Plants are of pivotal importance to sustain life on Earth because they supply oxygen, food, energy, and many valuable metabolites. All plant constituents, including secondary metabolites, some of which are used as flavors, fragrances, colorants, or pharmaceuticals, are ultimately derived from primary products of photosynthesis through multiple enzymatic steps encoded by the genome of each plant. However, our knowledge of how both primary and secondary metabolites are synthesized and which genes are involved is far from complete. A better understanding of metabolite synthesis and the regulation thereof will be increasingly important for improving the sustainability and efficiency of useful plant production. Recently, the availability of entire genome sequences of Arabidopsis thaliana and rice and the development of functional genomics tools have allowed the elucidation of metabolite syntheses by a systems biology approach (1–3). The mining and exploitation of the data obtained from genomics and the related research areas of genomewide transcriptomics, proteomics, and metabolomics will bring us into a new era of understanding of biological systems (Fig. 1).
Fig. 1.
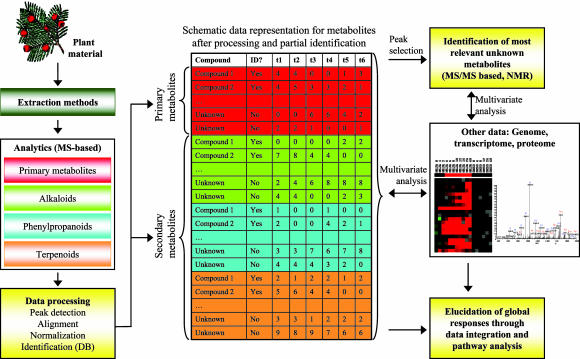
A schematic representation of a plant metabolomics platform using a systems biology approach. The platform is divided into multiple analytical approaches aimed at increased coverage of the metabolome. Data processing methods enable us to track both known and unknown metabolites, which can then be integrated from across multiple platforms. Multivariate analyses are used to find trends in data and select most relevant unknown compounds for further identification.
Sulfur is one of the essential nutrients for all plants, required to synthesize the key amino acids cysteine and methionine, which in turn are needed to produce functional proteins and many secondary metabolites (4). Approximately 90% of the reduced S is bound through these amino acids. Sulfur is also needed in the functional groups of coenzymes, such as biotin and CoA. Many parts of the world have low contents of sulfur in the soil. Although plants have adapted to live under sulfur-deficient conditions, the knowledge of how this adaptation is accomplished has been rather limited (5). Obviously, a better understanding of the mechanisms underlying this adaptation will allow us to improve crop yields in poor soils.
In this issue of PNAS, Hirai et al. (6) show significant progress exploring cellular processes by combining genomewide transcriptomics and metabolomics under deficiency of sulfur and nitrogen in the model plant A. thaliana. This contribution is one of the very first articles successfully bringing our current knowledge closer to understanding the important link between genomic data and the function of metabolites in plants. DNA array transcriptome analysis was combined with metabolite profiling and more specific targeted quantitative analysis. Both “-omics” approaches generated a huge amount of data. The authors had to develop novel bioinformatics tools to integrate the data sets and to generate gene-to-metabolite networks. Although the article mainly deals with primary metabolism, interesting conclusions were also obtained for secondary S-containing health beneficial metabolites, such as glucosinolates and alliins. An approach similar to that of Hirai et al. (6), but focusing entirely on secondary metabolites, was introduced recently by Goossens et al. (7), who combined cDNA-amplified fragment length polymorphism (AFLP) transcript profiling with targeted metabolite analysis to map the biosynthetic genes involved in alkaloid metabolism. Because sequence information for many medicinal plants is very limited, the cDNA-AFLP transcript profiling provided a very powerful tool to identify many candidate genes involved in the production of secondary metabolites. Functional analysis of these candidate genes will generate a lot of data and might help to find not only the biosynthetic genes of a particular plant pathway but also master regulators such as transcription factors that are involved in plant secondary metabolism in general. Such information is crucial for applications to overcome, e.g., the low product yield using cultivated plant cell cultures (8).
The general problems encountered when characterizing the plant metabolome are the highly complex nature and the enormous chemical diversity of the compounds. The metabolome cannot be computed from the genome (9). Plants produce ≈200,000 metabolites (10), many of which play specific roles in allowing adaptation to specific ecological niches. It has been estimated that 25–30% of the genes of Arabidopsis encode enzymes of metabolism (1). The range of chemical properties sets a challenge to the analytical tools both for profiling multiple metabolites in parallel and for quantitatively analyzing the selected ones. This has especially become obvious in secondary metabolite analysis, which is far more complex than metabolite profiling of primary metabolites. Metabolites have very different chemical natures, which influence their extractability in various solvents, pH requirements, and sensitivity for extraction conditions (e.g., temperature, pressure, time). As a consequence, if applying one general extraction system, it is very likely that many metabolites remain in the plant matrix and cannot be profiled. On the other hand, if the accuracy of the extraction systems is increased, fewer metabolites are analyzed.
One of the key challenges of metabolite profiling therefore is finding an optimal balance between the accuracy and coverage of metabolite measurements. This can be achieved by first splitting the metabolomics platform into subgroups of compounds sharing common chemical properties from the perspective of extraction conditions and chromatography (Fig. 1). A similar strategy has already been proposed in the domain of drug discovery (11). Advances in instrumentation for metabolite analyses are empowering us with the ability to increase the coverage of metabolites within a single analysis. For example, Hirai et al. (6) used the Fourier transform–ion cyclotron MS, with mass resolution >100,000 and accuracy <1 ppm, which enables analysis and separation of complex metabolite mixtures based on isotopic mass alone. More commonly, GC/MS- and liquid chromatography MS-based approaches have been applied in plant metabolomics applications (10).
Following analytical measurements, the role of data processing algorithms is to detect the peaks in spectral data, match the corresponding peaks across multiple samples, and correct the peak intensities caused by instrumental variability (normalization). These methods enable us to track differentially the metabolite levels across multiple environmental conditions or time points, even if some of the compound identities are not known. Although progress has been made in our ability to differentially track large numbers of peaks (11, 12), advances are still needed to integrate spectral analysis with prior (compound) information and improve quantitative estimates by combining traditional analytical approaches with new statistical algorithms. The latter is particularly important if integrating metabolomics data across multiple platforms. In addition, multivariate analyses of metabolite data rely on some kind of distance (or correlation) measure between the compound profiles or with other data types. Poor normalization may therefore bias the correlation structure of data and lead to erroneous conclusions (13). Once processed, metabolite profile data can be represented as a matrix (Fig. 1) and can be combined with other types of data such as transcriptional profiles and explored for major trends and associations by reducing the dimensionality of data through linear or nonlinear mappings. Hirai et al. (6) used successfully the principal component analysis and batch-learning self-organizing maps for that purpose.
When studying plant secondary metabolites and their role in physiological responses to various environmental stress conditions, we are also interested in finding and identifying compounds that are either unknown or not previously analyzed, so there is insufficient data available from the profiling experiment alone for accurate identification. The data processing methods outlined above may play an essential role in elucidating the biological role of such compounds, and multivariate approaches combining the profiles of unknown compounds with known metabolite, transcriptional, proteomic, and phenotype information may help us to direct the process of identifying most relevant compounds, based on their correlations with known compounds and specific biological processes (Fig. 1). This is particularly important because the process of identification can be very difficult, and it is unlikely that all peaks found in spectral data can be identified with sufficient confidence. The study of plant secondary metabolites is a demanding task, and no single lab can pursue this alone. Therefore, one of the challenges ahead is the development of standards for data exchange, which can also help the construction of databases containing information relevant for compound identification from spectral data.
Advances in metabolomics and its integration into systems biology research are being made possible by combining expertise from biology, chemistry, instrumentation, computer science, physics, and mathematics. Given that the era of such true interdisciplinary cooperation is only starting, many exciting discoveries are to be expected in the coming years.
See companion article on page 10205.
References
- 1.The Arabidopsis Genome Initiative (2000) Nature 408, 796-826. [DOI] [PubMed] [Google Scholar]
- 2.Yu, J., Hu, S., Wang, J., Wong, G. K.-S., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. (2002) Science 296, 79-92. [DOI] [PubMed] [Google Scholar]
- 3.Goff, S. A., Ricke, D., Lan, T.-H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002) Science 296, 92-100. [DOI] [PubMed] [Google Scholar]
- 4.Komarnisky, L. A., Christopherson, R. J. & Basu, T. K. (2003) Nutrition 19, 54-61. [DOI] [PubMed] [Google Scholar]
- 5.Saito, K. (2000) Curr. Opin. Plant Biol. 3, 188-195. [PubMed] [Google Scholar]
- 6.Hirai, M. Y., Yano, M., Goodenowe, D. B., Kanaya, S., Kimura, T., Awazuhara, M., Arita, M., Fujiwara, T. & Saito, K. (2004) Proc. Natl. Acad. Sci. USA 101, 10205-10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., Lammertyn, F., Nuutila, A. M., Söderlund, H., Zabeau, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8595-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oksman-Caldentey, K.-M. & Inzé, D. (2004) Trends Plant Sci., in press. [DOI] [PubMed]
- 9.Trethewey, R. N. (2004) Curr. Opin. Plant Biol. 7, 196-201. [DOI] [PubMed] [Google Scholar]
- 10.Fiehn, O. (2002) Plant Mol. Biol. 48, 155-171. [PubMed] [Google Scholar]
- 11.van der Greef, J., Davidov, E., Verheij, E., Vogels, J. T. W. E., van der Heijden, R., Adourian, A. S., Marple, E. W., Oresic, M. & Naylor, S. (2003) in Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis, eds. Harrigan, G. G. & Goodacre, R. (Kluwer, Boston), pp. 171-198.
- 12.Wang, W., Zhou, H., Lin, H., Roy, S., Shaler, T. A., Hill, L. R., Norton, S., Kumar, P., Anderle, M. & Becker, C. H. (2003) Anal. Chem. 75, 4818-4826. [DOI] [PubMed] [Google Scholar]
- 13.Atchinson, J. (2003) The Statistical Analysis of Compositional Data (Blackburn, Caldwell, NJ).