Biol. 18, 221-245. [DOI] [PubMed] [Google Scholar]
- 7.Scidmore, M. A., Rockey, D. D., Fischer, E. R., Heinzen, R. A. & Hackstadt, T. (1996) Infect. Immun. 64, 5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackstadt, T., Rockey, D. D., Heinzen, R. A. & Scidmore, M. A. (1996) EMBO J. 15, 964-977. [PMC free article] [PubMed] [Google Scholar]
- 9.Carabeo, R. A., Mead, D. J. & Hackstadt, T. (2003) Proc. Natl. Acad. Sci. USA 100, 6771-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackstadt, T. (1999) in Chlamydia, ed. Stephens, R. (Am. Soc. Microbiol., Washington, DC), pp. 101-138.
- 11.Rzomp, K. A., Scholtes, L. D., Briggs, B. J., Whittaker, G. R. & Scidmore, M. A. (2003) Infect. Immun. 71, 5855-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fudyk, T., Olinger, L. & Stephens, R. S. (2002) Infect. Immun. 70, 6444-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carabeo, R. A. & Hackstadt, T. (2001) Infect. Immun. 69, 5899-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carabeo, R. A., Grieshaber, S. S., Fischer, E. & Hackstadt, T. (2002) Infect. Immun. 70, 3793-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carabeo, R. A., Grieshaber, S. S., Hasenkrug, A., Dooley, C. & Hackstadt, T. (2004) Traffic 5, 418-425. [DOI] [PubMed] [Google Scholar]
- 16.Vallance, B. A., Chan, C., Robertson, M. L. & Finlay, B. B. (2002) Can. J. Gastroenterol. 16, 771-778. [DOI] [PubMed] [Google Scholar]
- 17.Fawaz, F., van Ooij, C., Mutka, S. & Engel, J. (1997) Infect. Immun. 65, 5301-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birkelund, S., Bini, L., Pallini, V., Sanchez-Campillo, M., Liberatori, S., Ostergaard, S., Holm, A. & Christiansen, G. (1997) Electrophoresis 18, 563-567. [DOI] [PubMed] [Google Scholar]
- 19.Boyd, A. P., Sory, M. P., Iriarte, M. & Cornelis, G. R. (1998) Mol. Microbiol. 27, 425-436. [DOI] [PubMed] [Google Scholar]