Abstract
Multidrug resistance among Gram-negative bacteria is conferred by three-component membrane pumps that expel diverse antibiotics from the cell. These efflux pumps consist of an inner membrane transporter such as the AcrB proton antiporter, an outer membrane exit duct of the TolC family, and a periplasmic protein known as the adaptor. We present the x-ray structure of the MexA adaptor from the human pathogen Pseudomonas aeruginosa. The elongated molecule contains three linearly arranged subdomains; a 47-Å-long α-helical hairpin, a lipoyl domain, and a six-stranded β-barrel. In the crystal, hairpins of neighboring MexA monomers pack side-by-side to form twisted arcs. We discuss the implications of the packing of molecules within the crystal. On the basis of the structure and packing, we suggest a model for the key periplasmic interaction between the outer membrane channel and the adaptor protein in the assembled drug efflux pump.
Gram-negative bacteria like Escherichia coli and Pseudomonas aeruginosa use three-component membrane pumps to export large protein toxins and small noxious chemicals, including antibacterial drugs, from the cell (1–4). They are therefore central to both pathogenicity and multidrug resistance. Pumps comprise diverse energized inner membrane transporters, including proton antiporters and ATPases, that form a constitutive complex with cognate periplasmic adaptor proteins (5–7). Substrate-laden inner membrane complexes recruit an outer membrane exit duct of the TolC family to assemble an active efflux pump that spans the entire bacterial cell envelope, simultaneously crossing both inner and outer membranes and the intervening periplasm (5).
X-ray crystallography at a 2.1-Å resolution (8) revealed that TolC is a trimeric 12-stranded α/β-barrel, comprising a 100-Å-long α-helical barrel that projects across the periplasmic space, embedded in the outer membrane by a contiguous 40-Å-long β-barrel. This structure establishes a 140-Å-long single pore open to the outside medium, but closed at its periplasmic entrance. Transition to the transient open state is achieved by an iris-like mechanism in which entrance α-helices undergo an untwisting realignment (9), suggested to be stabilized by interaction with the inner membrane transporter and adaptor protein. The architecture of an inner membrane drug efflux transporter has been revealed by the 3.5-Å resolution crystal structure (10) of the E. coli AcrB proton antiporter, a trimer with a 70-Å-long periplasmic domain and 50-Å-long transmembrane domain. The similarity in size and symmetry between the top of the periplasmic domain of AcrB and the bottom of the α-helical barrel of TolC has prompted the suggestion that these domains could make contact (10). Nevertheless, genetic analysis shows that assembly of functional drug efflux and protein export pumps requires the third component, the adaptor protein (6, 11). Indeed, direct interactions between all three components of efflux pumps have been detected by cross-linking studies, in both protein export (5) and antibiotic efflux systems (12).
Adaptor proteins form a broad family of conserved molecules, each of which operates with a cognate inner membrane transporter. The AcrA adaptor functions in complex with the AcrB transporter in E. coli whereas MexA and MexB form a homologous system in P. aeruginosa. Adaptors are anchored to the inner membrane by a single transmembrane helix, or by an N-terminal lipid modification. They contain a large periplasmic domain, which is predicted by sequence alignment studies to contain long α-helices (13). Whereas the adaptor proteins vary in length and content, the presence of these α-helices and a β-sandwich domain are thought to be common features (13).
It is envisaged that the adaptor has a key dynamic function, effecting a substrate-responsive coupling of opened TolC to the energy-providing transporter (1, 5). Several models have been suggested to explain this coupling. In the membrane fusion protein model, the two putative α-helices of the adaptor are predicted to function like a viral fusion protein, with coiled-coil formation driving the inner and outer membranes of the bacteria into close proximity, bringing together the opened exit duct and inner membrane transporter (6, 14). An alternative suggestion is that the α-helices of the adaptor protein form a stable coiled-coil (13). The transient open state of the TolC exit duct could then be established by putative α-helical hairpins of the adaptor repacking against the α-helical barrel of TolC (8). Here, we describe the crystal structure of the MexA adaptor from P. aeruginosa. We discuss the structure and the packing of molecules in the crystal and suggest a model for the role of the adaptor protein in the assembly of the efflux pump.
Methods
Protein Expression and Purification. Genomic DNA from the P. aeruginosa strain PAO1 was obtained from American Type Culture Collection (ATCC). The mexa gene (accession no. PA0425) was amplified by PCR by using Pfu Turbo polymerase (Stratagene). Two primers were used to amplify residues 24–383, thereby omitting the periplasmic signal sequence at the N terminus. In addition, Cys-24 was mutated to Ser, to prevent attachment of a fatty acid (forward primer, GGAATTCCATATGTCCGGAAAAAGCGAGGCGCCG; reverse primer, CGCGGATCCGCGTCAGCCCTTGCTGTCGGTTTT). This PCR produced a DNA fragment of 1,080 bp with unique restriction sites as the 5′ end (NdeI) and the 3′ end (BamHI). After restriction enzyme digestion, the fragment was ligated into pET15b (Novagen). This protocol generated a construct containing a gene for the Δ(1–23),C24S mutant of MexA with an N-terminal hexahistidine tag attached via a thrombin cleavage site.
Protein labeled with selenomethionine was produced by using the method of Ramakrishnan and Biou (15). The construct was transformed into the methionine-requiring auxotroph strain B384(DE3) and grown in minimal medium containing selenomethionine as the sole methionine source. Cells were grown at 30°C until the culture reached an absorbance at 600 nm of 0.8. They were induced by the addition of 50 μM isopropyl-β-d-thiogalactopyranoside and were harvested after 3 h and frozen in liquid nitrogen.
For purification, cells were resuspended in 20 mM MgCl2 and 20 mM Tris (pH 8.0) and lysed by three passages through a French press. After centrifugation at 125,000 × g for 20 min, the lysate was loaded onto a Q-Sepharose column (Amersham Pharmacia Biotech). MexA did not bind to the column. Unbound material was pooled, and imidazole was added to a final concentration of 5 mM. After addition of 2 ml Ni-NTA agarose (Qiagen, Valencia, CA), the mixture was incubated for 3 h with stirring at room temperature. Beads were washed twice with 50 ml of 20 mM MgCl, 20 mM Tris (pH 8.0), and 20 mM imidazole and resuspended in thrombin cleavage buffer (20 mM Tris, pH 8.3/150 mM NaCl/2.5 mM CaCl2). Seventy-five units of thrombin (Sigma) was added, and the protein was incubated overnight at 4°C. Cleavage efficiency was in excess of 90%. Beads were removed by centrifugation, and the protein was concentrated by using a Centricon Plus-20 (Millipore) to a volume of <1 ml. Gel filtration on a Superdex 200 gel filtration column (Amersham Pharmacia Biotech), with a buffer containing 50 mM NaCl and 20 mM Tris (pH 8.0), resulted in a single peak with a molecular weight consistent with a monomer of MexA. This protein was concentrated to 10 mg/ml by using a Centricon Plus-20 for crystallization.
Crystallization and Data Collection. Crystals were grown by using the hanging-drop vapor diffusion method. Droplets contained 2 μl of protein, 2 μl of well solution [50 mM ammonium sulfate/10% polyethylene glycol (PEG) 3350], and 0.5 μl of 0.1 mM n-tetradecyl-β-d-maltoside. Crystals grew to full size of 0.4 × 0.4 × 0.4 mm in 48 h at 18°C. Before freezing, they were dehydrated by transfer into a 10-μl droplet of 10% PEG 3350, 0.4 M ammonium sulfate, and 5 mM 2-mercaptoethanol for 5 min. This treatment increased diffraction limits from 9 Å to 3 Å resolution and reduced mosaicity. To prevent cracking, cryoprotectant was added slowly. A mixture of 30% glycerol, 0.4 M ammonium sulfate, 5 mM 2-mercaptoethanol, and 10% PEG 3350 was added to the droplet containing the crystal over 20–30 min to bring the final concentration of glycerol to in excess of 25%. Crystals were frozen in nylon loops by plunging into liquid nitrogen. Multiple anomalous diffraction (MAD) data were collected at 100 K from a single crystal at beamline ID29 at the European Synchrotron Radiation Facility (Grenoble, France).
Structure Determination and Refinement. Data were processed by using MOSFLM and SCALA from the ccp4 suite (16). The crystal belonged to space group P21 with cell dimensions a = 130.5 Å, b = 183.6 Å, c = 213.3 Å, and β = 107.4°. There were 13 molecules in the asymmetric unit, with a solvent content of 65%. The initial sites of 18 selenium atoms were found by using shelxd (17) and refined by using sharp (18) to a figure of merit of 0.31. The residual map showed the location of the remaining 8 sites. Density modification increased the overall figure of merit to 0.84. The resultant experimental map had interpretable, continuous density for residues 29–259 (mature protein); MexA has 360 residues in total. The reminder of the molecule is not ordered in the crystal. Model building was done in o with each monomer built individually (19), and the model was refined by using the remote data of the selenomethionine-substituted protein, in cns by using noncrystallographic symmetry restraints (20). The final model contained residues 29–259 of all 13 molecules in the asymmetric unit, with 86% of residues in the most favorable region of the Ramachandran plot and none in the disallowed region. The final Rcryst factor and Rfree were 27.3% and 28.5%, respectively (Table 1).
Table 1. Summary of statistics.
| Remote | Peak | Inflection | |
|---|---|---|---|
| Data collection and processing | |||
| Wavelength, Å | 0.9322 | 0.9792 | 0.9793 |
| Resolution, Å | 95–3.0 | 95–3.0 | 95–3.0 |
| (3.15–3.0) | (3.15–3.0) | (3.15–3.0) | |
| Reflections | 722,611 | 654,138 | 691,044 |
| Unique reflections | 189,293 | 188,683 | 189,056 |
| Completeness, % | 97.8 (90.6) | 97.9 (93.8) | 98.8 (97.9) |
| Multiplicity | 3.8 (3.4) | 3.5 (2.8) | 3.7 (3.2) |
| I/σ(I) | 9.7 (2.0) | 8.5 (1.5) | 8.5 (1.5) |
| Rmerge | 9.1 (45.1) | 9.4 (46.5) | 9.6 (65.5) |
| Refinement | |||
| Resolution (Å) | 500–3.0 | ||
| Reflections | 189,530 | ||
| Rcryst | 27.3% | ||
| Rfree (from 5% of reflections) | 28.5% | ||
| No. of protein atoms in model | 23,049 | ||
| No. of solvent atoms in model | 78 | ||
| rmsd bond angles | 0.0088 Å | ||
| rmsd bond lengths | 1.47 Å | ||
Data in parentheses show statistics for the 3.15- to 3.00-Å resolution range. rmsd, rms deviation.
Results and Discussion
The Structure of the MexA Monomer. Various adaptors from E. coli and P. aeruginosa were expressed in a soluble form with the N-terminal Cys residue (residue 1 of the protein after removal of the periplasmic signal sequence) mutated to Ser to prevent addition of the fatty acid. Both MexA and its homologue AcrA are functional in the absence of this membrane linkage (14, 21) and behave as monomers in solution. Crystallization trials were successful for MexA, and data were collected from crystals of selenomethionine-substituted protein to a 3.0-Å resolution. Multiple anomalous diffraction phasing allowed determination of an electron density map. The asymmetric unit of the MexA crystal contained 13 molecules. The mature processed form of MexA comprises 360 residues. Residues 29–259 of each monomer could be built in the structure. These residues correspond to residues Thr-52-Leu-282 of unprocessed MexA containing the N-terminal periplasmic signal sequence. Therefore 28 N-terminal and 101 C-terminal residues of the mature protein were not ordered in the crystal. However, they were present after crystallization, as determined by SDS/PAGE. Both isothermal titration calorimetry (ITC) interaction analysis and domain-swapping experiments suggest that the C-terminal domain encompassing the missing residues determines interaction with the inner membrane component (13, 22).
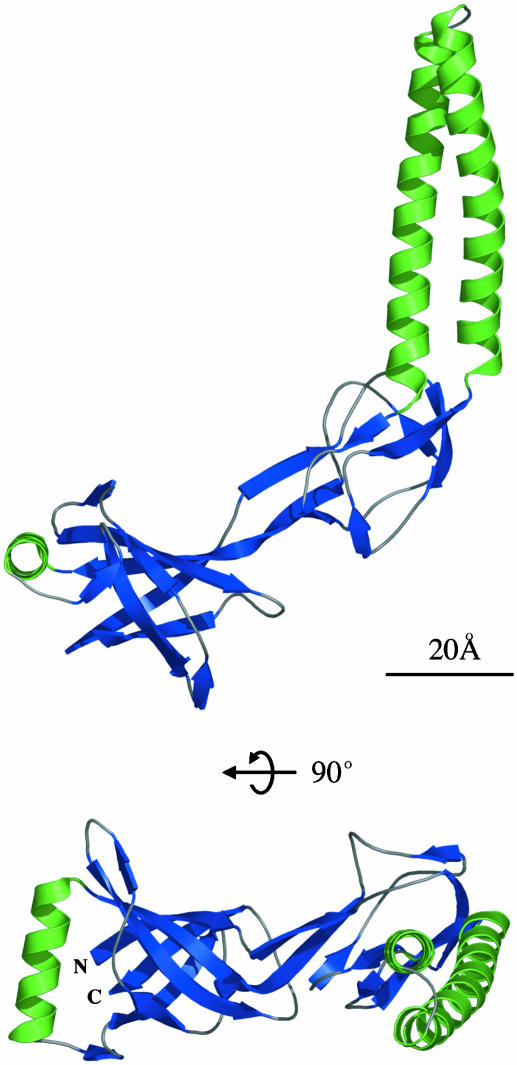
Each MexA monomer forms an elongated structure with a length of 89 Å and a maximum width of 35 Å (Fig. 1). It consists of three linearly arranged subdomains: a β-barrel, a lipoyl domain, and a 47-Å-long α-helical hairpin. The β-barrel domain contains six anti-parallel β-strands with a single α-helix situated at one entrance to the barrel. A similar topology, with a small β-barrel or pair of β-sheets capped at one end by an α-helix, is often found in domains involved in ligand binding. The flavin adenine dinucleotide (FAD)-binding domains of enzymes such as flavodoxin reductase (23) bind FAD in a cleft between two strands of the β-barrel, close to the α-helix. Other domains, such as the pleckstrin homology domain (24), odorant-binding domains (25), and the isomerase FKBP (26) bind ligands at the entrance to the barrel, opposite to the α-helix. Because one adaptor protein, EmrA, has been reported to bind drug molecules directly (27), it is possible that the β-barrel domain may take part in binding of efflux substrates.
Fig. 1.
Two perpendicular views of the Cα trace of the MexA monomer [residues Thr-29(N)-Leu-259(C), numbering from the mature protein]. The molecule is colored by secondary structure, with α-helices green and β-strands blue. In both cases, the α-helical hairpin is to the right, the lipoyl domain central, and the β-barrel to the left.
As predicted by Johnson and Church (13), the second domain is a β-sandwich that is structurally homologous to lipoyl and biotinyl domains. The carbonyl chain has a root mean square deviation (rmsd) of only 1.4 Å from that of the biotinyl domain of acetyl-CoA carboxylase (28) and an rmsd of 1.6 Å from that of the lipoyl domain of pyruvate dehydrogenase (29). The β-sandwich domain of pyruvate dehydrogenase consists of two interlocking lipoyl motifs, each containing four β-strands. These are linked by a short loop. The MexA lipoyl domain similarly comprises two interlocking lipoyl motifs of four β-strands. These two motifs are conserved throughout the family of adaptor proteins but are separated by variable lengths of intervening sequence. In MexA, the linking sequence is 64 residues long and forms the third (coiled-coil) domain.
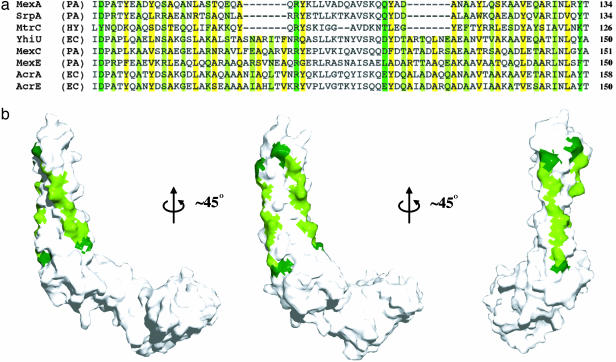
This third domain is a 47-Å-long α-helical hairpin. Whereas its C-terminal helix is straight, the N-terminal helix shows a left-handed superhelical twist (Fig. 1 Lower). The core of the hairpin is formed by residues in the a and d positions of the helical heptad repeats, which contain many large, hydrophobic side-chains. These residues are conserved throughout the family of periplasmic adaptors (13) (Fig. 2a). The exposed faces of the two helices, directly opposite the core of the hairpin, also contain conserved residues. Ala is found predominantly in the f position whereas the c position contains mainly hydrophilic residues such as Ser or Glu. When plotted on the molecular surface (Fig. 2b), these conserved small residues form stripes along the exposed sides of the hairpin. At either end of these stripes lie large hydrophilic residues with the potential to engage in hydrogen bonding (Fig. 2a). At the top of the hairpin lie Arg-96 and Gln-109 whereas Asp-72 and Tyr-134 are found at its base.
Fig. 2.
Crystallographic interactions between neighboring MexA molecules are mediated by conserved residues down the exposed surfaces of the MexA hairpin. (a) An alignment of the hairpin regions of eight drug efflux adaptors, adapted from Johnson and Church (13). Residues lying in the a and d positions of the hairpin are highlighted in yellow. Small residues on the exposed surfaces in position c and f of the hairpin are light green, with dark green indicating large hydrogen-bonding residues at the ends of these exposed surfaces. PA, P. aeruginosa; HY, Helicobacter pylori; EC, E. coli. The residue numbers are for mature proteins, after removal of the periplasmic signal sequences. (b) Three views of a surface representation of the MexA monomer. Small conserved residues are again colored light green whereas the larger hydrogen-bonding residues at either end of the conserved region are dark green.
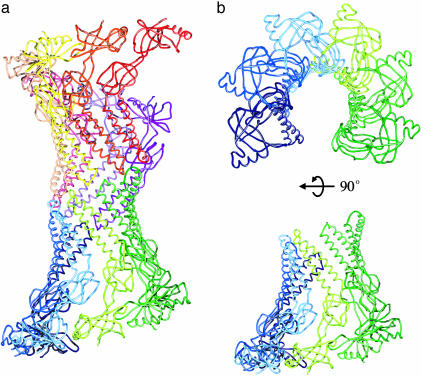
Side-by-Side Packing of MexA in the Crystal. Whereas MexA behaves as a monomer in solution, molecules pack within the crystal to form a complex cylinder of α-helices with domains rich in β-sheet extending outwards from the central axis (Fig. 3a). This cylinder is formed from two arcs, containing six and seven monomers. Each arc resembles a broken, twisted funnel (Fig. 3b) with α-helical hairpins packing to form the narrow funnel stem. The β-sheet-containing domains interact with their neighbors by a network of hydrogen bonds to form the wider part of the funnel. The loops at the top of the hairpins of the two arcs pack together in a head-to-head arrangement.
Fig. 3.
The arrangement of MexA molecules within the crystal. (a)Cα traces of the 13 molecules in the asymmetric unit are shown as lines with each molecule a different color. This complex cylinder is formed from two arcs of 6 or 7 molecules. (b) The Cα trace of the lower arc of 6 MexA molecules viewed from the top and side.
This arrangement of molecules within the asymmetric unit of the crystal reveals the propensity of MexA monomers to pack tightly side-by-side, with interaction interfaces formed by stripes of conserved residues that lie on the exposed faces of the α-helical hairpins (Fig. 2). The small, hydrophilic side chains interact with those of the neighboring hairpins through “knobsin-holes” packing characteristic of the arrangement of residues in the core of a conventional coiled-coil (30, 31).
A Model for the Role of MexA in an Assembled Efflux Pump. Several models have been suggested for the role of the adaptor protein in the assembly of tripartite efflux pumps (13). The membrane fusion model requires the adaptor to span the periplasm in the inactive complex, with the N terminus anchored in the inner membrane and the C terminus interacting with the outer membrane component (32). The structure shows that residues 29 and 259 of MexA lie within 5 Å of each other in the β-barrel domain, positioning the C terminus in close proximity to the N terminus, not near the outer membrane. Experimental evidence that the C-terminal domain contacts the inner membrane components (12, 22) also excludes it from interaction with the outer membrane. Furthermore, α-helical hairpin formation would need to occur reversibly, with the two long α-helices interacting to draw together the inner and outer membranes. The structure of MexA does not support this model, as it shows that a stable hairpin is formed from a large intervening sequence between the lipoyl motifs. For the two helices of this hairpin to come apart, as required in the membrane fusion protein model for MexA function, unfolding of both the β-sandwich and β-barrel domains would be necessary. A model of MexA function that requires reversible disruption of three stable domains is unlikely.
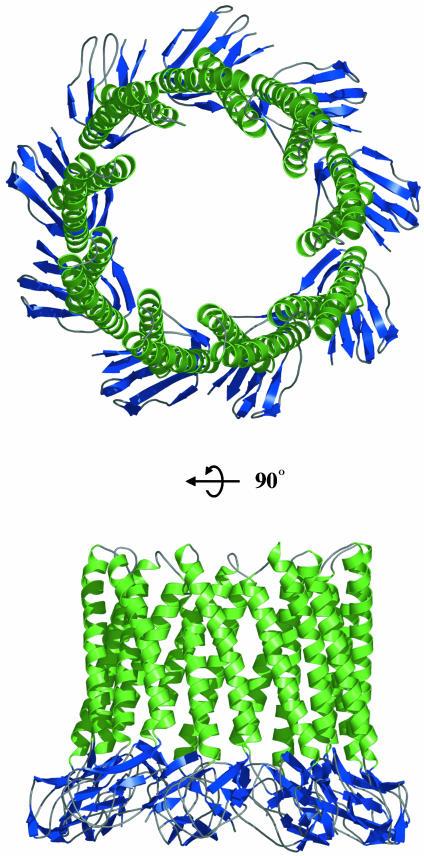
The structure of MexA and the parallel packing of molecules within the crystal suggest an alternative model for the arrangement of the adaptor protein in the assembled efflux apparatus (Fig. 4). Whereas two head-to-head arcs of molecules are observed in the asymmetric unit of the crystal, this arrangement is unlikely in vivo, where MexA is attached to the inner membrane by means of a fatty acid linkage. However, a single arc could interact with the inner membrane by means of the fatty acid at the N terminus, positioning α-helical hairpins that extend into the periplasm. The propensity of MexA to pack side-by-side allows modeling of a ring of MexA molecules. A ring formed from nine MexA monomers has a curvature similar to that observed in the crystal packing. In addition, this arrangement would match the known 3-fold symmetry of the outer membrane exit duct, TolC, and the inner membrane transporter, AcrB. Within this ring, the α-helical hairpins interact closely by means of the stripes of conserved small residues to form an α-barrel. Indeed, an algorithm that searched the sequence database for α-helical barrel-forming proteins identified members of the periplasmic adaptor family as likely to assemble into such a barrel (33). Whereas such a schematic model of a MexA oligomer is plausible, it is not yet supported by biochemical information, and future data may establish a different oligomeric form, probably based on 3-fold symmetry. In contrast to the TolC and AcrB trimeric structures, there is uncertainty about the oligomeric state of the adaptor component. Electron microscopy indicated an outline of a spiral-shaped cylindrical structure (34) that is not dissimilar to the arcs seen in the 3D crystal packing but interpreted as a dimeric form, whereas a hexamer has been suggested based on comparative cellular levels of the component proteins (35). Cross-linking of in vivo complexes using the short arm chemical cross-linker disuccinimidyl glutarate (DSG) has identified adaptor trimers in both the drug efflux and protein export systems (5, 7).
Fig. 4.
A model for the assembly of the α-helical hairpins of the elongated domain of MexA into an α-helical barrel. A ring is modeled from nine such molecules, with α-helices shown in green and the β-strands of the lipoyl domain in blue.
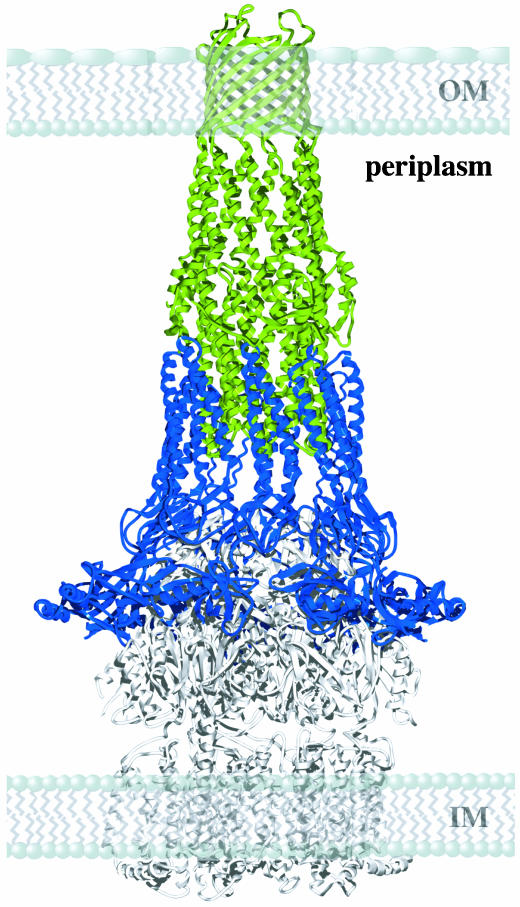
The modeled ring of nine MexA monomers is sufficiently large to form a sheath around the open state model of TolC (Fig. 5). During dynamic assembly of the active efflux pump, the hairpins of MexA could pack against the coiled-coils of the α-helical barrel of the TolC homologue OprM, inducing or stabilizing the open state (36). This ring could also surround the inner membrane transporter and so provide a seal to stabilize the complex, generating a conduit that spans the entire periplasm. The conserved nature of residues that mediate interactions between helical hairpins of neighboring MexA molecules (13) suggests that the formation of such a ring might be a conserved mechanism for the function of adaptor proteins.
Fig. 5.
A qualitative model for the interaction of a ring of MexA molecules with the α-helical barrel of TolC. The model shows both membranes and the periplasmic space. A ring of nine MexA molecules is shown in blue, and the open state model of TolC is green. A possible location for the inner membrane transporter is indicated by the structure of AcrB in white. IM and OM represent the locations of the inner and outer membranes. Not in the structure are the 28 residues of the N terminus and 101 residues of the C terminus of the mature protein. The missing N-terminal residues would be sufficient to bridge the gap between residue 29 of the model and the fatty acid attachment to the inner membrane at residue 1.
Whereas MexA has four heptad repeats in each helix, other adaptor proteins have five or six heptads and will form longer hairpins. In addition, adaptor proteins have different domains at the C terminus, thereby accommodating inner membrane transporters that differ in the size of their periplasmic loops. For example, whereas the protein transporter HlyB has far smaller periplasmic domains than MexB, the cognate periplasmic adaptor HlyD is larger than MexA (13). The elongated structure and modular nature of periplasmic adaptors allows them to use a conserved domain to mediate interactions with conserved outer membrane exit ducts, while using distinct modules to interact with diverse inner membrane transporters (1, 13).
The crystal structures of MexA and the TolC exit duct indicate a model for the key periplasmic interactions that lie at the core of an assembled efflux pump. This assembly would result in a transient 870-kDa transenvelope complex that presents a continuous export pathway over 270 Å in length, allowing the bacterial cell to expel drugs, protein toxins, and other molecules without release into the periplasmic space.
Acknowledgments
We thank J. Li for considerable help with data collection and structure determination and G. Schertler for substantial support and advice. Thanks also to P. Edwards for help with data collection, T. Warne for assistance with protein purification, J. Love for help with figure preparation, and R. Henderson, D. Stock, P. Evans, R. Williams, K. Nagai, and A. Crowther for valuable discussions. We also thank G. Leonard and the staff at beamline ID29 of the European Synchrotron Radiation Facility, Grenoble, France. We thank the Medical Research Council and the Biotechnology and Biological Sciences Research Council for support. M.K.H. was partially funded by Human Frontier Science Program Grant RGP 0054/2002.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1T5E).
References
- 1.Anderson, C., Hughes, C. & Koronakis, V. (2000) EMBO Rep. 1, 313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido, H. (2000) Trends Microbiol. 8, 481-483. [DOI] [PubMed] [Google Scholar]
- 3.Poole, K. (2001) Curr. Opin. Microbiol. 4, 500-508. [DOI] [PubMed] [Google Scholar]
- 4.Murakami, S. & Yamaguchi, A. (2003) Curr. Opin. Struct. Biol. 13, 443-452. [DOI] [PubMed] [Google Scholar]
- 5.Thanabalu, T., Koronakis, E., Hughes, C. & Kononakis, V. (1998) EMBO J. 17, 6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zgurskaya, H. I. & Nikaido, H. (1999) Proc. Natl. Acad. Sci. USA 96, 7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zgorskaya, H. I. & Nikaido, H. (2000) J. Bacteriol. 182, 4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. (2000) Nature 405, 914-919. [DOI] [PubMed] [Google Scholar]
- 9.Andersen, C., Koronakis, E., Bokma, E., Eswaran, J., Humphreys, D., Hughes, C. & Koronakis, V. (2002) Proc. Natl. Acad. Sci. USA 99, 11103-11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murakami, S., Nakashima, R., Yamashita, E. & Yamaguchi, A. (2002) Nature 419, 587-593. [DOI] [PubMed] [Google Scholar]
- 11.Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H. & Hearst, J. E. (1995) Mol. Microbiol. 16, 45-55. [DOI] [PubMed] [Google Scholar]
- 12.Touzé, T., Eswaran, J., Bokma, E., Koronakis, E., Hughes, C. & Koronakis, V. (June 16, 2004) Mol. Microbiol., 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed]
- 13.Johnson, J. M. & Church, G. M. (1999) J. Mol. Biol. 287, 695-715. [DOI] [PubMed] [Google Scholar]
- 14.Zgurskaya, H. I. & Nikaido, H. (1999) J. Mol. Biol. 285, 409-420. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan, V. & Biou, V. (1997) Methods Enzymol. 276, 538-557. [PubMed] [Google Scholar]
- 16.Collaborative Computational Project Number 4. (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 17.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 58, 1772-1779. [DOI] [PubMed] [Google Scholar]
- 18.La Fortelle, E. D. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 20.Brunger. A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama, H., Maseda, H., Kamiguchi, H. & Nakae, T. (2000) J. Biol. Chem. 275, 4628-4634. [DOI] [PubMed] [Google Scholar]
- 22.Elkins C. A. & Nikaido H. (2003) J. Bacteriol. 185, 5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingelman, M., Bianchi, V. & Eklund, H. (1997) J. Mol. Biol. 268, 147-157. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson, K. M., Lemmon, M. A., Schlessinger, J. & Sigler, P. B. (1995) Cell 83, 1037-1046. [DOI] [PubMed] [Google Scholar]
- 25.Vincent, F., Spinelli, S., Ramoni, R., Grolli, S., Pelosi, P., Cambillau, C. & Tegoni, M. (2000) J. Mol. Biol. 300, 127-139. [DOI] [PubMed] [Google Scholar]
- 26.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1991) Science 252, 839-842. [DOI] [PubMed] [Google Scholar]
- 27.Borges-Walmsley, M. I., Beauchamp, J., Kelly, S. M., Jumel, K., Candlish, D., Harding, S. E., Price, N. C. & Walmsley, A. R. (2003) J. Biol. Chem. 278, 12903-12912. [DOI] [PubMed] [Google Scholar]
- 28.Athappilly, F. K. & Hendrickson, W. A. (1995) Structure 3, 1407-1419. [DOI] [PubMed] [Google Scholar]
- 29.Berg, A., Vervoort, J & de Kok, A. (1997) Eur. J. Biochem. 244, 352-360. [DOI] [PubMed] [Google Scholar]
- 30.Crick, F. (1953) Acta Crystallogr. 6, 689-697. [Google Scholar]
- 31.Calladine, C. R., Sharff, A. & Luisi, B. (2001) J. Mol. Biol. 305, 603-618. [DOI] [PubMed] [Google Scholar]
- 32.Zgurskaya, H. I. & Nikaido, H. (2000) Mol. Microbiol. 37, 219-225. [DOI] [PubMed] [Google Scholar]
- 33.Sharff, A., Fanutti, C., Shi, J., Calladine, C. & Luisi, B. (2001) Eur. J. Biochem. 268, 5011-5026. [DOI] [PubMed] [Google Scholar]
- 34.Avila-Sakar, A. J., Misaghi, S., Wilson-Kubalek, E. M., Downing, K. H., Zgurskaya, H., Nikaido, H. & Nogales, E. (2001) J. Struct. Biol. 136, 81-88. [DOI] [PubMed] [Google Scholar]
- 35.Narita, S., Eda, S., Yoshihara, E. & Nakae, T. (2003) Biochem. Biophys. Res. Commun. 308, 922-926. [DOI] [PubMed] [Google Scholar]
- 36.Andersen, C., Koronakis, E., Bokma, E., Eswaran, J., Humphreys, D., Hughes, C. & Koronakis, V. (2002) Proc. Natl. Acad. Sci. USA 99, 11103-11108. [DOI] [PMC free article] [PubMed] [Google Scholar]