Abstract
Objective
Gender differences in immune response and the rate of disease progression in HIV-infected individuals have been reported but the underlying mechanism remains unclear, in part due to the lack of relevant animal models. Here, we report a novel nonhuman primate (NHP) model for investigation of sex disparity in HIV disease progression.
Design/Methods
Viral load and rate of disease progression were evaluated in rhesus macaques infected intrarectally with lineage-related subtype C R5 SHIVs. Cytokine/chemokine levels in rectal swab eluates, and bacterial species adherent to the swabs and in the feces were determined.
Results
SHIV infected female rhesus macaques progressed faster to AIDS than males, recapitulating the sex bias in HIV-1 disease in humans. There were no significant differences in the levels of soluble immune mediators in the rectal mucosa of naïve female and male macaques. However, an exploratory longitudinal study in six infected macaques indicates that the females mounted an earlier and more robust proinflammatory skewed rectal immune response to infection. Moreover, expansion of Proteobacteria that increase in other intestinal inflammatory disorders was significantly higher in the rectal mucosa of female than male macaques during acute infection.
Conclusion
These findings suggest that sex differences in local innate immune activation and compositional shifts in the gut microbiota could be the drivers of increased disease susceptibility in females. Further studies with this novel NHP model of HIV infection could lead to innovative research on gender differences, which may have important therapeutic implications for controlling disease in infected men as well as women.
Keywords: SHIV pathogenesis, sex disparity, regional innate immunity, gut microbiome response
Introduction
The gender of a host can significantly affect susceptibility to infections [1, 2]. Sex disparities have been reported for the prevalence of autoimmune [3] and infectious diseases [4-6]. Gender bias has also been observed in HIV-1 infection. Women with the same HIV-1 viral burden as men have a 1.6-fold higher risk of developing AIDS [7, 8]. Higher immune activation occurs in chronically-infected women than men [9, 10]. This finding may explain the increased risk for disease progression in women, but the mechanism is not well understood.
The gut-associated lymphoid tissue is a major site of acute CD4+ T cell depletion and early viral replication in HIV/SIV infection [11]. Disruption of the gut immune homeostasis is a hallmark of HIV/SIV infection and is central to disease pathogenesis [12]. The gastrointestinal tract also harbors the bulk of the human body’s microorganisms [13], with growing recognition that specific composition of the gut microbiota has a profound effect on host health and metabolic functions [14]. The gut microbiome also plays a pivotal role in maintaining mucosal and systemic immune cell homeostasis, and is important for the induction of host innate responses and systemic immunity [15, 16]. Notably, recent evidence indicates the innate immune response during acute infection impacts HIV/SIV disease progression [17]. However, without an animal model, it is difficult to dissect the association between innate immune activation and microbial changes during acute infection of HIV/SIV with gender differences in disease progression.
We now report the development and characterization of a nonhuman primate (NHP) model that replicates the gender differences in AIDS progression in humans. The sex bias disease susceptibility seen in rhesus macaques (RMs) infected intrarectally (IR) with R5 SHIVs does not appear to be due to a difference in the local basal innate immune environment. Rather, an exploratory longitudinal study in six infected animals revealed sex differences in gut innate immune and rmicrobiota response to infection. These findings add to the growing awareness that gender is an important source of biological, immunological, and microbial variation. They further highlight the importance of including female animals in preclinical research per recent NIH policy [18].
Materials and Methods
Animal welfare statement
The study was carried out in strict accordance with the Guide for the Care and use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the Tulane National Primate Research Center (TNPRC).
SHIV infection of rhesus macaques
All IR inoculations were carried out in 4-14 years old rhesus monkeys (Macaca mulatta) of Indian descent bred and housed at the TNRPC. Animals were confirmed to be serologically and virus negative for simian type D retrovirus, and serologically negative for SIV and simian T-cell lymphotropic virus prior to infection. They were also screened for the presence of the Mamu-A*01, Mamu-B*08 and Mamu-B*17 class I allele previously shown to be associated with spontaneous control of pathogenic SIVmac239 replication by standard PCR with allele-specific primers [19]. Macaques received a single 5 × 103 50% tissue culture infectious dose (TCID50) of the lineage-related subtype C R5 SHIVC109P3 or SHIVC109P3N viruses [20]. Plasma viremia was quantified by real-time RNA PCR (Washington National Primate Research Center) and absolute CD4+ and CD8+ cell counts were monitored in TruCount tubes (BD Biosciences, Palo Alto, CA). Animals exhibiting clinical signs of AIDS (peripheral blood CD4+ T-cell depletion (<200/mm3), greater than 25% loss of body weight, or combinations of the following conditions: diarrhea unresponsive to treatment, opportunistic infections, peripheral lymph node atrophy, and abnormal hematology) were euthanized by intramuscular administration of telazol and buprenorphine followed by an overdose of sodium pentobarbital.
Specimen collection and preparation
Rectal samples were obtained by gently inserting Merocel Eye Spear (Beaver-Visitec, Waltham, MA) into the rectum for 5 minutes to absorb mucosal fluid. The specimens were immediately placed on ice and stored in a −80°C freezer. For analysis, 0.8 ml cold, sterile phosphate-buffered saline (PBS) was added to the swabs, and the recovered eluates were centrifuged. Supernatants were collected and cryopreserved for chemokine/cytokine analysis. For the study in six animals to monitor longitudinal immune and microbial responses, rectal samples were not obtained on the day of IR challenge with SHIVC109P3 to minimize irritation at the inoculation site. Supernatants of the rectal eluates collected two weeks before and at various time points post-challenge were used for immunological studies and the pellets were subjected to DNA extraction for microbiome analysis. Fresh stool under the cage pan was also collected over time for the six animals and stored frozen. For analysis, one ml cold PBS was added to approximate 2 grams of frozen stool, vortexed briefly, and cleared by centrifugation.
Multiplex cytokine/chemokine assay
Cytokines and chemokines in rectal secretions and plasma samples were analyzed using a MAGPIX ® multiplexing instrument (EMD Millipore, Billerica, MA) and MILLIPLEX map Non-Human primate cytokine multiplex kits (cat# PCYTMG-40K-PX23) from Millipore (Billerica, MA) according to the manufacturer’s instructions. Results were analyzed using Millipore Analyst software.
DNA extraction and PCR amplification
Total DNA was extracted from the particulate fraction of the swab and fecal specimens collected using the MoBio Power Soil II Kit (MoBio, Carlsbad, CA). DNA concentrations and purity were determined using a NanoDrop 2000 UV-Vis Spectrophotometer (NanoDrop products, Wilmington, DE). 16S rRNA genes were amplified using a primer set spanning the V3 and V4 regions, as previously described [21]. The forward primer 347F was modified by a 5'-addition of a 6-mer nucleotide barcode to serve as a multiplex identifier (MID) and a two-base linker sequence (AG); The reverse primer 803R was tagged with a different MID. PCR amplicons were purified using Agencourt AMPure XP (Beckman Coulter, Inc., Indianapolis, IN), and multiplexed by pooling at equal molarity and then ligated to Illumina adaptors of PE Read 1 and Read 2 sequencing primers (PE Read 1 sequencing primer 5' ACACTCTTTCCCTACACGACGCTCTTCCGATCT and PE Read 2 sequencing primer 5' CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT) for MiSeq sequencing (Illumina, San Diego, CA).
Amplicon-based data analysis
Sequence reads were trimmed based on quality score 30 and maximum number of ambiguities 2. The high quality reads were then merged paired ended into longer sequences, and the merged sequences with lengths between 350 bp and 600 bp were selected for downstream analysis. Selected reads were de-multiplexed into samples according to the barcodes using software QIIME with default parameters. In all, 5,139,359 high quality sequences were analyzed. Each sample yielded an average of 22,741 (± 9,531) sequences, ranging from 8,747 to 47,174. Operational taxonomic units (OTUs) clustering was done at a 97% similarity threshold. Taxonomic identities were assigned using the RDP classifier included in the QIMME with a confidence threshold of 0.8. Relative abundances of the taxa at the phylum-, class- and genus-levels were calculated.
Statistics
Survival for the infected male and female RMs was compared using a log-rank test (GraphPad Prism 5). Differences in survival were considered significant when p < 0.05. Comparison of cytokine/chemokine levels in the rectal secretions of uninfected female and male macaques was performed using Mann–Whitney U test, and statistical analysis of the response in the two groups to viral infection was performed with SAS 9.3 (SAS Institute, Inc, Cary, NC). Briefly, each cytokine response at each time point was normalized by dividing its value by its corresponding pre-infection value. A log-binomial regression model of this binary variable using a repeated measures approach with each repeat defined by a cytokine-time-point combination was generated, and generalized equations and SAS proc genmod were used to estimate the repeated measures model, assuming an exchangeable correlation structure [22]. Independent variables considered were gender and time since infection, as well as an interaction term. Model coefficients were exponentiated to estimate the relative risk of an elevated (>±2.0) cytokine value for females vs. males at each time point; 95% confidence intervals for which the lower bound exceeded 1 correspond to statistical significance at the 0.05 level. Significance of differences in peak viremia (log transformed) and differences in a specific taxon between two groups of samples were calculated based on relative abundance using Mann–Whitney U test.
Results
R5 SHIV intrarectally infected female rhesus macaques progressed faster to disease than males
We recently reported the generation of lineage-related, mucosally transmissible subtype C R5 SHIVC109 viruses that are capable of AIDS development, induction of neurological disease and coreceptor switching in rhesus macaques [20]. We found no significant differences in the ability of the viruses to initiate and establish an infection with IR inoculation in male and female RMs (Figure 1). All eleven female macaques and nine of ten male macaques developed systemic infection following a single high dose challenge (Figure 1A). The sexes were comparable in age. None of the males expressed the restrictive Mamu-A*01, Mamu-B*08 and Mamu-B*17 class I alleles, but one female was Mamu-A*01 positive and another was Mamu-B*17 positive. Plasma viremia peaked at 2-3 weeks post-infection (wpi) in all infected monkeys, and no significant differences in magnitude were observed between the sexes (Figure 1B). However, a rapid progressor (RP) phenotype characterized by sustained high viral loads and weak or absence of anti-viral antibody response was seen only in females. Accordingly, the tempo of clinical AIDS development was faster in the infected females than males (p = 0.0105; Figure 1C).
Figure 1. Sex disparity in R5 SHIV disease progression.
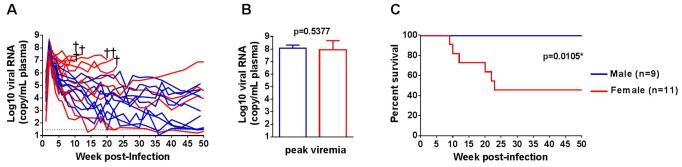
Comparison of virus replication (A), peak viremia (B), and survival (C) of female (red) and male (blue) RMs infected IR with lineage-related subtype C R5 SHIVs. Dotted line in (A) marks the limit of detection for viral load (30 RNA copies/mL plasma). †, indicates animals euthanized due to clinical AIDS.
Comparable basal innate immune milieu in the rectal mucosa of male and female rhesus macaques
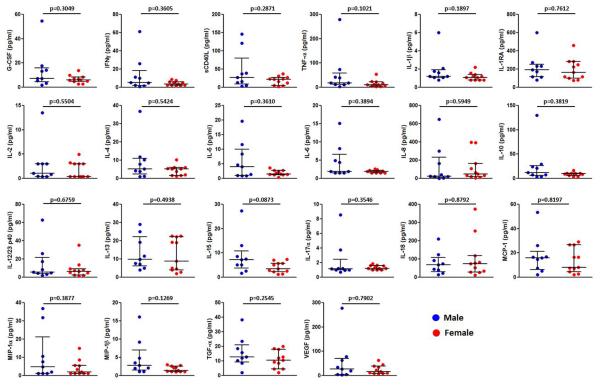
Women have been reported to have higher levels of immune activation and inflammation in the gut environment than men, which may predispose women to inflammation-associated diseases [23]. We first investigated the possibility of sex-based differences in the gut mucosal environment of RMs prior to infection by comparing the levels of chemokines, cytokines, and growth factors in rectal swab eluates of naive female (n=11) and male (n=9) monkeys. There were no significant differences between the sexes in the levels of soluble immune mediators measured (Figure 2). These findings suggest that the faster disease progression in female RMs was not due to a more inflamed mucosal immune milieu before infection.
Figure 2. Basal rectal mucosal innate immune mediator levels in male and female RMs.
Immune mediators in rectal swab elutes from naïve male (n=9) and female (n=11) macaques were analyzed by multiplex assays. Data represent median and interquartile range. Significance values were calculated with non-parametric 2-tailed Mann-Whitney test. P<0.05 was considered significant.
Acute mucosal inflammatory innate immune and microbiome response is more robust in R5 SHIV infected females than males
Mounting evidence points to a role of acute regional innate immunity in the control or development of disease [17]. Accordingly, we conducted a pilot study using six animals challenged IR with SHIVC109P3 to measure chemokine/cytokine levels over time in the rectal swab eluates. Moreover, given the known interplay between innate immunity and resident bacterial communities in the gut [14-16], we also investigated shifts in the gut microbiota of infected animals. None of the six animals expressed the class I alleles reported to be associated with spontaneous virologic control of pathogenic SIV replication. Initial viremia similar to acute HIV infection (106-107 RNA copies/ml plasma) was seen in both infected females (n=3) and male (n=3) macaques (Supplemental Figure 1A). Virus replication declined post-peak, establishing various set points. CD4+ T cell loss accompanied acute infection in the viremic animals, with levels rebounded close to pre-infection values in most of the infected animals by 10 wpi (Supplemental Figure 1B). The exception was DF73, the RP female macaque with high chronic viremia. Precipitous drop in peripheral CD4+ T cell numbers was seen in this animal at 10 wpi, with clinical AIDS development and euthanasia five weeks later. In contrast, and despite comparable viral load and absence of detectable antiviral antibody response in ER57, this male monkey remained clinically healthy during the 30 week observation period.
Longitudinal immune assessments indicated a more rapid and robust local innate immune response in the infected females than males (Figure 3). Sex differences in upregulation of innate immunity were noticeable at day 3 post-infection, and were sustained during the acute (d3 - 28) and post-acute (d35 - 175) stages of infection. As a group, the pro-inflammatory cytokine response (e.g., sCD40L, IL-12, IFN-γ and TNF-α) and inflammasome response (IL-1β) were more robust in females than males. Moreover, the marked induction of IFN-γ and IL-12 in two of the three infected females at day 3 after inoculation suggests early activation of T cells and dendritic cells at the site of virus infection. The peripheral innate immune response overall was less robust in all infected macaques, with no notable differences between the sexes, thus, highlighting the importance of local tissue sampling and analysis (Supplemental Figure 2). The use of a more sensitive detection method, such as gene expression analysis may reveal higher levels of acute immune activation in the periphery and sex-based differences. Although the cohort was small, the number of longitudinal specimens collected per animal was large, providing sufficient statistical power for comparisons of mucosal innate responses between the sexes. During the acute phase (d3-28), the three females were 2.6 times as likely [relative risk (RR); 95% CI =2.1, 3.2] as the three males to have elevated (≥2) cytokine levels. At all subsequent post-infection time points (d35 – 175), the females were more than 3 times as likely to have elevated cytokine levels (RR = 3.4; 95% CI = 2.7, 4.2).
Figure 3. Gender differences in rectal immune response to R5 SHIV IR challenge.
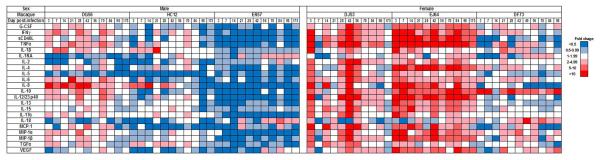
Fold change in analyte levels over time in rectal swab eluates of SHIVC109P3-IR infected male (n=3) and female (n=3) macaques. Color scale for down- (in blue) and up- (in red) regulation is shown on the right.
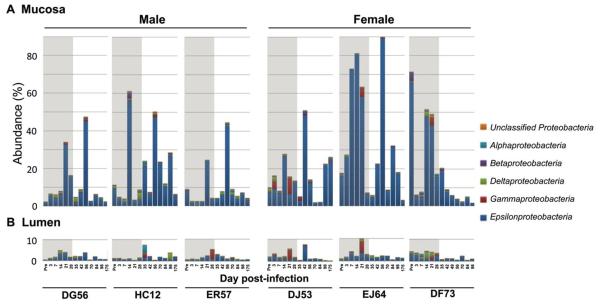
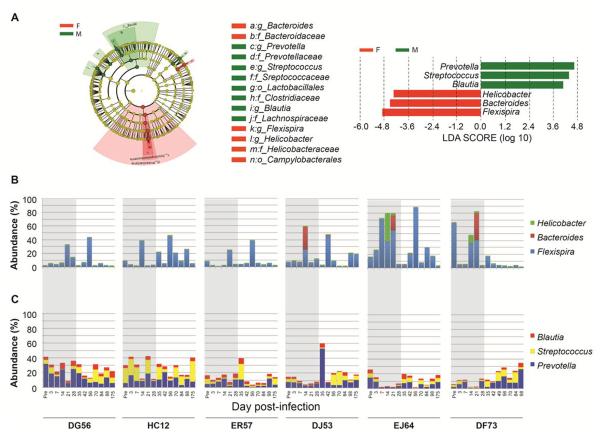
Analysis of bacterial community structures adherent to the mucosal surface in rectal swab eluates and of transitory bacteria in the form of feces in the lumen showed microbial community composition changes in all infected animals. Changes were more prominent in the rectal mucosa than in the lumen (Figure 4), which could help explain why early studies failed to show an association between pathogenic SIV infection and changes in the luminal bacteria at the family level [24, 25]. The main change in the rectal mucosa was enrichment along the Proteobacteria-Epsilonproteobacteria lineage, and the scale of this enrichment was significantly higher in females than males, especially during acute infection (Mann–Whitney U test, 31.15% vs. 12.77%, p<0.01 for Proteobacteria and 28.1% vs. 11.1%, p<0.01 for Epsilonproteobacteria, respectively). We employed linear discriminant analysis (LDA) effect size (LEfSe)[26] to identify bacterial taxa that might be informative of differences in mucosal bacterial community responses between males and females during the acute phase. Six genera were differentially represented (Figure 5A). Flexispira (p<0.01), Bacteroides (p<0.001), and Helicobacter (p<0.01) were significantly enriched in female mucosal samples (Figure 5B), while Prevotella (p<0.001), Streptococcus (p<0.01), and Blautia (p<0.001) were overly represented in male mucosal samples (Figure 5C).
Figure 4. Gender differences in microbial response to R5 SHIV IR challenge.
Comparison between mucosal and luminal Proteobacteria abundance with stratification at the class level. Rectal swabs and fecal samples were collected prior to and following intrarectal R5 SHIVC109P3 infection of male (DG56, HC12, ER57) and female (DJ53, EJ64, DF73) RMs. Shaded area demarks the pre- and acute phase of infection.
Figure 5. Gender specific microbial communities in infected animals.
Bacterial taxa informative of differences in mucosal bacterial communities between infected male and female RMs were identified by LEfSe. Differential representation of bacterial lineages, as highlighted by small circles and by shading, between male (DG56, HC12, ER57) and female (DJ53, EJ64, DF73) RMs during acute stage (d3 - 28) is mapped to a taxonomic cladogram constructed with known bacterial taxa (A). The circles represent phylogenetic levels from phylum to genus inside out. Diameter of each circle is proportional to taxon abundance; red, females; green, males. Histogram of bacterial genera enriched (LDA scores >4) during acute infection is also shown; red, female RMs; green, male RMs. Histograms showing temporal changes in relative abundance of genera significantly enriched in female and male rhesus macaques during the course of infection (B and C). Flexispira (11.69% in females vs. 3.42% in males), Bacteroides (0.37% vs. 0.09%), and Helicobacter (0.63% vs. 0.11%) were significantly enriched in female mucosal samples (B), while Prevotella (4.10% vs. 10.87%), Streptococcus (0.80% vs.2.20%), and Blautia (1.52% vs. 3.78%) were overly represented in male mucosal samples (C). Shaded area demarks the pre- and acute phase of infection.
Discussion
In this study, we presented evidence for a novel NHP model of sex inequality in disease risk that recapitulates the sex bias in HIV-1 disease progression in humans (Figure 1). Gender-based differences in basal immune activation and inflammation in the gut environment of macaques do not appear to be contributing factors (Figure 2), but a previously unrecognized sex disparity in local innate immunity and microbial response to SHIV infection was observed. While there is some degree of heterogeneity between these outbred animals within each sex, an earlier, stronger and proinflammatory skewed chemokine/cytokine response was seen in the rectal mucosa of two of the three SHIV-infected females (Figure 3), consistent with previous reports in humans that females generally mount a more potent innate immune response, as well as a stronger Th1-biased adaptive immune response, than do males following infection or vaccination [27]. Moreover, there were notable changes in the gut microbiota during the acute phase of SHIV infection of both sexes, but the magnitude of the microbial community shift as well as enrichment of adherent Proteobacteria in the rectal mucosa of female macaques was significantly higher than the males (Figure 4). At the taxa level, Flexispira, Bacteroides and Helicobacter predominated in female but not male mucosal samples (Figure 5). Mechanistically, increased expression of IL-1β and other inflammatory cytokines (e.g., IFN-γ, TNF-α) has been shown to disrupt epithelial barrier integrity [28, 29], and is associated with microbial translocation and chronic immune activation, markers of HIV/SIV disease progression [30-32]. In humans, HIV infection is associated with an increase of Proteobacteria accompanied by decreases of Firmicutes and Bacterrioidia [33-36], similar to the profile observed in inflammatory gastrointestinal conditions [34, 37-39]. Notably, the increase in Proteobacteria in HIV patients is associated with markers of mucosal immune disruption, T cell activation, and chronic inflammation. A positive association between Bacteroidales and CD8+ T cell activation had also been reported [40], and in mouse models, Helicobacter hepaticus-induced inflammatory bowel disease is more severe in females than in males [41]. Our finding that innate immune mediators and bacterial species associated with gut immune activation and damage are augmented early in the rectal mucosa of infected female than male macaques, therefore, is of interest, and is consistent with a more inflammatory tone in the rectal mucosa of infected females than males, which could contribute to faster disease progression in females.
In summary, nearly half of the more than 30 million adults newly infected with HIV worldwide are women, and unprotected anal receptive intercourse is practiced by members of this sex [42, 43]. A better understanding of the host innate immune and microbial interactions that might contribute to the sex disparity in disease seen in this novel NHP model of HIV infection therefore could inform the development of comprehensive and effective health care of infected women.
Supplementary Material
Acknowledgements
This work was supported by US National Institutes of Health grants R01AI046980 (CCM), R56AI084765 (CCM), R01AI110372 (TC and ZP), U01CA18237 (ZP), UH3CA140233 (ZP), R01CA159036 (ZP), and R03CA159414 (LY), and by the Department of Veterans Affairs, Veterans Health Administration. The New York University Genome Technology Center is partially supported by grant P30CA016087 from the US National Cancer Institute. Additional support was provided by the Tulane National Primate Research Center Base grant OD011104 and by the NIH grant P51 00010425 grant to the Virology Core at the Washington National Primate Research Center, Seattle, WA.
Footnotes
Author contributions: WR, ZP, TC and CCM conceived of and designed the study; WR, LY, AG, JS, KR, JB performed the experiments; WR, YM, LY, ZP, TC and CCM analyzed data; YM and AD, with help from ZP and TC performed the statistical analysis; WR, ZP, TC and CCM prepared the manuscript, with help from all the other authors.
Conflict of Interest: The authors declare no conflict of interests.
References
- 1.Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Washburn TC, Medearis DN, Jr., Childs B. Sex Differences in Susceptibility to Infections. Pediatrics. 1965;35:57–64. [PubMed] [Google Scholar]
- 3.Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 4.Garenne M. Sex differences in measles mortality: a world review. Int J Epidemiol. 1994;23:632–642. doi: 10.1093/ije/23.3.632. [DOI] [PubMed] [Google Scholar]
- 5.Neyrolles O, Quintana-Murci L. Sexual inequality in tuberculosis. PLoS Med. 2009;6:e1000199. doi: 10.1371/journal.pmed.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varkevisser CM, Lever P, Alubo O, Burathoki K, Idawani C, Moreira TM, et al. Gender and leprosy: case studies in Indonesia, Nigeria, Nepal and Brazil. Lepr Rev. 2009;80:65–76. [PubMed] [Google Scholar]
- 7.Farzadegan H, Hoover DR, Astemborski J, Lyles CM, Margolick JB, Markham RB, et al. Sex differences in HIV-1 viral load and progression to AIDS. Lancet. 1998;352:1510–1514. doi: 10.1016/S0140-6736(98)02372-1. [DOI] [PubMed] [Google Scholar]
- 8.Prins M, Meyer L, Hessol NA. Sex and the course of HIV infection in the pre- and highly active antiretroviral therapy eras. Aids. 2005;19:357–370. doi: 10.1097/01.aids.0000161765.75663.27. [DOI] [PubMed] [Google Scholar]
- 9.Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JJ, Woods M, Lindsay RJ, Doyle EH, Griesbeck M, Chan ES, et al. Higher expression of several interferon-stimulated genes in HIV-1-infected females after adjusting for the level of viral replication. J Infect Dis. 2013;208:830–838. doi: 10.1093/infdis/jit262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 2008;1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douek DC, Roederer M, Koup RA. Emerging concepts in the immunopathogenesis of AIDS. Annu Rev Med. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 14.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 17.Katsikis PD, Mueller YM, Villinger F. The cytokine network of acute HIV infection: a promising target for vaccines and therapy to reduce viral set-point? PLoS Pathog. 2011;7:e1002055. doi: 10.1371/journal.ppat.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clayton JA, Collins FS. Policy: NIH to balance sex in cell and animal studies. Nature. 2014;509:282–283. doi: 10.1038/509282a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren W, Mumbauer A, Gettie A, Seaman MS, Russell-Lodrigue K, Blanchard J, et al. Generation of lineage-related, mucosally transmissible subtype C R5 simian-human immunodeficiency viruses capable of AIDS development, induction of neurological disease, and coreceptor switching in rhesus macaques. J Virol. 2013;87:6137–6149. doi: 10.1128/JVI.00178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, Desantis TZ, et al. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol. 2010;16:4135–4144. doi: 10.3748/wjg.v16.i33.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang K-YaZ, Scott L. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 23.Sankaran-Walters S, Macal M, Grishina I, Nagy L, Goulart L, Coolidge K, et al. Sex differences matter in the gut: effect on mucosal immune activation and inflammation. Biol Sex Differ. 2013;4:10. doi: 10.1186/2042-6410-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein SL, Poland GA. Personalized vaccinology: one size and dose might not fit both sexes. Vaccine. 2013;31:2599–2600. doi: 10.1016/j.vaccine.2013.02.070. [DOI] [PubMed] [Google Scholar]
- 28.Hirao LA, Grishina I, Bourry O, Hu WK, Somrit M, Sankaran-Walters S, et al. Early mucosal sensing of SIV infection by paneth cells induces IL-1beta production and initiates gut epithelial disruption. PLoS Pathog. 2014;10:e1004311. doi: 10.1371/journal.ppat.1004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, Graham WV, Wang Y, Witkowski ED, Schwarz BT, Turner JR. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am J Pathol. 2005;166:409–419. doi: 10.1016/s0002-9440(10)62264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 32.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 39.Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis CL, Ma ZM, Mann SK, Li CS, Wu J, Knight TH, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livingston RS, Myles MH, Livingston BA, Criley JM, Franklin CL. Sex influence on chronic intestinal inflammation in Helicobacter hepaticus-infected A/JCr mice. Comp Med. 2004;54:301–308. [PubMed] [Google Scholar]
- 42.Karim SS, Ramjee G. Anal sex and HIV transmission in women. Am J Public Health. 1998;88:1265–1266. doi: 10.2105/ajph.88.8.1265-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misegades L, Page-Shafer K, Halperin D, McFarland W, Survey YWSSIGYWs Anal intercourse among young low-income women in California: an overlooked risk factor for HIV? AIDS. 2001;15:534–535. doi: 10.1097/00002030-200103090-00017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.