Abstract
Freshwater lakes represent large methane sources that, in contrast to the Ocean, significantly contribute to non-anthropogenic methane emissions to the atmosphere. Particularly mixed lakes are major methane emitters, while permanently and seasonally stratified lakes with anoxic bottom waters are often characterized by strongly reduced methane emissions. The causes for this reduced methane flux from anoxic lake waters are not fully understood. Here we identified the microorganisms and processes responsible for the near complete consumption of methane in the anoxic waters of a permanently stratified lake, Lago di Cadagno. Interestingly, known anaerobic methanotrophs could not be detected in these waters. Instead, we found abundant gamma-proteobacterial aerobic methane-oxidizing bacteria active in the anoxic waters. In vitro incubations revealed that, among all the tested potential electron acceptors, only the addition of oxygen enhanced the rates of methane oxidation. An equally pronounced stimulation was also observed when the anoxic water samples were incubated in the light. Our combined results from molecular, biogeochemical and single-cell analyses indicate that methane removal at the anoxic chemocline of Lago di Cadagno is due to true aerobic oxidation of methane fuelled by in situ oxygen production by photosynthetic algae. A similar mechanism could be active in seasonally stratified lakes and marine basins such as the Black Sea, where light penetrates to the anoxic chemocline. Given the widespread occurrence of seasonally stratified anoxic lakes, aerobic methane oxidation coupled to oxygenic photosynthesis might have an important but so far neglected role in methane emissions from lakes.
Introduction
Freshwater lakes occupy only a small portion of the Earth's surface and yet they contribute more non-anthropogenic methane to the atmosphere (up to 16% Bastviken et al., 2004) than the Ocean (<1% Rhee et al., 2009). The lower concentrations of sulphate in lake waters (mostly <0.2 mM) compared with ocean waters (28 mM) lead to enhanced degradation of organic material by methanogenesis, which is one of the main causes for higher methane emission from continental compared with marine aquatic systems. Additionally, aerobic and anaerobic methane oxidation processes act as a very efficient methane filter in marine sediments and water column (for example, Hinrichs and Boetius, 2002; Reeburgh, 2007). In contrast, the full variety of processes responsible for methane removal in lacustrine environments is not understood nor are the key microbial players known. Mixed lakes with oxygenated water columns exhibit high rates of methane oxidation at the sediment/water interface (for example, Lidstrom and Somers, 1984). Permanently or seasonally stratified lakes, on the other hand, show the highest rates of methane oxidation at and around the oxic/anoxic transition zone in the water column (for example, Blees et al., 2014). In all cases, the main methane-removing process from limnic systems is considered aerobic methane oxidation, fuelled by oxygen diffusion or intrusion events from the oxic water column.
Methanotrophic microorganisms have a key role in reducing methane fluxes in oxic as well as in anoxic aquatic systems. Both bacteria and archaea possess the capacity to oxidize methane. At circumneutral pH, aerobic oxidation of methane is performed by methane-oxidizing bacteria (MOB) belonging to alpha- and gamma-proteobacteria (for a review, see Hanson and Hanson, 1996 and Trotsenko and Murrell, 2008). The biochemical pathway for methane oxidation employed by all known MOB involves soluble and/or particulate methane monooxygenase enzymes (sMMO and pMMO, respectively). This pathway is also utilized by the bacterium Candidatus Methylomirabilis oxyfera that can couple aerobic oxidation of methane to nitrite reduction to N2 (Raghoebarsing et al., 2006; Ettwig et al., 2008). Interestingly, this bacterium is found in anoxic habitats, because it has the capacity to generate oxygen intracellularly from NOx (Ettwig et al., 2010). True anaerobic oxidation of methane (AOM) is performed by anaerobic methanotrophic archaea of which three different clades are known to date (ANME-1, -2 and -3). Most commonly reported AOM in the environment is directly coupled to reduction of sulphate and occurs in sulphate-rich marine sediments (Hinrichs and Boetius, 2002; Knittel and Boetius, 2009; Milucka et al., 2012). Recently, it has been proposed that ANME-2 archaea belonging to a new family Candidatus ‘Methanoperedenaceae' might also be capable of linking AOM to nitrate reduction to nitrite (Haroon et al., 2013). The biochemical pathway of anaerobic oxidation of methane utilized by the ANME is fundamentally different from the aerobic one and employs a set of nine enzymes, with methyl-coenzyme M reductase (MCR) being the key enzyme-activating methane (Scheller et al., 2010). AOM coupled to reduction of metal oxides (iron and manganese) was also proposed to occur in the ocean (Beal et al., 2009; Wankel et al., 2012) and in lakes (Sivan et al., 2011), but the mechanisms behind these processes and the responsible microorganisms have not yet been conclusively identified.
We investigated methane oxidation in the permanently stratified water column of Lago di Cadagno. This meromictic lake, unlike non-stratified lakes, hosts a very efficient methane filter and high methane fluxes from the anoxic hypolimnion are nearly completely consumed just below the oxycline. The aim of our study was to identify the biogeochemical processes and microorganisms responsible for the methane consumption.
Materials and methods
Study site
Lago di Cadagno is a meromictic lake located in the Piora valley in the southern Alps of Switzerland (1923 m altitude). Further lake and water characteristics are listed in Supplementary Material and Methods.
Sampling
Sampling was performed from a fixed platform. A SBE 19 CTD probe (Sea-Bird Electronics, Bellevue, WA, USA) equipped with oxygen and pH sensor (SBE 18 pH sensor, Sea-Bird Electronics, measurement range 0–14, accuracy 0.1 pH units) was used to measure conductivity, temperature, light transmission, pH, dissolved oxygen and turbidity. Additionally, micro-optodes (types PSt1 and TOS7 with 50- and 140-μm tip diameter, respectively, white optical isolation, PreSens, Regensburg, Germany) were used for online measurements of oxygen down to ca. 300 nmol l−1 (2009) and ca. 20 nmol l−1 (2011, 2013) to determine the absolute zero oxygen depth. Samples for Winkler titration were collected in order to correct for the offset in the oxygen sensor. Water samples for nutrient and chemical analyses were retrieved with a sampling system containing 12 × 60 ml plastic syringes that could be triggered from the platform (see Kirf et al., 2014). Water was aliquoted into respective reagents immediately after retrieval of the syringe sampler. Samples for dissolved gas analysis were preserved with NaOH (pH>12) or Cu(I)Cl and closed with butyl septa avoiding air bubbles in the bottles. Samples for sulphate and sulphide were sampled into 5% ZnCl2, samples for ammonium were sampled into a working reagent containing orthophthaldialdehyde and samples for iron and manganese (unfiltered as well as filtered through a 0.45-μm filter) were sampled into ferrozine reagent and 1 M HCl, respectively. For manganese concentrations, water (2 ml) was acidified onsite with 45-μl ultrapure concentrated nitric acid (65%, VWR) and then kept frozen until analysis using inductively coupled plasma optical emission spectroscopy (Spectro Analytical Instruments, Kleve, Germany). Water samples for the incubation experiments were collected with Niskin bottles (General Oceanics Inc., Miami, FL, USA) or a pump system and immediately aliquoted into serum bottles, using clean rubber tubing. Each bottle was filled from bottom to top, avoiding aeration and letting bottles overflow before closing them, headspace-free, with butyl stoppers. All bottles were kept cold and dark during transport to the laboratory.
Chemical analyses
Sulphide was measured spectrophotometrically according to Cline (1969). Ammonium was measured fluorometrically as described by Holmes et al. (1999). Sulphate was analysed on a 761 Compact ion chromatograph (Metrohm, Filderstadt, Germany) equipped with a Metrosep A SUPP 5 column. Iron and manganese species (total and dissolved (that is, reduced)) were measured photometrically using the ferrozine assay (Stookey, 1970) and inductively coupled plasma-mass spectrometry (NEPTUNE Plus Multicollector ICP-mass Spectrometer; ThermoFisher Scientific, Bremen, Germany), respectively.
Natural abundance methane carbon isotopes measurements
Methane carbon isotopes were measured for water depths of 9, 10, 11, 12 and 13 m where water was retrieved by Niskin bottles and aliquoted into serum bottles as described. Bottles were kept in the dark at close to in situ temperatures (7 °C), then killed with HgCl2 after 0, 50, 114, 162 and 210 h and measured for methane concentrations and carbon isotopic composition. Methane concentrations were measured on an Agilent 6890 gas chromatograph with a flame ionization detector (Agilent, Basel, Switzerland), a Carboxen 1010 PLOT column (Supelco, Sigma-Aldrich, Steinheim, Germany) and helium as carrier gas. Initial oven temperature was 40 °C and detector temperature was 200 °C. The temperature was kept constant at 40 °C for 5 min and then raised to 110 °C at a rate of 10 °C min−1. Standards (15 p.p.m., 1000 p.p.m. and 1%) used for calibration were supplied from Scott Specialty Gases (Breda, The Netherlands) and accuracy was ±3%. Dissolved methane concentrations were calculated using the solubility data from Wiesenburg and Guinasso (1979).
The methane isotopic composition was determined with a preconcentration unit (Trace Gas, Isoprime, Manchester, UK) connected to a mass spectrometer (Isoprime). Units are in δ notation: δ13C=((13C/12Csample)/(13C/12C standard)−1) × 1000, and values are referenced against the Vienna Pee Dee Belemnite standard. The fractionation factor α for methane oxidation was calculated by using the methane isotopic composition and methane concentration at different time points (for incubations) or different depths (in situ) with the following formula:
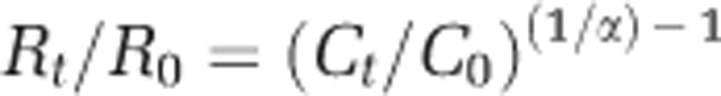 |
where R is the isotope ratio and C the concentration at the initial time point (0) and at subsequent time point or depths (t). The fractionation factor α for Lago di Cadagno was calculated based on the increase of δ13C–CH4 from an average value of −70.2‰ below 11 m (11–13.5 m) to an average value of −40.9‰ above 9.25 m (8–9.25 m) (Figure 1c).
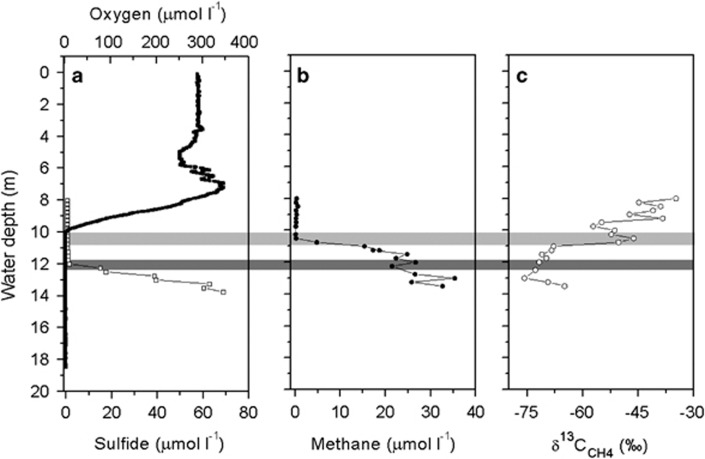
Figure 1.
Typical water column parameters of Lago di Cadagno in August 2009. Chemical profiles of (a) oxygen (full line), sulphide (empty squares) and (b) methane (full circles) concentrations; and (c) the distribution of methane carbon isotopes (empty circles). Zones of methane and sulphide oxidation are highlighted in light and dark grey, respectively.
In vitro incubations with 13C-labelled methane
Experiment 1 (dark, light, oxygen, iron, manganese and photosynthetic inhibitor-addition experiments)
For the 13CH4 incubations, lake water was collected from specific water depths with a peristaltic pump on deck connected to a home-build CTD system equipped with trace oxygen optodes (Kirf et al., 2014) and immediately filled via a tubing directly into 250 ml serum bottles, as described above. The bottles were subsequently bubbled with helium for 15 min before the addition of 10 ml of lake water saturated with 13C-labelled methane (99%, Campro Scientific, Berlin, Germany). The final concentration of methane in the incubation was ca. 60 μmol l−1 (that is, 13C atom %=99). To the CH4-supplemented bottle, a ferrihydrite stock solution was added to a final concentration of ∼100 μM FeIII for the iron-addition experiment, and birnessite stock solution was added to a final concentration of ∼100 μM MnIV for the manganese-addition experiment. For the oxygen-addition experiment, 10 ml of CH4-supplemented water was replaced with oxygenated non-filtered lake water from the respective depth. For the photosynthetic inhibitor-addition experiment, we followed the standard DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea) inhibition assay, previously used also in Lago di Cadagno (Camacho et al., 2001). DCMU (dissolved in 50% ethanol) was added to a final concentration of 20 μmol l−1. The 13CH4 incubations were started immediately after sampling. Water was transferred from serum bottles into 12-ml Exetainers (Labco, High Wycombe, Buckinghamshire, UK) following the filling procedure described in Holtappels et al. (2011). The Exetainers were incubated for up to 24 h at in situ temperatures in the dark or light. A dimmed luminescent tube with a photosynthetic active radiation (PAR) of ∼0.5 μEinstein, which is comparable to in situ light intensity at the chemocline (PAR=0.2-2 μEinstein), was used for the light incubations in the laboratory. Individual Exetainers were killed off after 0, 3, 6, 12 and 24 h by adding HgCl2 to stop biological activity. Subsequently, the samples were stored at room temperature until analysis. 13CO2 production was determined for five individual incubation vessels per treatment after sample acidification by gas chromatography-isotope ratio mass spectrometry (GC-IRMS; Isoprime). For each depth and treatment, the rates of methane oxidation and s.es. were determined from the slope of the linear correlation between 13CO2 production in the individual Exetainers over time.
Experiment 2 (nitrite- and sulphate-addition experiments)
For the strictly anoxic 13CH4 incubations with sulphate and nitrite, 1 litre of lake water was collected from specific water depths with a Niskin bottle, transferred into clean Duran bottles as described above, closed headspace-free with a butyl stopper and transported in the dark at 4 °C to the MPI laboratory (Bremen, Germany). In an anoxic hood (Mecaplex, Grenchen, Switzerland), three sterile serum bottles (156 ml) were filled with lake water (120 ml) and supplied with 1 bar 13C-labelled methane (99%, Campro Scientific) and 1 bar 12C-methane (Air Liquide, Duesseldorf, Germany). The final concentration of methane in each incubation bottle was ca. 3 mmol l−1 (13C atom %=50). Into one of the CH4-amended bottles, sulphate was added from a sterile anoxic stock to a final concentration of 2 mmol l−1. Another CH4-amended bottle was supplemented with 15N-labelled nitrite to a final concentration of 20 μmol l−1 from a sterile anoxic stock (100 mmol l−1). A third bottle was incubated in parallel without any additions. All bottles were incubated at 4 °C in the dark on a shaker (40 r.p.m.). Each bottle was subsequently sampled (in a sterile and anoxic way) for 13CO2, 15N2, sulphate and sulphide at 0, 11, 15, 21, 28 and 37 days of incubation. Samples were sterile filtered to stop biological activity and prepared for 13CO2 and 15N2 measurements performed on a GC-IRMS (Isoprime). Sulphate (using ion chromatograph) and sulphide measurements (photometric) were performed as described above.
Experiment 3 (nanometer-scale secondary ion mass spectrometry (nanoSIMS))
Serum bottles with water samples from Lago di Cadagno (120 ml; prepared as described above) were degassed for 15 min with Helium (PanGas, Dagmersellen, Switzerland) and subsequently supplied with 10 ml of lake water (non-filtered, from the respective depth) that was degassed and subsequently saturated (∼1.5 mmol CH4 l−1) with 13C-labelled methane (99%, Campro Scientific). The final concentration of methane in the incubation was ca. 125 μmol l−1 (13C atom %=99). The samples were incubated under nearly in situ conditions by hanging the bottles into the lake at 10 m water depth. A sample for nanoSIMS analysis was taken after 5 days of incubation (see below).
Catalysed reporter deposition–fluorescence in situ hybridization (CARD–FISH) and cell marking with laser microdissection system
CARD–FISH procedure was performed with following probes: ANME I-350, ANME II-538, Mgamma84+Mgamma705 mix, Alfa968, and Malfa450. A detailed description is listed in Supplementary Material and Methods. Cell marking was performed as described previously (Musat et al., 2008; Milucka et al., 2012).
Nanometer-scale secondary ion mass spectrometry
Image acquisition and data processing
Filters containing hybridized cells were analysed using a NanoSIMS 50L (Cameca, Paris, France). Secondary ions 12C−, 13C−, 19F−, 12C14N− and 32S− were recorded using five electron multipliers. Obtained ion-count images were processed using Look@NanoSIMS (Polerecky et al., 2012). Details of nanoSIMS data acquisition and processing are listed in Supplementary Material and Methods. Altogether, 12 fields of view containing 19 gamma-MOB cells were analysed in the sample from the chemocline (10 m depth), and 10 fields of view containing 39 gamma-MOB were analysed in the sample from anoxic depth (13 m depth).
Methane assimilation rate calculations
For the methane carbon assimilation rates of gamma-MOB, the biovolume was determined assuming a coccoid cell and a mean cell size of 2 μm, as determined by nanoSIMS (equals ca. 4.2 μm3). For the size-to-biomass conversion, a calibration factor of 6.4 fmol C μm−3 was used (Musat et al., 2008). Subsequently, the cellular methane carbon assimilation rates were calculated as the 13C/12C enrichment of the respective cells incubated with 13CH4, multiplied by the average cellular biomass, and divided by 13C-labelling % of the dissolved methane and incubation time (5 days).
Diffusive flux calculation
Diffusive fluxes (J) of CH4 across the hypolimnion–epilimnion interface were calculated assuming steady state by using the maximum concentration change (∂C) over a specific depth range (∂x; that is, 11.20–11.00 m) applying the Fick's first law: J=−D∂C/∂x.
Iron and manganese fluxes were calculated from concentrations of their dissolved forms over those of 20 cm of the water column at the base of the oxic zone where concentrations changed dramatically. Turbulent fluxes were calculated assuming steady-state conditions. For the flux calculations, a turbulent diffusion coefficient (that is, Eddy diffusivity D) of 1.6 × 10−6 m2 s−1 was used, which was obtained from high-resolution temperature profiles and SF6 tracer experiments (Uhde, 1992; Wüest, 1994). The same turbulent diffusion coefficient was used to calculate fluxes for all other dissolved substrates (O2, CH4, SO42−) into the zone of methane consumption in the upper part of the anoxic chemocline. For method description of the heat budget method, see Powell and Jassby (1974).
Functional gene analyses
DNA was extracted from water samples collected during two sampling campaigns. In August 2006, water samples were collected from 10.5 m (LC6), 13.5 m (LC12) and 17 m (LC14) water depths (as described by Halm et al., 2009). During this campaign, the chemocline was found at 13.5 m. In August 2009, water samples were collected from 10 m and 13 m. During this campaign, the chemocline was found at 10 m. The genes encoding the subunit A (that is, beta subunit) of particulate methane monooxygenase (pmoA) were PCR amplified with the primers A186-A689 (Holmes et al., 1995) and A189-mb661 (Costello and Lidstrom, 1999). The genes encoding alpha subunit of the soluble methane monooxygenase (mmoX) were targeted using primer pairs mmof882-mmor1403 (McDonald et al., 1995), mmoXA-mmoXB (Auman and Lidstrom, 2002) and mmoX206f-mmoX886r (Hutchens et al., 2004). Methyl-coenzyme M reductase genes (mcrA) were targeted using the primers ME1-ME2 (Hales et al., 1996) and MCRf-MCRr (Springer et al., 1995). 16 rRNA of Type II putative alpha-MOB was targeted using primers Type IIF/R (Chen et al., 2007). Additionally, nested PCR using primer pairs A189_b-cmo682 and cmo182-cmo568 specific for Candidatus Methylomirabilis oxyfera (Luesken et al., 2011) was performed on samples from August 2009. For further details on 16S and functional gene analyses, see Supplementary Material and Methods.
Results and discussion
Oxidation of methane at the chemocline of Lago di Cadagno
Typical water column parameters of Lago di Cadagno in summer (late August) are shown in Figure 1 (data from 2009) and Supplementary Figure S1 (data from 2011). Oxygen concentrations dropped from ca. 300 μmol l−1 in the surface waters below the detection limit of the oxygen sensor (1 μmol l−1) at 10 m depth (Figure 1). The absence of oxygen from waters <10.9 m was confirmed by high-resolution amperometric microsensors (2009) and micro-optode measurements (2011; detection limit <20 nmol l−1; Kirf et al., 2014). The bottom waters contained substantial concentrations of hydrogen sulphide (ca. 40 μmol l−1 at 13 m), which was completely oxidized at the chemocline at ca. 12 m depth (Figure 1a), likely due to the activity of sulphide-oxidizing anoxygenic phototrophic bacteria. Sulphur bacteria such as Chromatium okenii, Chlorobium clathratiforme and Lamprocystis purpurea were shown to dominate the chemocline of Lago di Cadagno (>70% of all cells; Habicht et al., 2011; Musat et al., 2008; Storelli et al., 2013), and high densities of these bacteria were also found during our sampling campaigns (data not shown). Sulphide depletion coincided with the disappearance of ammonium (Supplementary Figure S1), presumably due to the assimilatory activity of the abundant purple and green sulphur bacteria.
Methane concentration profiles showed a sharp decrease in the upper part of the anoxic chemocline, from 15 μmol l−1 at 11 m to 0.2 μmol l−1 above 10.5 m depth (Figure 1b). The total flux of methane consumed within the anoxic chemocline was 9.8 mmol methane m−2 d−1. The decrease in methane concentrations was accompanied by a strong increase in methane carbon isotope values (δ13C-CH4; Figure 1c) from −75 to −65‰ in the anoxic layer to −35‰ at 8.5 m depth, suggesting microbial methane oxidation at the chemocline. Despite their fundamentally different biochemistry, both aerobic and anaerobic oxidation of methane result in a narrow range of fractionation factors for methane carbon (αC; 1.003–1.039 and 1.009–1.039, respectively; Holler et al., 2009; Reeburgh et al., 2006). The fractionation factor α for Lago di Cadagno calculated from in situ δ13C values (Figure 1c) was 1.012 and thus within the range of fractionation factors reported so far for methane oxidation. A lower αC value for methane (1.005) was determined from in vitro anoxic incubation from 13 m depth, whereas the αC from incubations from 11 and 12 m depth was higher (1.035 and 1.024, respectively) and thus in line with values reported for aerobic (1.009–1.039; Reeburgh et al., 2006), as well as sulphate-reducing (1.012–1.039, Holler et al., 2009; 1.003–1.039, Templeton et al., 2006) and nitrite-reducing (1.030, Rasigraf et al., 2012) AOM enrichment cultures.
Abundant aerobic MOB in the anoxic waters of Lago di Cadagno
Despite the geochemical evidence for high methane-oxidizing activity, no anaerobic methane-oxidizing archaea ANME-1 or -2 were detected in samples from the anoxic zone (that is, from 10 and 13 m) by fluorescence in situ hybridization (CARD–FISH) with any of the used probes. Likewise, no methyl-coenzyme M reductase genes (mcrA; encoding the key enzyme for archaeal methane oxidation) were detected at the investigated depths. Instead, the methane-rich anoxic and even sulphidic waters of Lago di Cadagno contained abundant aerobic alpha- and gamma-proteobacterial MOB (Figures 2 and 3). It is unlikely that the dense population of MOB (up to 0.5 million cells per ml) in the apparently anoxic chemocline settled down from the oxic zone, as MOB abundances were evenly distributed throughout both zones. The MOB activity throughout the chemocline provides further evidence for the autochthonous nature of the population of MOB in the anoxic waters (see section ‘Active growth of gamma-MOB on methane').
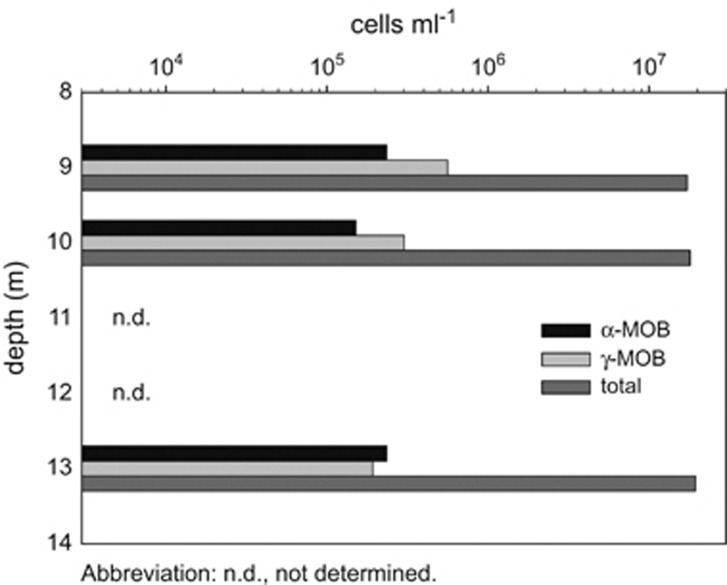
Figure 2.
Total cell counts and the abundance of alpha- and gamma-MOB in Lago di Cadagno. The abundance of aerobic methanotrophic bacteria was comparable in oxic and fully anoxic water and comprised ∼1% of the total cell counts. Anaerobic archaeal methane oxidizers were not detected in the investigated depths. Note the logarithmic scale.
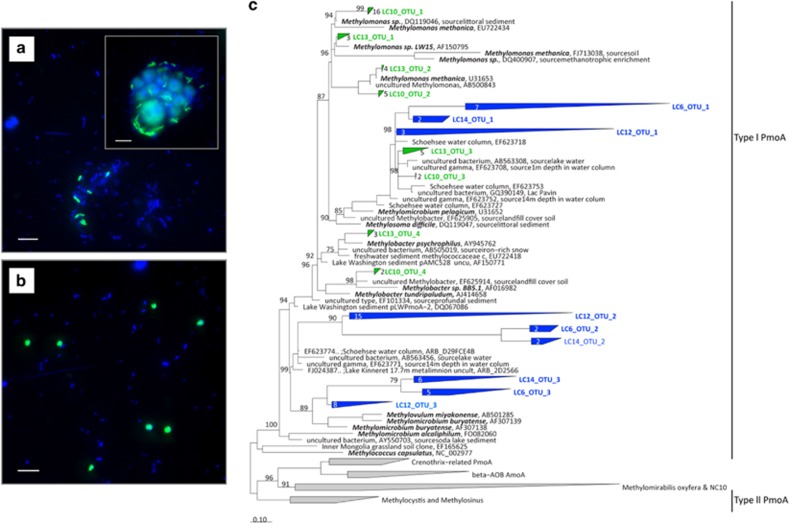
Figure 3.
Aerobic MOB in Lago di Cadagno. (a) Alpha-proteobacterial methanotrophs in the chemocline visualized by DAPI (4,6-diamidino-2-phenylindole; blue) and fluorescence in situ hybridization with Malpha450 probe (green). The cells are oval and often associated with particles and algae (inset). (b) Gamma-proteobacterial methanotrophs in the chemocline visualized by DAPI (blue) and fluorescence in situ hybridization with Mgamma84+Mgamma750 probe mix (green). The cells are coccoid and free-living. (c) Phylogenetic tree showing the sequences for particulate methane monooxygenase gene (pmoA) retrieved from different depths of Lago di Cadagno sampled in 2006 (blue) and 2009 (green). The pmoA sequences from 2006 originate from oxic (LC6), chemocline (LC12) and anoxic (LC14) water; the pmoA sequences from 2011 originate from chemocline (LC10) and anoxic (LC13) water. Clustering of operational taxonomic units (OTUs) indicate sequences with similarities of >90%. Bootstrap values >70% are indicated at the branch nodes. The scale bar represents the number of changes per nucleotide position. Note that only gamma-proteobacterial pmoA sequences were retrieved. Scale bar in panels (a) and (b) represents 5 μm.
Up to 2.4 × 105 cells per ml of aerobic, putative alpha-proteobacterial methanotrophs (targeted by MAlpha450 probe; Figure 2) were detected in the fully anoxic water at 13 m water depth by CARD-FISH. This corresponds to ca. 1.2% of total cell counts at this depth (Figure 2). Interestingly, the putative alpha-MOB were as abundant in the oxic water at 9 m as in the anoxic water, where they comprised ca. 1.4% of total cells. The putative alpha-MOB were oval, ca. 1–2 μm in size and were attached to particles and algae (Figure 3a). Furthermore, gamma-proteobacterial methanotrophs (targeted by probe mix Mgamma84+Mgamma705) were also detected in the anoxic water. The abundance of gamma-MOB at 13 m depth was comparable to that of the alpha-MOB, but gamma-MOB were more abundant than the putative alpha-MOB in the oxic waters at 9 m and anoxic waters at 10 m (Figure 2). Gamma-MOB were round, ca. 2 μm in diameter and free living (Figure 3b).
Consistent with CARD-FISH analyses, genes encoding for aerobic particulate methane monooxygenase (pmoA) could be amplified from the oxic waters, the anoxic chemocline and even the underlying sulphidic waters. All retrieved pmoA sequences belonged to gamma-proteobacterial Type I methanotrophs (Figure 3c). Despite using three different primer pairs and samples from two independent sampling campaigns, no alpha-proteobacterial Type II pmoA sequences were retrieved. Similarly, no pmoA genes of Candidatus Methylomirabilis oxyfera were retrieved. These observations concur with those from Lake Rotsee and Lake Lugano, where only Type I gamma-MOB pmoA genes were found (Schubert et al., 2010; Blees et al., 2014).
The inability to detect Type II MOB-related pmoA genes appears at first sight at odds with our CARD-FISH results, which indicate an abundance of putative alpha-MOB. In principle, this discrepancy could be explained by unspecific binding of the Malpha450 FISH probe, insufficient coverage of the applied pmoA primers or insufficient sequencing depths. Other studies have resolved similar contradictory results by attributing a methylotrophic rather than a methanotrophic lifestyle to the putative alpha-MOB (Wang et al., 2004; Nercessian et al., 2005). A recent study investigating the evolution of alpha-MOB showed that several members of this group were not methanotrophs but rather facultative methylotrophs and non-methylotrophic chemoorganotrophs growing on substrates, such as sugars, alcohols and organic acids (Tamas et al., 2014). Thus the putative alpha-MOB in Lago di Cadagno might not grow on methane but rather on other organic substrates. Consistent with this hypothesis, we could confirm the presence of putative alpha-MOB in the anoxic waters of Lago di Cadagno (Supplementary Figure S2) by using primers specific for 16S rRNA Type II alpha MOB (Type IIF/R; Chen et al., 2007). These retrieved alpha MOB-type sequences are closely related to several facultative methylotrophic and non-methylotrophic chemoorganotrophic organisms.
All obtained Type I pmoA sequences retrieved from the oxic water, anoxic chemocline and sulphidic bottom water were closely related and belonged to the family Methylococcaceae (Figure 3c). The nearest cultured representatives were known gamma-proteobacterial aerobic methane oxidizers, such as Methylomonas spp., Methylobacter spp., Methylomicrobium spp. and Methylovulum miyakonense. Related were also many environmental sequences from freshwater lake sediments, including Lake Washington (Nercessian et al., 2005), Lake Constance (Bussmann et al., 2004) and Lake Pavin (Biderre-Petit et al., 2011). Despite using two different primer pairs, no true soluble methane monooxygenase-encoding genes (mmoX) were found, consistent with reports on the absence of soluble monooxygenase in most Type I methanotrophs (Bowman, 2006). The absence of detectable Type II mmoX genes is in line with the fact that the putative alpha-MOB in Lago di Cadagno might not be growing on methane.
Active growth of gamma-MOB on methane
Despite being obligate aerobes, some MOB are capable of surviving long periods of anoxia (Roslev and King, 1995; Hanson and Hanson, 1996). We used nanoSIMS to confirm that the abundant aerobic MOB found in the anoxic water column were actively growing in the apparent absence of oxygen. For this, water samples were incubated under in situ conditions with 13C-labelled methane, and single cells were analysed for methane assimilation.
Although both alpha- and gamma-MOB were found by CARD-FISH, only gamma-MOB showed a strong 13C enrichment with an average 13C/12C ratio of 0.22 (that is, 18 atomic percent (AT%) 13C) at 13 m and 1.1 (that is, 52 AT % 13C) at 10 m (Figure 4 and Supplementary Figure S3). Although the oxidation of 13CH4 in our incubations would produce 13CO2 that could subsequently be assimilated into the biomass as well, the amount of produced 13CO2 was considered negligible due to the short incubation time (that is, ∼0.2 AT% 13C-enrichment of dissolved inorganic carbon after 5 days). This conclusion is further supported by the lack of detectable 13C in the cells of autotrophic bacteria (such as the abundant green sulphur bacteria Chlorobium clathratiforme) in the same incubation (Figure 4).
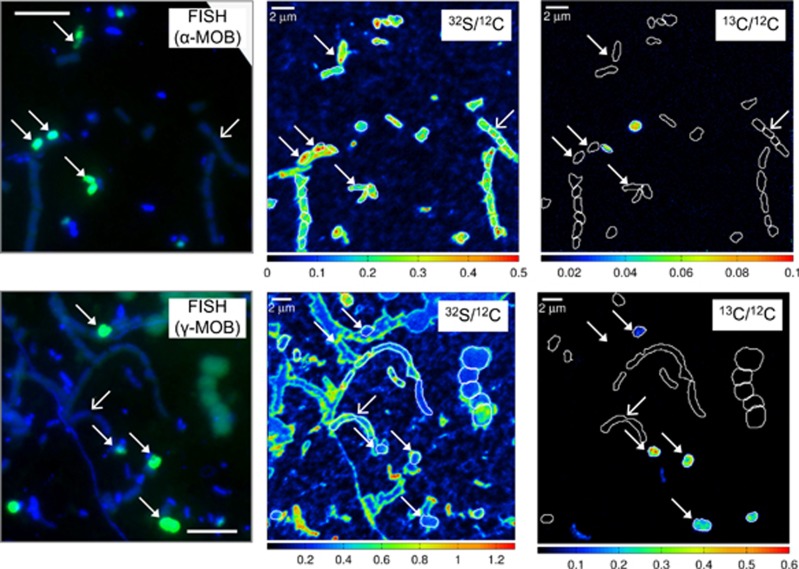
Figure 4.
Methane assimilation by single cells of alpha (upper panel) and gamma (bottom panel) MOB from 13 m depth. Full arrows indicate hybridized MOB, empty arrows indicate cells of Chlorobium clathratiforme, identified based on morphology. Scale bar on fluorescent images represents 5 μm. Corresponding 32S/12C and 13C/12C nanoSIMS images show distribution of cell biomass and assimilation of 13C-labelled methane, respectively. Note different scales of nanoSIMS ratios. Scale bar on nanoSIMS images represents 2 μm.
The uptake of labelled methane by gamma-MOB was observed in the anoxic chemocline (at 10 m, Supplementary Figure S4) as well as in the sulphidic zone of the water column (at 13 m, Figure 4). The methane assimilation rates were ca. five times higher at the chemocline than at 13 m depth (5.9 fmol C per day vs 1.2 fmol C per day) but both could support growth of gamma-MOB with average doubling times of ca. 5 and 16 days, respectively. These calculated doubling times are substantially longer than the reported doubling times of cultivated relatives (for example, Methylococcus capsulatus) under laboratory conditions that are in the range of several hours (for example, Patel and Hoare, 1971). This is likely a result of sub-optimal growth conditions due to varying supply of methane and oxygen as well as the low in situ temperatures in the environment. The average recorded 13C/12C ratio of putative alpha-MOB (that is, hybridized with Malpha450 probe) in the incubation from 13 m depth (0.011) was close to natural 13C abundance values of bulk biomass, indicating that in our in vitro incubations putative alpha-MOB did not assimilate detectable amounts of methane carbon. The average recorded 13C/12C ratio of putative alpha-MOB in the incubation from 10 m depth (0.026) was higher than natural abundance 13C but not different from 13C-enrichment in other (unidentified) bacteria. Because of the utilization of serine cycle for biomass formation, up to 50% of the cell material of putative alpha-MOB might be CO2 derived (Trotsenko and Murrell, 2008). However, even with this consideration in mind the observed uptake of methane-derived 13C was too low to prove active growth of putative alpha-MOB in the investigated depths. Our nanoSIMS results are thus consistent with our molecular studies, which also failed to detect alpha-proteobacterial pmoA in situ (Figure 3c). These results are also consistent with previous studies of other freshwater lakes (for example, Blees et al., 2014), rice-field soils and wetlands (Noll et al., 2008; Qiu et al., 2008; Bodelier et al., 2013) where predominantly gamma-MOB were found to be active.
Our nanoSIMS analyses provide clear evidence that the aerobic gamma-MOB were actively growing at and below the anoxic chemocline of Lago di Cadagno. As such, they are the most likely candidates responsible for methane oxidation in these zones. A previous study investigating active diazotrophic communities within the chemocline and hypolimnion of Lago di Cadagno has also reported active nitrogenase (nifH) gene expression by Type I methanotrophs (Halm et al., 2009). This further supports an active role of these obligately aerobic organisms in the carbon and nitrogen cycling in the anoxic waters of Lago di Cadagno.
A proposed mechanism for methane oxidation at the chemocline of Lago di Cadagno
The abundance of active obligate aerobic gamma-MOB in the anoxic water of Lago di Cadagno poses the question how oxygen-dependent oxidation of methane can operate in an apparently anoxic environment. A bacterium, Candidatus Methylomirabilis oxyfera, has recently been reported to perform aerobic methane oxidation under apparently anoxic conditions where molecular oxygen (together with N2) is produced intracellularly from NOx (Ettwig et al., 2010). Nutrient analyses showed that nitrite was non-detectable throughout the whole water column of Lago di Cadagno (data not shown), and the production of nitrite from nitrate was also unlikely as nitrate could not be detected <12.5 m depth (Halm et al., 2009). We also could not retrieve pmoA sequences belonging to Candidatus Methylomirabilis oxyfera. Nonetheless, we further investigated the potential for nitrite-driven methane oxidation in the hypolimnion of Lago di Cadagno with in vitro substrate-addition experiments. The additions of nitrite (20 μmol l−1 final concentration) to water collected from the anoxic depth did not stimulate methane oxidation but rather led to systematically lower methane-oxidation rates when compared with untreated controls (Supplementary Figure S5). Moreover, the consumption of methane and nitrite followed a fixed ratio of 1 mol CO2 per 2 mol N2 produced (Supplementary Figure S5), which was substantially lower than the ratio of 3:4 expected from the stoichiometry of nitrite-dependent methane oxidation. Based on these multiple independent lines of evidence, we conclude that nitrite-driven methane oxidation (if occurring at all) does not have a major role in methane removal at the chemocline of Lago di Cadagno. The consumption of nitrite in our incubation is more likely a result of canonical denitrification as described by Halm et al. (2009).
Flux calculations (Supplementary Figure S6) showed that total iron (−0.7 mmol m−2 d−1) and manganese (−1.2 mmol m−2 d−1) fluxes were much too low to oxidize methane (−9.8 mmol m−2 d−1) diffusing into the chemocline. Furthermore, additions of oxidized iron (in the form of ferrihydrite) and manganese (in the form of birnessite) substantially decreased or completely inhibited methane oxidation in our in vitro incubations (data not shown). In depths where methane oxidation occurred, only sulphate was present in sufficient amounts to oxidize the available methane. We could not detect methane-oxidizing archaea responsible for canonical sulphate-dependent AOM in our samples. Further in vitro experiments also showed that the amendment of sulphate reduced the rates of methane oxidation (Supplementary Figure S5). Moreover, the observed sulphide production was not linked to methane oxidation in the 1:1 stoichiometry (that is, 1 mol HS produced for 1 mol CH4 oxidized; Supplementary Figure S5) expected for AOM coupled to sulphate reduction. Given the abundance of sulphate-reducing bacteria in these depths (up to 23% of the total bacterial community; Peduzzi et al., 2003), we speculate that the consumption of sulphate in our anoxic incubations might be a result of ongoing organoclastic sulphate reduction.
In contrast to amendments of anaerobic electron acceptors, additions of oxygen to our incubations led to an approximately fivefold increase in methane-oxidation rates at both suboxic and anoxic depths (1.12 μmol l−1 day-1 at 11 m and 0.12 μmol l−1 day−1 at 13 m depth, Figure 5). This is intriguing, as oxygen measurements performed by us (Figure 1 and Supplementary Figure S1) and others (Wagener et al., 1990; Camacho et al., 2001) repeatedly failed to detect oxygen in the water at the chemocline. It is unlikely that the methane-oxidation rates observed in situ could be sustained through occasional oxygen intrusions, because Lago di Cadagno has been reported to remain strongly stratified, with the last potential mixing event in winter 1998/1999 (Tonolla et al., 2005). Given the high abundance of oxygenic photosynthetic algae (Figure 3a) and diatoms (Supplementary Figure S7) at and below the chemocline (Bossard et al., 2001), we propose that aerobic methane oxidation (as well as other aerobic processes) can be sustained in these anoxic waters via in situ production of oxygen by photosynthesis, similar to what has been hypothesized for surface wetland sediments (King, 1990). The light conditions at the chemocline appear suitable for active photosynthesis because up to 10% of the light in the photic zone is still available at the chemocline (Halm et al., 2009; Habicht et al., 2011). Notably, chlorophyll a measurements performed by us and others (Camacho et al., 2001; Habicht et al., 2011) show a chlorophyll a maximum at the chemocline (15.4 μg l−1 in August 2013), overlapping with the zone where oxygen disappeared. Indeed, high rates of oxygenic photosynthesis at the chemocline were measured previously (Camacho et al., 2001).
Figure 5.
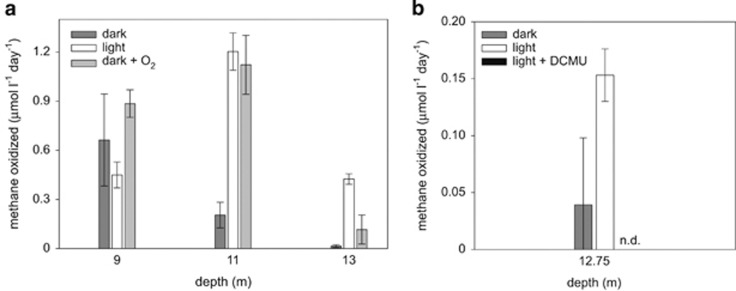
In vitro methane-oxidation rates in the dark, light, in the presence of oxygen and with DCMU, a photosynthesis inhibitor. (a) Incubation in light or addition of oxygen to incubations from anoxic depths increases the rates of methane oxidation. (b) After addition of DCMU, the rates of methane oxidation are not detectable (ND). Rates in panel (a) and (b) were measured during two different sampling campaigns in 2011 and 2013, respectively.
Additional in vitro incubations in the light were set up to confirm the effect of active photosynthesis on methane oxidation. These incubations were performed during the sampling campaign in August 2011 when the oxycline was found at 11 m (Supplementary Figure S1) and waters <11.5 m were anoxic (as confirmed by micro-optode measurements; Kirf et al., 2014). Control incubations in the dark showed the highest methane-oxidation rates (up to 0.7 μmol l−1 day−1) at an oxic depth (9 m), which abruptly ceased after ca. 30 h (data not shown), possibly due to the depletion of available oxygen. Incubation experiments from the oxic zone at 9 m depth did not show a significant light stimulation. Coinciding with the lower in situ oxygen concentrations, less methane was consumed in the dark incubations from the anoxic chemocline at 11 m (0.2 μmol l−1 day−1). At this depth, a fivefold stimulation of methane oxidation by light was observed. The rates (1.2 μmol l−1 day−1) in these anoxic incubations were similar to those obtained in the oxygen addition experiment (Figure 5a). In the sulphidic waters at 13 m, no methane oxidation was measured in the dark. Just like the addition of oxygen, light treatment also strongly enhanced rates of methane oxidation (up to 0.4 μmol l−1 day−1) in our anoxic incubations from 13 m depth (Figure 5 and Supplementary Figure S8). Differences in the overall methane-oxidation kinetics between the treatments most likely resulted from different oxygen availability during the course of the experiments (Supplementary Figure S8). In the oxygen treatment, oxygen was added as a single pulse at the beginning of the experiment, while the flux of dissolved oxygen resulting from photosynthesis was likely continuous during the light treatment.
A several fold increase in methane-oxidation rates in the light was also observed for the anoxic chemocline during a separate campaign to Lago di Cadagno in 2013 (Figure 5b). During this campaign, an additional experiment with DCMU, a photosynthetic inhibitor, was performed. DCMU addition resulted in non-detectable methane-oxidation rates in the light (Figure 5b).
Our molecular data combined with the results of the in vitro experiments indicate that methane removal at the anoxic chemocline of Lago di Cadagno is due to true aerobic oxidation of methane coupled to oxygenic photosynthesis. To the best of our knowledge, this is the first time that a coupling of these processes has been directly shown for a natural anoxic environments. Interestingly, microbial consortia of photosynthetic algae and MOB are employed in oxygen-deficient wastewater treatment plants to remove methane (Bahr et al., 2011; van der Ha et al., 2011, 2012). In the environment, this mechanism might occur in permanently and seasonally stratified lakes as well as marine basins such as the Black Sea, where light penetrates to the anoxic chemocline. In these anoxic waters, reduced mixing due to stratification leads to the accumulation of methane from the methanogenic sediments. Stratification, however, also reduces oxygen transport to the zone of methane oxidation. In situ oxygen production by photosynthesis overcomes this O2 limitation and as such provides ideal conditions for aerobic methane oxidizers. In these anoxic environments, oxygenic photosynthesis most likely also fuels other microaerobic processes, such as microbial sulphide and iron oxidation. We hypothesize that such an in situ production of non-detectable oxygen concentrations might have had an important role for life in the anoxic oceans of the early Earth (Holland, 2006).
Our combined results show that aerobic methane oxidation tightly coupled to oxygenic photosynthesis is a very efficient methane filter, almost completely consuming the methane flux in the permanently stratified Lago di Cadagno. Recent studies (Schubert et al., 2012 and references within) show that the vast majority of methane stored in the anoxic waters of seasonally stratified lakes is also effectively oxidized at the chemocline. Given the widespread occurrence of fully and seasonally stratified anoxic lakes, aerobic methane oxidation coupled to oxygenic photosynthesis might have an important but so far neglected role in regulating methane emissions from lakes.
Acknowledgments
We thank the personnel of Piora Centro Biologia Alpina, especially Mauro Tonolla, for making it possible to use the laboratory facilities and housing. We also thank Daniela Franzke, Gabi Klockgether, Gaute Lavik, Niculina Musat, Meri Eichner, Christiana Lungu, Frank Schreiber, Marc Strous and Martin Koenneke for their assistance in the field and in the laboratory. Tim Ferdelman and David Bastviken are thanked for discussions. Martin Schmid is thanked for calculating the diffusion coefficient and Beat Müller for flux calculations. Funding has been provided by the Swiss Federal Institute of Aquatic Science (Eawag), the Max Planck Society and Swiss National Science Foundation grants (20021_135299, 20020_153091, 128707).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Auman AJ, Lidstrom ME. Analysis of sMMO-containing Type I methanotrophs in Lake Washington sediment. Environ Microbiol. 2002;4:517–524. doi: 10.1046/j.1462-2920.2002.00323.x. [DOI] [PubMed] [Google Scholar]
- Bahr M, Stams AJM, De la Rosa F, García-Encina PA, Muñoz R. Assessing the influence of carbón oxidation-reduction in algal-bacterial photobioreactos. Appl Microbiol Biotechnol. 2011;90:1527–1536. doi: 10.1007/s00253-011-3204-8. [DOI] [PubMed] [Google Scholar]
- Bastviken D, Cole J, Pace M, Tranvik L. Methane emissions from lakes: dependence of lake characteristics, two regional assessments, and a global estimate. Global Biogeochem Cyc. 2004;18:1–12. [Google Scholar]
- Beal EJ, House CH, Orphan VJ. Manganese- and iron-dependent marine methane oxidation. Science. 2009;325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- Biderre-Petit C, Jezequel D, Dugat-Bony E, Lopes F, Kuever J, Borrel G, et al. Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol Ecol. 2011;77:533–545. doi: 10.1111/j.1574-6941.2011.01134.x. [DOI] [PubMed] [Google Scholar]
- Blees J, Niemann H, Wenk CB, Zopfi J, Schubert CJ, Kirf MK, et al. Micro-aerobic bacterial methane oxidation in the chemocline and anoxic water column of deep south-Alpine Lake Lugano (Switzerland) Limnol Oceanogr. 2014;59:311–324. [Google Scholar]
- Bodelier PLE, Meima-Franke M, Hordijk CA, Steenbergh AK, Hefting MM, Bodrossy L, et al. Microbial minorities modulate methane consumption through niche partitioning. ISME J. 2013;7:2214–2228. doi: 10.1038/ismej.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard P, Gammeter S, Lehmann C, Schanz F, Bachofen R, Bürgi HR, et al. Limnological description of the Lakes Zürich, Lucerne, and Cadagno. Aquat Sci. 2001;63:225–249. [Google Scholar]
- Bowman JP.2006The methanotrophs - the families Methylococcacae and MethylocystaceaeIn: Balows AT, Trüper HG, Dworkin M, Harder W, Schleifer KH, (eds)The Prokaryotes Springer: Berlin, Heidelberg, Germany; New York, NY, USA; 266–289. [Google Scholar]
- Bussmann I, Pester M, Brune A, Schink B. Preferential cultivation of type II methanotrophic bacteria from littoral sediments (Lake Constance) FEMS Microbiol Ecol. 2004;47:179–189. doi: 10.1016/S0168-6496(03)00260-5. [DOI] [PubMed] [Google Scholar]
- Camacho A, Erez J, Chicote A, Florin M, Squires MM, Lehmann C, et al. Microbial microstratification, inorganic carbon photoassimilation and dark carbon fixation at the chemocline of the meromictic Lake Cadagno (Switzerland) and its relevance to the food web. Aquat Sci. 2001;63:91–106. [Google Scholar]
- Chen Y, Dumont MG, Cebron A, Murrell JC. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environ Microbiol. 2007;9:2855–2869. doi: 10.1111/j.1462-2920.2007.01401.x. [DOI] [PubMed] [Google Scholar]
- Cline JD. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- Costello AM, Lidstrom ME. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol. 1999;65:5066–5074. doi: 10.1128/aem.65.11.5066-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, op den Camp HJM, et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol. 2008;10:3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Habicht KS, Miller M, Cox RP, Frigaard NU, Tonolla M, Peduzzi S, et al. Comparative proteomics and activity of a green sulfur bacterium through the water column of Lake Cadagno, Switzerland. Environ Microbiol. 2011;13:203–215. doi: 10.1111/j.1462-2920.2010.02321.x. [DOI] [PubMed] [Google Scholar]
- Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. Isolation and identification of methanogen-specific DNA from blanket bog feat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halm H, Musat N, Lam P, Langlois R, Musat F, Peduzzi S, et al. Co-occurrence of denitrification and nitrogen fixation in a meromictic lake, Lake Cadagno (Switzerland) Environ Microbiol. 2009;11:1945–1958. doi: 10.1111/j.1462-2920.2009.01917.x. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- Hinrichs KU, Boetius A.2002The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistryIn: Wefer G, Hebbeln DB, Jørgensen BB, Schlüter M, van Weering TCE, (eds). Ocean Margin Systems Springer-Verlag: Berlin, Germany; 457–477. [Google Scholar]
- Holland HD. The oxygenation of the atmosphere and oceans. Phil Trans R Soc B. 2006;361:903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler T, Wegener G, Knittel K, Boetius A, Brunner B, Kuypers MMM, et al. Substantial 13C/12C and D/H fractionation during anaerobic oxidation of methane by marine consortia enriched in vitro. Environ Microbiol Rep. 2009;1:370–376. doi: 10.1111/j.1758-2229.2009.00074.x. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- Holmes RM, Aminot A, Kerouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- Holtappels M, Lavik G, Jensen MM, Kuypers MMM.201115 N-Labeling experiments to dissect the contributions of heterotrophic denitrification and anammox to nitrogen removal in the OMZ waters of the OceanIn: Klotz MG, (ed). Methods in Enzymology (Research on Nitrification and Related Processes, Part A) Academic Press: Burlington, NJ, USA; 223–251. [DOI] [PubMed] [Google Scholar]
- Hutchens E, Radajewski S, Dumont MG, Mcdonald IR, Murrell JC. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol. 2004;6:111–120. doi: 10.1046/j.1462-2920.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Ann Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- King GM. Regulation by light of methane emissions from a wetland. Nature. 1990;345:513–515. [Google Scholar]
- Kirf MK, Dinkel C, Schubert CJ, Wehrli B. Submicromolar oxygen profiles at the oxic-anoxic boundary of temperate lakes. Aquat Geochem. 2014;20:39–57. [Google Scholar]
- Lidstrom ME, Somers L. Seasonal study of methane oxidation in Lake Washington. Appl Environ Microbiol. 1984;47:1255–1260. doi: 10.1128/aem.47.6.1255-1260.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, Zhu BL, van Alen TA, Butler MK, Diaz MR, Song B, et al. pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol. 2011;77:3877–3880. doi: 10.1128/AEM.02960-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IR, Kenna EM, Murrell JC. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environl Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, et al. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature. 2012;491:541–546. doi: 10.1038/nature11656. [DOI] [PubMed] [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Bacterial populations active in metabolism of C-1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol. 2005;71:6885–6899. doi: 10.1128/AEM.71.11.6885-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Frenzel P, Conrad R. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol Ecol. 2008;65:125–132. doi: 10.1111/j.1574-6941.2008.00497.x. [DOI] [PubMed] [Google Scholar]
- Patel RN, Hoare DS. Physiological studies of methane and methanol-oxidizing bacteria - oxidation of C-1 compounds by Methylococcus capsulatus. J Bacteriol. 1971;107:187–192. doi: 10.1128/jb.107.1.187-192.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduzzi S, Tonolla M, Hahn D. Vertical distribution of sulfate-reducing bacteria in the chemocline of Lake Cadagno, Switzerland, over an annual cycle. Aquat Microb Ecol. 2003;30:295–302. [Google Scholar]
- Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM. Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- Powell T, Jassby A. The estimation of vertical eddy diffusivities below the thermocline in lakes. Water Resour Res. 1974;10:191–198. [Google Scholar]
- Qiu QF, Noll M, Abraham WR, Lu YH, Conrad R. Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ. ISME J. 2008;2:602–614. doi: 10.1038/ismej.2008.34. [DOI] [PubMed] [Google Scholar]
- Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- Rasigraf O, Vogt C, Richnow HH, Jetten MSM, Ettwig KF. Carbon and hydrogen isotope fractionation during nitrite-dependent anaerobic methane oxidation by Methylomirabilis oxyfera. Geochim Cosmochim Acta. 2012;89:256–264. [Google Scholar]
- Reeburgh WS, Tyler SC, Carroll J. Stable carbon and hydrogen isotope measurements on Black Sea water-column methane. Deep-Sea Res Pt II. 2006;53:1893–1900. [Google Scholar]
- Reeburgh WS. Oceanic methane biogeochemistry. Chem Rev. 2007;107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- Rhee TS, Kettle AJ, Andreae MO. Methane and nitrous oxide emissions from the ocean: a reassessment using basin-wide observations in the Atlantic. J Geophys Res. 2009;114:1–20. [Google Scholar]
- Roslev P, King GM. Aerobic and anaerobic starvation metabolism in methanotrophic bacteria. Appl Environ Microbiol. 1995;61:1563–1570. doi: 10.1128/aem.61.4.1563-1570.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature. 2010;465:606–608. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- Schubert CJ, Diem T, Eugster W. Methane emissions from a small wind shielded lake determined by Eddy covariance, flux chambers, anchored funnels, and boundary model calculations: a comparison. Environ Sci Technol. 2012;46:4515–4522. doi: 10.1021/es203465x. [DOI] [PubMed] [Google Scholar]
- Schubert CJ, Lucas FS, Durisch-Kaiser E, Stierli R, Diem T, Scheidegger O, et al. Oxidation and emission of methane in a monomictic lake (Rotsee, Switzerland) Aquat Sci. 2010;72:455–466. [Google Scholar]
- Sivan O, Adler M, Pearson A, Gelman F, Bar-Or I, John SG, et al. Geochemical evidence for iron-mediated anaerobic oxidation of methane. Limnol Oceanogr. 2011;56:1536–1544. [Google Scholar]
- Springer E, Sachs MS, Woese CR, Boone DR. Partial gene sequences for the a subunit of methyl-coenzyme M reductase (McrI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol. 1995;45:554–559. doi: 10.1099/00207713-45-3-554. [DOI] [PubMed] [Google Scholar]
- Stookey LL. Ferrozine-a new spectrophotometric reagent for iron. Anal Chem. 1970;42:779–781. [Google Scholar]
- Storelli N, Peduzzi S, Saad MM, Frigaard NU, Perret X, Tonolla M. CO2 assimilation in the chemocline of Lake Cadagno is dominated by a few types of phototrophic purple sulfur bacteria. FEMS Microbiol Ecol. 2013;84:421–432. doi: 10.1111/1574-6941.12074. [DOI] [PubMed] [Google Scholar]
- Tamas I, Smirnova AV, He Z, Dunfield PF. The (d)evolution of methanotrophy in the Beijerinckiaceae—a comparative genomics analysis. ISME J. 2014;8:369–382. doi: 10.1038/ismej.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AS, Chu KH, Alvarez-Cohen L, Conrad ME. Variable carbon isotope fractionation expressed by aerobic CH4-oxidizing bacteria. Geochim Cosmochim Acta. 2006;70:1739–1752. [Google Scholar]
- Tonolla M, Peduzzi R, Hahn D. Long-term population dynamics of phototrophic sulfur bacteria in the chemocline of Lake Cadagno, Switzerland. Appl Environ Microbiol. 2005;71:3544–3550. doi: 10.1128/AEM.71.7.3544-3550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotsenko Y, Murrell JC.2008Metabolic aspects of aerobic obligate methanotrophyIn: Laskin AI, Sariaslani S, Gadd G, (eds). Advances in Applied Microbiology Elsevier: Amsterdam, the Netherlands; 183–229. [DOI] [PubMed] [Google Scholar]
- Uhde MAC.1992. Mischungsprozesse im hypolimnion des meromiktischen Lago di Cadagno: eine untersuchung mit hilfe natürlicher und künstlicher tracer. Unpublished MSc Thesis, Albert Ludwigs Universität Freiburg. Library Eawag: Duebendorf, Switzerland; p90 [Google Scholar]
- van der Ha D, Bundervoet B, Verstraete W, Boon N. A sustainable, carbon neutral methane oxidation by a partnership of methane oxidizing communities and microalgae. Water Res. 2011;45:2845–2854. doi: 10.1016/j.watres.2011.03.005. [DOI] [PubMed] [Google Scholar]
- van der Ha D, Nachtergaele L, Kerckhof M, Rameiyant D, Bossier P, Verstraete W, et al. Conversion of biogas to bioproducts by algae and methane oxidizing bacteria. Environ Sci Technol. 2012;46:13425–13431. doi: 10.1021/es303929s. [DOI] [PubMed] [Google Scholar]
- Wagener S, Schulz S, Hanselmann K. Abundance and distribution of anaerobic protozoa and their contribution to methane production in Lake Cadagno (Switzerland) FEMS Microbiol Ecol. 1990;74:39–48. [Google Scholar]
- Wang P, Wang F, Xu M, Xiao X. Molecular phylogeny of methylotrophs in a deep-sea sediment from a tropical west Pacific warm pool. FEMS Microbiol Ecol. 2004;47:77–84. doi: 10.1016/S0168-6496(03)00252-6. [DOI] [PubMed] [Google Scholar]
- Wankel SD, Adams MM, Johnston DT, Hansel CM, Joye SB, Girguis PR. Anaerobic methane oxidation in metalliferous hydrothermal sediments: influence on carbon flux and decoupling from sulfate reduction. Environ Microbiol. 2012;14:2726–2740. doi: 10.1111/j.1462-2920.2012.02825.x. [DOI] [PubMed] [Google Scholar]
- Wiesenburg DA, Guinasso NL. Equilibrium solubilities of methane, carbon monoxide, and hydrogen in water and sea-water. J Chem Eng Data. 1979;24:356–360. [Google Scholar]
- Wüest A. Interactions in lakes: biology as source of dominant physical forces. Limnologica. 1994;24:93–104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.