Abstract
5-Fluorouracil (5-FU) is the standard chemotherapy for certain high risk stage 2 and all stage 3 and 4 human colorectal cancer patients. However, patients often develop chemoresistance to 5-FU. We have identified verticillin A from Verticillium-infected wild mushrooms as a potent anti-cancer agent that effectively suppresses 5-FU-resistant human colon cancer cells. Interestingly, a sublethal dose of verticillin A also acts as a potent sensitizer that overcomes human colon carcinoma cell resistance to FasL- and TRAIL-induced apoptosis. To identify verticillin A-regulated genes, we performed a genome-wide gene expression analysis and identified 1287 genes whose expression levels were either up- or down-regulated 1.5 fold. Forty-six of these genes have known function in regulation of apoptosis, and ninety genes have function in cell cycle regulation. Our recent study has identified verticillin A as a selective histone methyltransferase inhibitor. These identified genes are thus potential molecular targets for epigenetic-based therapy to overcome human colon cancer 5-FU resistance. The entire dataset is deposited in the NIH GEO database; accession number GSE51262.
Keywords: Cell cycle arrest, Histone methyltransferase, Colon cancer, Apoptosis, H3K9 methylation
| Specifications | |
|---|---|
| Organism/cell line/tissue | Cell line: Human colon carcinoma cell line LS411N-5FU-R. LS411N-5FU-R cell line was generated by culturing LS411N cells in the presence of 5-FU. LS411N cell line was established from Dukes' type B colon carcinoma tissue of a 37 year old patient. LS411N-5FU-R cells grow in the presence of 5-FU as high as 2.0 mg/ml in the culture medium. |
| Sex | Male colon cancer patient. |
| Sequencer or array type | Affymetrix Human Gene 2.0 ST Array |
| Data format | Raw and analyzed |
| Experimental factors | Cells were cultured in the absence (control) or presence of 50 nM verticillin A for 3 days. The gene expression level of treated cells was compared to the untreated cells. |
| Experimental features | |
| Consent | Exempted human colon cancer cell lines. |
| Sample source location | LS411N cell line was obtained from ATCC. |
1. Direct link to deposited data
2. Experimental design, materials and methods
The human colon carcinoma cell line LS411N was obtained from ATCC (Manassas, VA). Verticillin A was purified either from mushroom or fungus fermentation as previously described [1], [2]. To identify verticillin A-regulated genes, the 5-FU-resistant LS411N-5FU-R cells were cultured in the absence (control) or presence of verticillin A (50 nM) for three days. Total RNA was then isolated from cells using TRIzol according to the manufacturer's instructions (Life Technologies) as previously described [3], [4]. Biotinylated cDNAs were prepared according to the standard Ambion and Affymetrix protocol from 250 ng total RNA (The Ambion WT Expression Kit and GeneChip Terminal Labeling Kit, Affymetrix). Following labeling, cDNAs were hybridized for 16 h at 45 °C on Affymetrix Human Gene 2.0 ST Array. GeneChips were washed and stained in the Affymetrix Fluidics Station 450. GeneChips were scanned using the Affymetrix GeneChip Scanner 3000. Intensities of arrays have been quantile-normalized using Partek Genomic Suite (v6.6). Differential expressions were calculated using ANOVA of Partek package.
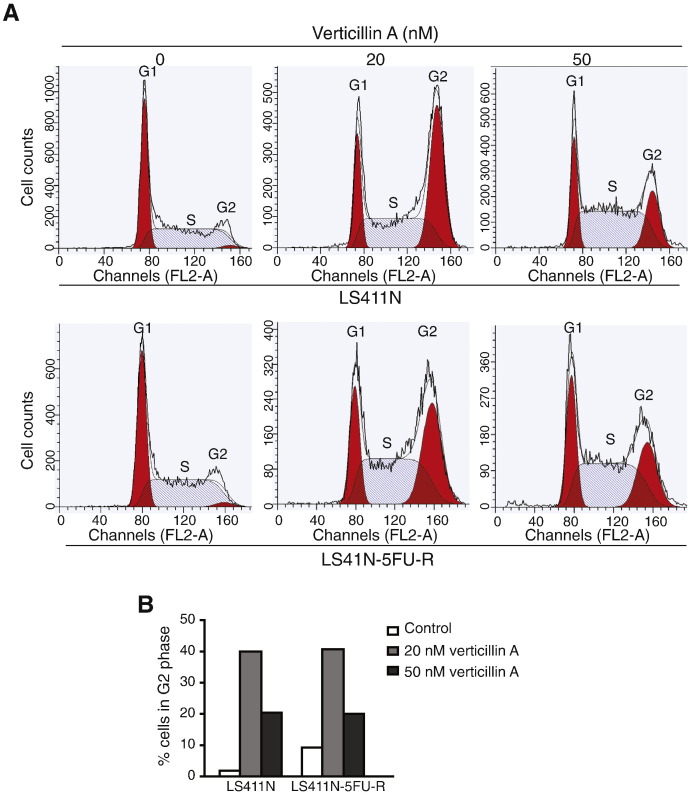
The entire data set was analyzed for differentially expressed genes. Using a 1.5 fold change and p value less than 0.01, we have identified 1287 genes whose expression levels were either up- or down-regulated 1.5 fold. Among these genes, forty-six have known function in regulation of apoptosis (Table 1), and ninety have function in cell cycle regulation (Table 2). Consistent with the altered gene expression in cell cycle regulation, functional analysis revealed that a sublethal dose of verticillin A induces cell cycle arrest at G2 phase (Fig. 1A and B). Comparison of effects of verticillin A on cell cycle arrest between the parent LS411N and the LS411N-5FU-R cells indicate that development of resistance to 5-FU does not alter human colon carcinoma cell sensitivity to verticillin A in induction of cell cycle arrest (Fig. 1B). Induction of cell cycle arrest is a mechanism by which chemotherapeutic agents suppress tumor development. Our observation that verticillin A induces cell cycle arrest in 5-FU-resistant human colon carcinoma cells as effectively as in the parent cells (Fig. 1B) suggests that verticillin A is potentially an effective agent for 5-FU-resistant cancer suppression [5], [6]. In summary, our data indicate that verticillin A-regulated genes, including these involved in apoptosis and cell cycle regulation, are potential molecular targets for epigenetic-based therapy [7], [8] to overcome human colon cancer 5-FU resistance.
Table 1.
Apoptosis regulatory genes.
| Gene symbol | RefSeq | p value | Fold change |
|---|---|---|---|
| SORD | ENST00000267814 | 0.00061649 | − 1.87556 |
| ZMAT3 | ENST00000311417 | 0.000557593 | 1.57282 |
| ING3 | ENST00000315870 | 0.000102115 | − 1.51561 |
| CLU | ENST00000316403 | 0.00658658 | 1.88366 |
| ZADH2 | ENST00000322342 | 0.00116724 | − 1.45788 |
| MALT1 | ENST00000348428 | 8.70E− 05 | − 1.59075 |
| DNAJB4 | ENST00000370763 | 0.000141036 | 1.57462 |
| GADD45A | ENST00000370986 | 0.00036571 | − 1.62913 |
| SMOX | ENST00000379460 | 0.000453598 | − 1.7213 |
| FAS | NM_000043 | 0.00562506 | 1.64701 |
| PMP22 | NM_000304 | 0.0117398 | 1.80063 |
| TP53 | NM_000546 | 3.20E− 06 | − 1.71073 |
| OR51B5 | NM_001005567 | 5.42E− 05 | 1.47933 |
| MYBL1 | NM_001080416 | 0.000313286 | 1.60353 |
| CRYZ | NM_001130042 | 1.48E− 06 | 1.48424 |
| JDP2 | NM_001135049 | 0.00306543 | − 1.60104 |
| BIRC3 | NM_001165 | 1.43E− 05 | 2.0818 |
| EMP1 | NM_001423 | 8.59E− 08 | 3.35724 |
| EMP3 | NM_001425 | 0.0010598 | 1.45705 |
| CNTN1 | NM_001843 | 0.0074873 | − 1.56199 |
| EDNRA | NM_001957 | 0.0279268 | − 1.6314 |
| FKBP4 | NM_002014 | 9.38E− 05 | 1.46074 |
| FYN | NM_002037 | 0.000281551 | − 1.71704 |
| LIF | NM_002309 | 0.0155574 | − 1.45691 |
| MAGEB2 | NM_002364 | 0.00564587 | 1.57381 |
| MDM2 | NM_002392 | 4.74E− 05 | 1.96004 |
| MYBL2 | NM_002466 | 0.000287216 | 1.46171 |
| ROBO1 | NM_002941 | 0.000219031 | − 1.87573 |
| TXN | NM_003329 | 2.62E− 06 | 1.47208 |
| CBX4 | NM_003655 | 0.00057116 | − 1.65897 |
| BAG3 | NM_004281 | 0.000615002 | 1.68888 |
| CEBPB | NM_005194 | 3.37E− 05 | − 1.55513 |
| HDAC5 | NM_005474 | 0.00951344 | − 1.57188 |
| DNAJB1 | NM_006145 | 0.000604263 | 1.46166 |
| FKBP9 | NM_007270 | 0.000399435 | − 1.49977 |
| SHC2 | NM_012435 | 0.015437 | − 1.62921 |
| CAPN6 | NM_014289 | 0.000292268 | − 1.62879 |
| KRCC1 | NM_016618 | 0.000625115 | − 1.51118 |
| SHC3 | NM_016848 | 9.40E− 05 | − 1.79462 |
| FKBP10 | NM_021939 | 0.00031242 | − 1.59534 |
| TMEM47 | NM_031442 | 1.92E− 06 | − 2.58264 |
| IL20RB | NM_144717 | 3.47E− 06 | − 2.65592 |
| UNC5B | NM_170744 | 0.00192834 | − 1.52166 |
| IFNE | NM_176891 | 0.00165318 | − 2.26639 |
| RASSF3 | NM_178169 | 0.000187772 | − 1.47727 |
| TYMS | NM_001071 | 0.000178443 | 1.46165 |
Table 2.
Cell cycle regulatory genes.
| Gene symbol | RefSeq | p value | Fold change |
|---|---|---|---|
| VRK1 | ENST00000216639 | 0.00109304 | 1.48503 |
| GINS2 | ENST00000253462 | 0.000631431 | 1.55066 |
| POLA2 | ENST00000265465 | 0.000337543 | 1.50477 |
| SKP2 | ENST00000274255 | 0.000407268 | 1.4572 |
| PLK2 | ENST00000274289 | 0.000151913 | 1.72602 |
| DUSP6 | ENST00000279488 | 7.20E− 06 | − 2.10422 |
| ACTR1B | ENST00000289228 | 0.00108964 | − 1.51729 |
| FGF19 | ENST00000294312 | 5.05E− 05 | − 2.13694 |
| CDC25A | ENST00000302506 | 0.000648382 | 1.56293 |
| ESCO2 | ENST00000305188 | 0.00119695 | 1.53802 |
| PSMD2 | ENST00000310118 | 0.000687665 | 1.49718 |
| DSCC1 | ENST00000313655 | 0.00380772 | 1.49838 |
| ING3 | ENST00000315870 | 0.000102115 | − 1.51561 |
| TUBB6 | ENST00000317702 | 0.000718394 | 1.62504 |
| NR2F1 | ENST00000327111 | 0.000840833 | − 1.73635 |
| KLHDC1 | ENST00000359332 | 0.00198403 | − 1.85646 |
| S100A10 | ENST00000368811 | 0.000119039 | 1.45917 |
| GADD45A | ENST00000370986 | 0.00036571 | − 1.62913 |
| JUN | ENST00000371222 | 4.77E− 05 | − 1.92902 |
| PLK3 | ENST00000372201 | 0.000951583 | 1.49099 |
| BICC1 | ENST00000373886 | 0.000196311 | − 1.5164 |
| TTK | ENST00000509894 | 0.000571488 | 1.49117 |
| PMP22 | NM_000304 | 0.0117398 | 1.80063 |
| TP53 | NM_000546 | 3.20E− 06 | − 1.71073 |
| HNRNPA1L2 | NM_001011724 | 0.0265379 | − 1.55219 |
| VEGFA | NM_001025366 | 7.18E− 06 | − 2.08509 |
| INCENP | NM_001040694 | 5.34E− 06 | 1.70379 |
| MYBL1 | NM_001080416 | 0.000313286 | 1.60353 |
| PABPC1L | NM_001124756 | 0.000496661 | − 1.98338 |
| JDP2 | NM_001135049 | 0.00306543 | − 1.60104 |
| FBXL17 | NM_001163315 | 0.00683003 | − 1.45608 |
| ELAVL2 | NM_001171197 | 0.0107444 | − 1.83695 |
| A1CF | NM_001198819 | 0.00310545 | − 1.53498 |
| CDKN1A | NM_001220778 | 0.0376178 | 1.47916 |
| CCNE1 | NM_001238 | 1.80E− 05 | 1.89998 |
| DTNA | NM_001390 | 0.000846729 | − 1.60955 |
| EMP1 | NM_001423 | 8.59E− 08 | 3.35724 |
| EMP3 | NM_001425 | 0.0010598 | 1.45705 |
| CNTN1 | NM_001843 | 0.0074873 | − 1.56199 |
| EGR1 | NM_001964 | 4.11E− 05 | − 2.05333 |
| FKBP4 | NM_002014 | 9.38E− 05 | 1.46074 |
| MYBL2 | NM_002466 | 0.000287216 | 1.46171 |
| PCNA | NM_002592 | 7.01E− 06 | 1.80038 |
| MAPK4 | NM_002747 | 0.00147303 | − 1.53275 |
| ROBO1 | NM_002941 | 0.000219031 | − 1.87573 |
| TXN | NM_003329 | 2.62E− 06 | 1.47208 |
| CDKL2 | NM_003948 | 0.0503581 | 1.57628 |
| ORC1 | NM_004153 | 1.65E− 05 | 1.93504 |
| BUB1 | NM_004336 | 4.67E− 07 | 1.70252 |
| ETV5 | NM_004454 | 4.20E− 05 | − 1.65774 |
| RPS6KA5 | NM_004755 | 9.56E− 06 | − 2.43124 |
| ETV1 | NM_004956 | 2.61E− 05 | − 1.74844 |
| E2F1 | NM_005225 | 0.000685259 | 1.62065 |
| JUND | NM_005354 | 0.000591427 | − 1.61353 |
| SKIL | NM_005414 | 6.76E− 05 | − 1.71684 |
| HDAC5 | NM_005474 | 0.00951344 | − 1.57188 |
| UST | NM_005715 | 0.00693733 | − 1.47903 |
| KIF20A | NM_005733 | 0.000183352 | 1.66921 |
| RASGRP1 | NM_005739 | 0.00120733 | − 1.49275 |
| SFN | NM_006142 | 2.54E− 05 | 2.33324 |
| POLH | NM_006502 | 2.21E− 05 | 2.1575 |
| BTG2 | NM_006763 | 4.54E− 05 | 1.54606 |
| ZFP36L2 | NM_006887 | 0.00908993 | − 1.48935 |
| RAPGEF4 | NM_007023 | 0.010428 | − 1.48297 |
| POLI | NM_007195 | 0.00109012 | − 1.57536 |
| FKBP9 | NM_007270 | 0.000399435 | − 1.49977 |
| HS2ST1 | NM_012262 | 2.62E− 05 | − 1.49078 |
| ESPL1 | NM_012291 | 0.00490513 | 1.47314 |
| SESN1 | NM_014454 | 0.0014303 | 1.5657 |
| RPS6KA6 | NM_014496 | 0.00411955 | − 1.6027 |
| HUNK | NM_014586 | 0.00831223 | − 1.45803 |
| MYO1D | NM_015194 | 0.00043732 | − 1.62238 |
| TUBE1 | NM_016262 | 0.000107931 | − 2.14386 |
| FKBP10 | NM_021939 | 0.00031242 | − 1.59534 |
| MOB3B | NM_024761 | 0.0171544 | − 1.48453 |
| DUSP16 | NM_030640 | 0.00020683 | − 1.46829 |
| SESN2 | NM_031459 | 2.47E− 05 | − 2.27913 |
| CCNB1 | NM_031966 | 0.000656847 | 1.54547 |
| GINS4 | NM_032336 | 0.000779725 | 1.67209 |
| BRSK1 | NM_032430 | 0.00851809 | − 1.63611 |
| CORO6 | NM_032854 | 0.000911746 | 1.49868 |
| CDKN2B | NM_078487 | 6.95E− 05 | − 2.14147 |
| MSI2 | NM_138962 | 0.00017894 | − 1.47847 |
| SLFN5 | NM_144975 | 8.48E− 06 | − 2.23313 |
| DBF4B | NM_145663 | 0.000273346 | 1.55948 |
| SIK1 | NM_173354 | 0.00486314 | 1.49766 |
| PTPDC1 | NM_177995 | 2.21E− 05 | − 2.35451 |
| WDR49 | NM_178824 | 0.0202432 | 1.5734 |
| AURKA | NM_198433 | 0.00160995 | 1.62216 |
| POLQ | NM_199420 | 3.09E− 05 | 1.62956 |
Fig. 1.
Verticillin A mediates cell-cycle progression in 5-FU-resistant human colon carcinoma cells. A. LS411N-5FU-R cells were cultured in the presence of Verticillin A at the indicated concentrations for 24 h. Cells were then stained with PI, and analyzed by flow cytometry. A. Cells as shown in A were quantified for percentages of cells in various phases the cell cycle (as shown). Columns, mean; bars, SD.
Acknowledgments
We thank Drs. Chang-Sheng Chang and Jeong-Hyeon Choi for their excellent technical assistance in the DNA microarray data analysis, and grant support from National Institutes of Health (CA182518 and CA185909 and VA Merit Review Award BX001962.
References
- 1.Liu F., Liu Q., Yang D., Bollag W.B., Robertson K., Wu P. Verticillin a overcomes apoptosis resistance in human colon carcinoma through DNA methylation-dependent upregulation of BNIP3. Cancer Res. 2011;71:6807–6816. doi: 10.1158/0008-5472.CAN-11-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueroa M., Graf T.N., Ayers S., Adcock A.F., Kroll D.J., Yang J. Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J. Antibiot. 2012;65:559–564. doi: 10.1038/ja.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paschall A.V., Zhang R., Qi C.F., Bardhan K., Peng L., Lu G. IFN regulatory factor 8 represses GM-CSF expression in T cells to affect myeloid cell lineage differentiation. J. Immunol. 2015;194:2369–2379. doi: 10.4049/jimmunol.1402412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paschall A.V., Zimmerman M.A., Torres C.M., Yang D., Chen M.R., Li X. Ceramide targets xIAP and cIAP1 to sensitize metastatic colon and breast cancer cells to apoptosis induction to suppress tumor progression. BMC Cancer. 2014;14:24. doi: 10.1186/1471-2407-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klajic J., Busato F., Edvardsen H., Touleimat N., Fleischer T., Bukholm I. DNA methylation status of key cell-cycle regulators such as CDKNA2/p16 and CCNA1 correlates with treatment response to doxorubicin and 5-fluorouracil in locally advanced breast tumors. Clin. Cancer Res. 2014;20:6357–6366. doi: 10.1158/1078-0432.CCR-14-0297. [DOI] [PubMed] [Google Scholar]
- 6.Montagnoli A., Moll J., Colotta F. Targeting cell division cycle 7 kinase: a new approach for cancer therapy. Clin. Cancer Res. 2010;16:4503–4508. doi: 10.1158/1078-0432.CCR-10-0185. [DOI] [PubMed] [Google Scholar]
- 7.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crea F., Nobili S., Paolicchi E., Perrone G., Napoli C., Landini I. Epigenetics and chemoresistance in colorectal cancer: an opportunity for treatment tailoring and novel therapeutic strategies. Drug Resist. Updat. 2011;14:280–296. doi: 10.1016/j.drup.2011.08.001. [DOI] [PubMed] [Google Scholar]