Abstract
Behavioral displays or physiological responses are often influenced by intrinsic and extrinsic mechanisms in the context of the organism's evolutionary history. Understanding differences in transcriptome profiles can give insight into adaptive or pathological responses. We utilize high throughput sequencing (RNA-sequencing) to characterize the neurotranscriptome profiles in both males and females across four strains of zebrafish (Danio rerio). Strains varied by previously documented differences in stress and anxiety-like behavioral responses, and generations removed from wild-caught individuals. Here we describe detailed methodologies and quality controls in generating the raw RNA-sequencing reads that are publically available in NCBI's Gene Expression Omnibus database (GSE61108).
Keywords: RNA-sequencing, Brain, Gene expression, Sex, Zebrafish
| Specifications | |
|---|---|
| Organism/cell line/tissue | Zebrafish brain |
| Sex | Male and female |
| Sequencer or array type | Illumina Genome Analyzer IIx |
| Data format | Raw: FASTQ files |
| Experimental factors | Strain (AB, Scientific Hatcheries, LSB, HSB), sex (male, female) |
| Experimental features | RNA-sequencing analysis of male and female zebrafish brains in four different strains (AB, Scientific Hatcheries, LSB, HSB), 17 weeks post-fertilization |
| Consent | All procedures approved by the North Carolina State University Institutional Animal Care and Use Committee |
| Sample source location | Experiments were conducted at North Carolina State University, Raleigh, North Carolina, USA |
1. Direct link to deposited data
The raw FASTQ files can be accessed through the Gene Expression Omnibus.
2. Experimental design, materials and methods
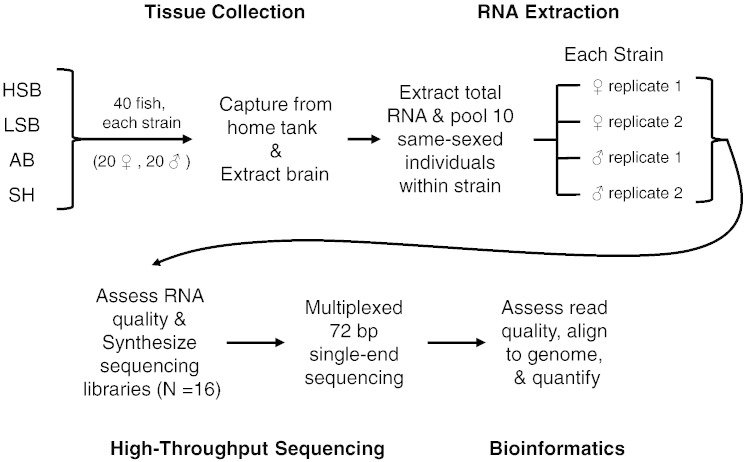
In this study we analyzed the whole-brain transcriptome profiles of male and female zebrafish (Danio rerio) in four different strains [1], [2]. In brief, 17 week old zebrafish were quickly sacrificed and whole-brains were removed and processed for RNA-sequencing. Sequencing reads were subsequently aligned, analyzed, and quantified using open-source software. We also conducted technical and biological validation and replication of the RNA-sequencing results using quantitative reverse-transcriptase PCR (for overview of procedures, see Fig. 1).
Fig. 1.
Workflow for collecting and processing the neurotranscriptome in each zebrafish strain.
2.1. Animal subjects
Zebrafish cohorts were generated and reared using previously described methods [3]. All fish were kept in mixed sex 100-liter tanks. Tanks were on a custom-built recirculating filtration system with water temperature kept at 28 °C and on a 12:12 light:dark cycle. Fish were fed twice daily with commercial feed (Tetramin). The AB and Scientific Hatcheries (SH) zebrafish strains originated from commercial suppliers (Zebrafish International Resource Center and Scientific Hatcheries, respectively). Although the AB and SH strains were bred in laboratory conditions for many generations at their respective stock centers, these strains were maintained in our laboratory for four and one generations, respectively. The two other strains (High Stationary Behavior (HSB); Low Stationary Behavior (LSB)) of zebrafish originated from approximately 200 wild caught individuals and were six generations removed from the wild (see [3] for additional selective breeding details).
2.2. Tissue collection
We collected whole brains from 160 individual zebrafish (n = 20 for each sex for each strain) that were 17 weeks post-fertilization. Between 09:00–12:00 we quickly removed fish from their home tanks, deeply anesthetized with tricaine methanesulfonate, followed by decapitation. Whole brains were removed within 3 min of being caught and placed in RNAlater (Ambion). After storing the samples at 4 °C overnight, we removed all RNAlater and stored brains at − 80 °C until RNA extraction. Sex was assigned by observation of testes or ovaries on dissection.
2.3. RNA isolation
We extracted total RNA using column purification (RNeasy Plus Mini Kit, Qiagen). Brains were homogenized for 3 min at maximum speed with 50–100 μl of zirconium oxide beads (Bullet Blender, Next Advance) in 0.6 ml of Buffer RLT (Qiagen) with 2-mercaptoethanol (Sigma). We then added 100 μl of chloroform, mixed, and incubated at room temperature for 5 min. We subsequently centrifuged the samples at 12,000 × g for 15 min at 4 °C. The supernatant was transferred to the RNeasy genomic DNA column (Qiagen) and then we proceeded according to the manufacturer's instructions. All samples were eluted with 30 μl of DEPC-treated water (Ambion).
2.4. RNA-sequencing library preparation and sequencing
For each strain we pooled 1 μg of total RNA from 10 same sex individuals into a biological replicate. This generated four biological replicates for each strain (two biological replicates for each sex). We analyzed the quantity and quality of the RNA for the 16 samples with a 2100 Bioanalyzer (Agilent). All samples were of high quality (RIN > 8.0, Table 1). Using 1 μg of total RNA from the pooled samples we generated cDNA libraries following the manufacturer's protocol (TruSeq RNA Sample Prep V2, Illumina). We ligated a unique Illumina Index adapter to each biological replicate to allow for multiplexing. After cDNA library synthesis we submitted samples to the Genomic Sciences Laboratory at North Carolina State University for 72 bp single-end RNA sequencing (Illumina GAIIx). We followed a balanced block design [4] and multiplexed all 16 samples and ran them across 16 lanes.
Table 1.
RNA characteristics of biological replicates as measured by a 2100 Bioanalyzer (Agilent).
| Sample name in GSE61108 | Strain | Sex | RNA concentration (ng/μl) | RNA integrity number |
|---|---|---|---|---|
| AB female rep1 | AB | Female | 69.88 | 8.5 |
| AB female rep2 | AB | Female | 70.58 | 8.5 |
| AB male rep1 | AB | Male | 67.76 | 8.5 |
| AB male rep2 | AB | Male | 73.16 | 8.6 |
| SH female rep1 | Scientific Hatcheries | Female | 66.12 | 8.6 |
| SH female rep2 | Scientific Hatcheries | Female | 90.58 | 8.5 |
| SH male rep1 | Scientific Hatcheries | Male | 118.38 | 8.7 |
| SH male rep2 | Scientific Hatcheries | Male | 105.76 | 8.7 |
| LSB female rep1 | Low Stationary Behavior | Female | 88.8 | 8.4 |
| LSB female rep2 | Low Stationary Behavior | Female | 69.88 | 8.4 |
| LSB male rep1 | Low Stationary Behavior | Male | 54.14 | 8.7 |
| LSB male rep2 | Low Stationary Behavior | Male | 57.52 | 8.4 |
| HSB female rep1 | High Stationary Behavior | Female | 71.48 | 8.5 |
| HSB female rep2 | High Stationary Behavior | Female | 89.62 | 8.7 |
| HSB male rep1 | High Stationary Behavior | Male | 82.3 | 8.5 |
| HSB male rep2 | High Stationary Behavior | Male | 75.62 | 8.5 |
2.5. Data processing
With reads that passed default quality controls (Illumina), we combined across lanes for each biological replicate. Total read counts varied between 34–65 million reads (Table 2). We utilized the open source software GSNAP [5] to align the reads to the zebrafish genome. We first built GSNAP genomic and GSNAP known and novel splice site databases using the Zv9 (release 71) D. rerio genome and gene sets, respectively (Ensembl). For each biological replicate we successfully aligned over 99% of the reads (assessed by SAMtools [6]) to the zebrafish genome using the default GSNAP parameters (Table 2).
Table 2.
Sequenced library characteristics.
| Sample name in GSE61108 | Strain | Sex | Read count | Reads aligning to zebrafish genome (%) |
|---|---|---|---|---|
| AB female rep1 | AB | Female | 63,333,522 | 99.2863542 |
| AB female rep2 | AB | Female | 48,539,389 | 99.194584 |
| AB male rep1 | AB | Male | 52,400,106 | 99.2919213 |
| AB male rep2 | AB | Male | 42,245,840 | 99.2484941 |
| SH female rep1 | Scientific Hatcheries | Female | 65,493,707 | 99.257547 |
| SH female rep2 | Scientific Hatcheries | Female | 60,919,457 | 99.2179313 |
| SH male rep1 | Scientific Hatcheries | Male | 59,127,323 | 99.2323989 |
| SH male rep2 | Scientific Hatcheries | Male | 44,528,195 | 99.2321562 |
| LSB female rep1 | Low Stationary Behavior | Female | 43,983,674 | 99.1853227 |
| LSB female rep2 | Low Stationary Behavior | Female | 49,943,584 | 99.2696399 |
| LSB male rep1 | Low Stationary Behavior | Male | 57,927,979 | 99.2060158 |
| LSB male rep2 | Low Stationary Behavior | Male | 53,242,350 | 99.2381685 |
| HSB female rep1 | High Stationary Behavior | Female | 55,298,353 | 99.2311091 |
| HSB female rep2 | High Stationary Behavior | Female | 34,150,835 | 99.2296909 |
| HSB male rep1 | High Stationary Behavior | Male | 60,575,809 | 99.2129366 |
| HSB male rep2 | High Stationary Behavior | Male | 44,987,343 | 99.2242418 |
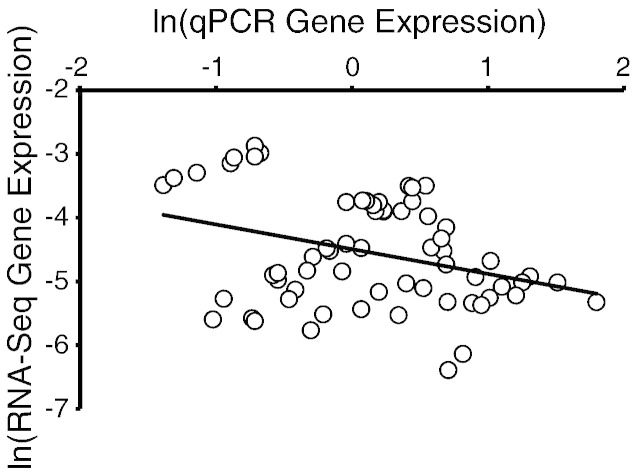
2.6. Validation and replication with quantitative reverse-transcriptase PCR
We performed both technical validation of RNA-sequencing libraries and independent biological replication (HSB and LSB strains) through quantitative reverse-transcriptase PCR (qPCR). We quantified the reads for each protein-coding gene by using the “union” mode in HTSeq [7] in all of our RNA-sequencing libraries. Read counts were then normalized to the library size in edgeR [8]. We selected eight genes (msmo1, oxt, gabbr1a, comta, sell, prodha, hsd11b2, gapdh) for technical validation and 14 genes (msmo1, oxt, gabbr1a, comta, sell, prodha, hsd11b2, gapdh, cyp19a1b, dio2, pmchl, cfos, gabbr1b, igf1) for independent biological replication (see [1], [2] for detailed primer characteristics and qPCR reaction parameters).
After normalizing each gene's expression to ef1a, an endogenous reference gene [9], we confirmed a significant correlation between gene expression measured by RNA-sequencing and qPCR. Using the same material from cDNA libraries that were submitted for RNA-sequencing, we found a significant correlation between normalized read count (RNA-sequencing quantification) and cycle threshold (qPCR quantification) for the eight genes examined (technical validation; n = 64, Spearman's ρ = − 0.278 p = 0.026; Fig. 2). Using independent samples (n = 9 for each sex in each of the LSB and HSB strains), we similarly observed a significant correlation between expression measurements from the two techniques (RNA-sequencing and qPCR) for 14 genes (independent biological replication; n = 56, Spearman's ρ = − 0.406 p = 0.002). Of note, we also observed consistent patterns of differential gene expression between sexes and stress coping styles (see [1], [2] for details).
Fig. 2.
Technical validation of RNA-sequencing results using qPCR. Each point represents a gene expression value for one of eight genes in each of the biological replicates in the HSB and LSB strains. Gene expression was normalized to an endogenous reference, ef1a, as measured in their respective quantification methods.
3. Conclusions
Zebrafish are a model system utilized in many developmental, toxicological, neuroscience, and biomedical studies [10], [11], [12], [13], [14], [15]. Understanding and accounting for genomic and transcriptomic variation will provide important additional insights. Here we describe in detail the procedures and methodologies in sequencing the whole-brain transcriptome of both male and female adult zebrafish in four different strains. The high quality RNA-sequencing results, which have been both technically and biologically validated, are available through the NCBI's GEO database (GSE61108). This dataset should be of use to studies in a variety of contexts (e.g. evolution, neuroscience, genetics, bioinformatics, and biomedicine).
Acknowledgements
We thank Brad Ring and John Davis for the assistance with fish husbandry. We are grateful to Cory Dashiell, Melissa Lamm, Melissa McLeod, Katie Robertson, Christopher Gabriel, and Noffisat Oki for the helpful discussions and technical assistance. We are grateful to Barrie Robison for the generous donation of the Scientific Hatcheries line of zebrafish. This study received support from the National Institutes of Health (1R21MH080500) to JG. This study is a contribution of the W.M. Keck Center for Behavioral Biology at North Carolina State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wong R.Y., McLeod M.M., Godwin J. Limited sex-biased neural gene expression patterns across strains in zebrafish (Danio rerio) BMC Genomics. 2014;15(1):905. doi: 10.1186/1471-2164-15-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong R.Y., Lamm M.S., Godwin J. Characterizing the neurotranscriptomic states in alternative stress coping styles. BMC Genomics. 2015;16:425. doi: 10.1186/s12864-015-1626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong R.Y., Perrin F., Oxendine S.E., Kezios Z.D., Sawyer S., Zhou L.R., Dereje S., Godwin J. Comparing behavioral responses across multiple assays of stress and anxiety in zebrafish (Danio rerio) Behaviour. 2012;149(10–12):1205–1240. [Google Scholar]
- 4.Auer P.L., Doerge R.W. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185(2):405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing S: the Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCurley A.T., Callard G.V. Characterization of housekeeping genes in zebrafish: male–female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong R.Y., Oxendine S.E., Godwin J. Behavioral and neurogenomic transcriptome changes in wild-derived zebrafish with fluoxetine treatment. BMC Genomics. 2013;14:348. doi: 10.1186/1471-2164-14-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson R.T., Macrae C.A. Systematic approaches to toxicology in the zebrafish. Annu. Rev. Pharmacol. Toxicol. 2012;52:433–453. doi: 10.1146/annurev-pharmtox-010611-134751. [DOI] [PubMed] [Google Scholar]
- 13.Grunwald D.J., Eisen J.S. Headwaters of the zebrafish — emergence of a new model vertebrate. Nat. Rev. Genet. 2002;3(9):717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 14.Stewart A.M., Braubach O., Spitsbergen J., Gerlai R., Kalueff A.V. Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014;37(5):264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira R.F. Mind the fish: zebrafish as a model in cognitive social neuroscience. Frontiers in neural circuits. 2013;7:131. doi: 10.3389/fncir.2013.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]