Abstract
Methanogenic hydrocarbon metabolism is a key process in subsurface oil reservoirs and hydrocarbon-contaminated environments and thus warrants greater understanding to improve current technologies for fossil fuel extraction and bioremediation. In this study, three hydrocarbon-degrading methanogenic cultures established from two geographically distinct environments and incubated with different hydrocarbon substrates (added as single hydrocarbons or as mixtures) were subjected to metagenomic and 16S rRNA gene pyrosequencing to test whether these differences affect the genetic potential and composition of the communities. Enrichment of different putative hydrocarbon-degrading bacteria in each culture appeared to be substrate dependent, though all cultures contained both acetate- and H2-utilizing methanogens. Despite differing hydrocarbon substrates and inoculum sources, all three cultures harbored genes for hydrocarbon activation by fumarate addition (bssA, assA, nmsA) and carboxylation (abcA, ancA), along with those for associated downstream pathways (bbs, bcr, bam), though the cultures incubated with hydrocarbon mixtures contained a broader diversity of fumarate addition genes. A comparative metagenomic analysis of the three cultures showed that they were functionally redundant despite their enrichment backgrounds, sharing multiple features associated with syntrophic hydrocarbon conversion to methane. In addition, a comparative analysis of the culture metagenomes with those of 41 environmental samples (containing varying proportions of methanogens) showed that the three cultures were functionally most similar to each other but distinct from other environments, including hydrocarbon-impacted environments (for example, oil sands tailings ponds and oil-affected marine sediments). This study provides a basis for understanding key functions and environmental selection in methanogenic hydrocarbon-associated communities.
Introduction
Since the dawn of the industrial age, anthropogenic activities have resulted in contamination of various soil, water and subsurface environmental systems with organic compounds including hydrocarbons. Methanogenic biodegradation of hydrocarbons is important in many environments such as contaminated groundwater, sediments and organic chemical waste disposal sites that are devoid of electron acceptors such as oxygen, nitrate, iron(III) and sulfate (Chang et al., 2005, Essaid et al., 2011; Lykidis et al., 2011). Methanogenic hydrocarbon biodegradation is also an important biogeochemical process in subsurface fossil energy reservoirs that, over geological time, has contributed to the formation of natural gas deposits and/or the conversion of light oil to heavy oil (Jones et al., 2008).
Hydrocarbon mineralization via methanogenesis involves the coordinated metabolism of syntrophic (fermentative) bacteria that catalyze the initial attack on hydrocarbon substrates with subsequent conversion to methanogenic substrates, and methanogens that produce CH4 (and CO2 and/or H2O) in a thermodynamically-interdependent manner (Gieg et al., 2014). Many previous studies have shown that microbial communities in methanogenic hydrocarbon-associated environments (for example, Chang et al., 2005; Kasai et al., 2005; Sun and Cupples, 2012; Wawrik et al., 2012; An et al., 2013) or enrichment cultures (Ficker et al., 1999; Fowler et al., 2012) can be highly diverse. Literature-wide compilations examining such communities (Gray et al., 2011; Strapoc et al., 2011; Kleinsteuber et al., 2012) have revealed that they frequently contain hydrogenotrophic (for example, Methanoculleus, Methanolinea, Candidatus Methanoregula, Methanospirillum), acetotrophic (for example, Methanosaeta), and/or methylotrophic methanogens (for example, Methanolobus; Wuchter et al., 2013). Deltaproteobacteria (for example, Syntrophus/Smithella, Desulfovibrio, Geobacter) and Firmicutes (for example, Desulfotomaculum, Desulfosporosinus, Pelotomaculum) are commonly abundant bacteria, and are often identified as the putative hydrocarbon-activating organisms (Gray et al., 2011; Kleinsteuber et al., 2012). Members of the Spirochaetes, Bacteroidetes, Chloroflexi and Betaproteobacteria are also frequently found, but generally in lower abundance (Strapoc et al., 2011; Kleinsteuber et al., 2012). Methanogenic hydrocarbon-degrading co-cultures, consisting of a single bacterium and archaeon, are difficult to obtain (Sieber et al., 2012), and therefore the specific roles of this wide diversity of organisms in methanogenic hydrocarbon metabolism remain cryptic. The specific community structure may depend on the hydrocarbon substrates, the available nutrients and/or specific biogeochemical characteristics. Genes encoding anaerobic hydrocarbon-degrading enzymes (for example, bssA, assA) have been detected in hydrocarbon-degrading methanogenic cultures (for example Aitken et al., 2013; Cheng et al., 2013; Fowler et al., 2012; Washer and Edwards, 2007) as well as genes involved in syntrophic processes such as H2 and formate transfer, and flagella, amino-acid and coenzyme biosynthesis (Kato et al., 2009; Walker et al., 2009). Despite recent reports of hydrocarbon-degrading co-cultures of sulfate reducers and methanogens (Callaghan et al., 2012; Lyles et al., 2014), the diversity of key enzymes and genes involved in methanogenic hydrocarbon metabolism are not yet well described.
In this study, we carried out metagenomic sequencing of three methanogenic cultures enriched from two different environments (an oil sands tailings pond and a hydrocarbon-contaminated aquifer) and on different hydrocarbon substrates. We examined the diversity of taxa and genes involved in various anaerobic hydrocarbon biodegradation pathways in the cultures to determine whether differences would be observed based on different inoculum sources and hydrocarbon substrates. Further, we used a comparative metagenomic approach to examine how certain metabolic genes and functions compared among the cultures themselves and with other environments including hydrocarbon-impacted sites. We hypothesized that the three cultures would be more functionally similar to each other than to other enrichment cultures and environments. We sought to identify the functions that distinguish these cultures from other hydrocarbon-impacted and anaerobic samples and enrichments to discern the hallmark features of methanogenic hydrocarbon-degrading communities.
Materials and methods
Methanogenic cultures and incubation conditions
The three methanogenic hydrocarbon-degrading cultures (naphtha-degrading culture (NAPDC), short chain alkane-degrading culture (SCADC) and toluene-degrading culture (TOLDC)) analyzed in this study were enriched from hydrocarbon-impacted environments on different hydrocarbons as the sole carbon and energy source(s) (Table 1). These cultures were maintained on the same substrates since their initial enrichment. Some metabolic features of the TOLDC and SCADC cultures have been reported (Fowler et al., 2012; Tan et al., 2013), while NAPDC is newly described here. NAPDC and SCADC were enriched (see Supplementary Information) from methanogenic mature fine tailings collected from Mildred Lake Settling Basin (MLSB), an oil sands tailings pond in Alberta, Canada that contains diverse low and high molecular weight hydrocarbons from unrecovered bitumen and naphtha (Siddique et al., 2006). NAPDC was enriched with 0.2% (v/v) naphtha (CAS no. 64742-49-0), a petroleum product comprising monoaromatic hydrocarbons and C6-C10 alkanes that is present in many oil sands tailings ponds; NAPDC cultivation methods are provided in Supplementary Information. SCADC was enriched on 0.1% v/v C6-C10 n-alkanes with minor proportions of 2-methylpentane, 3-methylpentane and methylcyclopentane, as described by Tan et al. (2013). TOLDC was established from aquifer sediments contaminated with gas condensate (containing predominantly C5-C15 hydrocarbons, Gieg et al., 1999) that was enriched solely on 0.01% (v/v) toluene (Fowler et al., 2012). The three cultures were incubated statically in the dark under mesothermic conditions (20–28 °C) and transferred to defined minimal salts freshwater medium at regular intervals (Table 1). Gas chromatography was used to monitor substrate depletion and methane production in the cultures (Fowler et al., 2012; Supplementary Information).
Table 1. Source of hydrocarbon-degrading methanogenic cultures and enrichment conditions.
| NAPDC | SCADC | TOLDC | |
|---|---|---|---|
| In situ conditions in environment | |||
| Sample environment | Mature fine tailings in oil sands tailings ponda | Mature fine tailings in oil sands tailings ponda | Gas condensate-contaminated aquifer sedimentsb |
| Location | Alberta, Canada | Alberta, Canada | Colorado, USA |
| Depth (m below surface) | 31 m | 35 m | 1.5 m |
| Bulk pH | 7–8 | 7–8 | 7–8 |
| Temperature | 12–20 °C | 12–20 °C | 15–20 °C |
| Redox condition | Anoxic | Anoxic | Anoxic |
| Incubation conditions for enrichment cultures | |||
| Initial inoculum % | 50% (v/v) | 50% (v/v) | 30% (v/v) |
| Number of transfers | 2 | 4 | >10 |
| Inoculum proportion transferred | 10% (v/v) | 50% (v/v) | 30% (v/v) |
| Substrate | 0.2% (v/v) Naphtha | 0.1% (v/v) C6-C10 n-alkanesc | 0.01% (v/v) Toluene |
| Culture medium | Anoxic mineral mediumd | Anoxic mineral mediumd | Anoxic mineral mediume |
| Initial medium pH | 7–8 | 7–8 | 7–8 |
| Initial headspace gas | 20/80% (v/v) CO2/N2 | 20/80% (v/v) CO2/N2 | 20/80% (v/v) CO2/N2 |
| Incubation temperature | 28 °C | 28 °C | 20–25 °C |
| Incubation time (before DNA extraction) | 120 days | 120 days | 150 days |
| Light exposure | Dark | Dark | Dark |
Abbreviations: NAPDC, naphtha-degrading culture; SCADC, short chain alkane-degrading culture; TOLDC, toluene-degrading culture.
Mildred Lake Settling Basin, Alberta, Canada (Siddique et al., 2007).
Ft. Lupton, Colorado, USA (Fowler et al., 2012).
Includes minor proportions of 2-methylpentane, 3-methylpentane and methylcyclopentane (Tan et al., 2013).
Per Widdel and Bak (1992) (anoxic freshwater mineral salts medium).
Per McInerney et al. (1979) (anoxic freshwater mineral salts medium).
DNA extraction for 16S rRNA gene pyrotag sequencing and metagenomic sequencing
DNA extraction from TOLDC and SCADC for amplification and pyrotag sequencing of 16S rRNA gene amplicons has been described (Fowler et al., 2012; Tan et al., 2013). Total DNA from NAPDC used for 16S rRNA gene pyrosequencing was extracted and PCR amplified performed as described by Foght et al. (2004) and Tan et al. (2013). All 16S rRNA gene pyrotag sequencing reads were generated using the same ‘universal' primer set, 926 F and 1392 R, targeting the V6-V8 regions of the 16S rRNA gene of bacteria and archaea (Engelbrektson et al., 2010).
For high throughput pyrosequencing of total DNA without amplification, 500 ml of TOLDC and NAPDC or 800 ml of SCADC were filtered, extracted with phenol-chloroform, and purified using cesium chloride centrifugation (Wright et al., 2009). Additionally, total DNA was obtained from two oil sands tailings ponds located in Alberta, Canada: MLSB (the same tailings pond used for enrichment of the NAPDC and SCADC cultures), and Tailings Pond 6 (TP6; Ramos-Padrón et al., 2011). These tailings samples were collected and handled as described by An et al. (2013) before total DNA isolation using the bead-beating methods described by Ramos-Padrón et al. (2011) for TP6 and Foght et al. (2004) for MLSB. DNA was sequenced on a half plate using the GS FLX Titanium Sequencing Kit XLR70 (Roche Diagnostics, Laval, QC, Canada) at McGill University and the Génome Québec Innovation Centre, Montreal, Canada. Detailed information about library construction is provided by An et al. (2013).
Downstream processing of 16S rRNA gene and metagenomic pyrosequencing reads
All 16S rRNA gene pyrotag data sets were submitted simultaneously to Phoenix 2 (Soh et al., 2013) for quality control, cluster analysis using the average neighbor joining method at 0.05 distance cutoff, and taxonomic classification using RDP classifier with SILVA Small Subunit rRNA Database release 108 (http://rdp.cme.msu.edu/). A lack of consensus for similarity thresholds used in cluster analysis exists (for example, 97% or 95%). In this study, a 95% similarity threshold for operational taxonomic unit (OTU) clustering was used to accommodate potential biases introduced through homopolymers and sequencing artifacts produced by 454 pyrotag sequencing (Kunin et al., 2010).
All metagenomic 454 pyrosequencing reads were subjected to quality control and de novo assembly using a Newbler v2.6-based pipeline developed in-house, as previously described (Tan et al., 2013). All quality-controlled metagenomic reads were submitted to MG-RAST for automated gene calling and annotation. Unassembled metagenomic reads were also subjected to taxonomic classification using the MG-RAST classifier with the following parameters: M5NR annotation, e-value ⩽1 × 10−10, alignment ⩾50 bp, identity ⩾60% (Meyer et al., 2008), and homology-based binning using SOrt-ITEMS (Haque et al., 2009). Additionally, unassembled reads were mapped to selected reference genomes (Supplementary Table S2) using Geneious Pro 7.05 (Biomatters Ltd., Auckland, New Zealand) with default settings. Metagenomic sequences from NAPDC, SCADC and TOLDC are available on the Short Read Archive under the SRA accession numbers SRX210874, SAMN01828453 and SRX210873. Those for the pyrotag data are SRX831148 (SCADC), SRX831147 (NAPDC) and SRX831099 (TOLDC).
Comparative analysis of functional categories in metagenomic data sets
Unassembled metagenomic sequences from NAPDC, SCADC and TOLDC (Metagenome Group 1; Supplementary Table S1) were annotated in MG-RAST V2 using SEED with the following parameters: e-value ⩽1 × 10 × 10; identity ⩾60% alignment length ⩾50 bp (Meyer et al., 2008). Additional metagenomes used in the comparison were chosen according to their environment type in MG-RAST. Both terrestrial and marine environments were selected, as the methanogenic cultures were derived from terrestrial environments previously associated with ancient marine environments before geological uplift. Eight additional metagenomes representing anaerobic dechlorinating enrichment cultures and hydrocarbon-impacted sediments (Metagenome Group 2; Supplementary Table S1) and 33 metagenomes from soil, sludge and marine and freshwater environments (Metagenome Group 3; Supplementary Table S1) were compared using principal component analysis based on the relative abundance of SEED functional categories (normalized against the total gene count of each metagenome) using R with the ade4 package (www.r-project.org). Metagenome Group 2 was further compared with Group 1 using STAMP (Parks and Beiko, 2010). For subsystem categories with rare counts (for example, ⩽0.1% of total counts), a fixed number of pseudocounts proportional to the total gene count was added to all categories across all groups used in comparisons to prevent selection of rare categories (Hug et al., 2012). For all comparisons, unassembled metagenomic reads were used unless stated otherwise.
Comparative analysis of anaerobic hydrocarbon degradation and other metabolic pathways
Specific genes encoding proteins involved in various anaerobic hydrocarbon-degrading pathways (Figure 2,Supplementary Table S3) and a variety of other metabolic pathways (Supplementary Figures S4 and S5) were sought in NAPDC, SCADC and TOLDC metagenomes using Hidden Markov Models (HMMs). Where available, Pfam profile HMMs (Finn et al., 2010) were used to identify protein families; in-house HMMs were made for genes for which no Pfam models existed and a minimum of four representative sequences were available in GenBank. To construct HMMs, amino-acid sequences of a gene were aligned using T-Coffee, and multiple alignments were manually curated using GeneDoc (Notredame et al., 2000). Edited alignments were used as an input to HMMER to create HMMs (Eddy, 1998). Models were then used to screen assembled metagenomic data sets (including singletons) with an e-value of <1 × 10−10. For genes with <4 representative sequences available, tBLASTn searches were performed on each of the metagenomes (e-value <1 × 10−10). Both HMM and BLAST-based methods were used, as each employs different methodologies in finding target genes, thereby minimizing the chances of missing genes of interest. Assembled sequences, which include singletons, were used for this analysis, as detection of full-length or near full-length genes was difficult using unassembled reads (since many of these can include intergenic regions or partial genes) and precluded gene quantitation. Therefore, due to differences in sequencing depth in the three metagenomes, and likelihood of key genes not being sequenced, we used ‘identification of the majority of genes in a pathway' (or enzyme-encoding subunits) as a more suitable indicator of the presence of a pathway or enzyme than individual gene abundance.
Phylogenetic analysis of putative fumarate addition enzymes
Illumina data from SCADC (Tan et al., 2013) were re-assembled de novo in the current study using CLC Genomics Workbench (CLC-Bio, Boston, MA, USA) with the default settings. Amino-acid sequences of genes encoding enzymes for fumarate addition to hydrocarbons (for example, assA, bssA and nmsA) were recovered from the SCADC Illumina sequence assembly using tBLASTn. All recovered sequences were aligned with fumarate addition genes detected in TOLDC and NAPDC metagenomes plus reference sequences from GenBank using MUSCLE V3.3 (Edgar, 2004). Poorly aligned regions were manually edited and indels were removed. Phylogenetic analysis was performed using PhyML (Guindon and Gascuel, 2003) with WAG model and 100 bootstrap replicates. Trees were visualized and edited in FigTree V14.0 (Rambaut, 2012).
Results and discussion
Features of the enrichment cultures and their metagenomes
In this study, we first compared the metagenomes of three hydrocarbon-degrading methanogenic enrichments that were derived from different locations, enriched for varying lengths of time, and/or amended with different hydrocarbon substrates. Such variations allowed us to determine whether varying selective pressures have an effect on the genetic potential for anaerobic hydrocarbon metabolism in methanogenic enrichment cultures.
As shown in Table 1, TOLDC was enriched from a hydrocarbon-impacted aquifer, and has been maintained solely on toluene as the carbon and energy source for >10 years (Fowler et al., 2012, 2014). SCADC and NAPDC were newer enrichments (~2 years old at time of analyses) that were derived from a methanogenic oil sands tailings pond, enriched on short-chain alkanes (SCADC), or a complex hydrocarbon mixture (naphtha) that contains both aromatic and aliphatic hydrocarbons (NAPDC). Supplementary Figure S1 shows that NAPDC consumed toluene, xylenes and iso-alkanes concomitant with methane production over 103 days. For all cultures, substrate-unamended controls produced low amounts of CH4 (Fowler et al., 2012; Supplementary Figure S1) indicating that the added hydrocarbons were the sources of methane.
Metagenomic 454 pyrosequencing of DNA from NAPDC, SCADC and TOLDC generated ~370 000, 670 000 and 550 000 quality-controlled reads, respectively (Table 2). Rarefaction analysis and assembly statistics revealed that TOLDC was more deeply sequenced than either NAPDC or SCADC, and that none of the cultures was sequenced to saturation (Supplementary Figure S2). The metagenomes assembled into approximately 8500 (NAPDC), 15 000 (SCADC) and 11 000 (TOLDC) contigs, with ~162 000, 327 000 and 179 000 singletons, respectively, and had GC contents of 49–53% (Table 2). The average assembled sequence lengths ranged from 1118 to 1863 bp but contigs varied greatly in length (200–28 000 bp). This likely reflects the relative abundance of different community members or the presence of closely related strains (for example, microdiversity), with more abundant members being better assembled due to a higher proportion of reads. The longest contigs ranged from 25.8 to 28.3 Kb, which is relatively short versus other comparable metagenomes (Hug et al., 2012), suggesting that these metagenomes reflect particularly biodiverse communities.
Table 2. Features of the metagenomes generated by 454 pyrosequencing of three hydrocarbon-degrading methanogenic enrichment cultures.
| NAPDC | SCADC | TOLDC | |
|---|---|---|---|
| Number of raw reads post-quality control (QC) | 368 209 | 667 134 | 550 247 |
| Unassembled reads post-QC (bp) | 130 215 413 | 230 716 341 | 215 982 194 |
| Mean read length post-QC (bp) | 353.65 | 345.83 | 392.52 |
| Newbler assembly | |||
| Total length of contigs (bp) | 9 473 370 | 17 382 962 | 20 280 655 |
| Number of contigs | 8471 | 15 274 | 10 888 |
| Range of contig lengths (bp) | 200–26 000 | 200–26 000 | 200–28 000 |
| Mean contig length (bp) | 1118 | 1138 | 1863 |
| Largest contig (bp) | 25 813 | 26 073 | 28 253 |
| Number of singletons | 161 851 | 326 382 | 179 067 |
| N50 | 1330 | 1343 | 2813 |
| Number of predicted proteins | 133 107 | 261 378 | 170 842 |
| Number of rRNA genesa | 23/54/155 | 46/135/267 | 29/104/189 |
| Number of tRNA genes | 816 | 1322 | 891 |
| MG-RAST data (assembled) | |||
| MG-RAST ID | 4492772.3 | 4492619.3 | 4492778.3 |
| Metagenome size (bp) | 64 676 632 | 127 644 733 | 89 513 841 |
| Average sequence length (bp) | 379±352 | 373±310 | 471±647 |
| Number of sequences | 170 322 | 341 656 | 189 955 |
| GC content (%) | 49±8% | 50±9% | 53±9% |
| Number of predicted ORFs | 162 642 | 325 794 | 187 741 |
| ORFs with predicted function | 94 562 | 184 014 | 105 547 |
| Alpha diversityb | 662 | 771 | 907 |
Abbreviations: NAPDC, naphtha-degrading culture; SCADC, short chain alkane-degrading culture; TOLDC, toluene-degrading culture.
5S, 16S and 23S, respectively.
Number of distinct species in a given metagenome sample as calculated by MG-RAST.
Community compositions of methanogenic hydrocarbon-degrading cultures
The microbial community compositions of NAPDC, SCADC and TOLDC were determined in two ways; by pyrotag sequencing of amplified 16S rRNA genes (Table 3), and by taxonomic profiling of unassembled metagenomic reads (Figure 1; Supplementary Figure S3). Overall, the total numbers of OTUs determined by 16S rRNA gene amplicon pyrosequencing (⩾95% similarity) were 153 (NAPDC), 320 (SCADC) and 147 (TOLDC), including 65, 167 and 65 singletons, respectively. This is consistent with species richness as predicted by rarefaction analysis using the unassembled metagenomic reads (Supplementary Figure S2). The microbial communities predicted based on shotgun metagenomic sequencing (Figure 1; Supplementary Figure S3; PCR-independent) were largely consistent with those detected by pyrotag sequencing (Table 3) at the intra-taxon level. Some inter-taxon discrepancies were observed when comparing pyrotag sequencing (OTU assignment) and taxonomic profiling of metagenomes (Supplementary Figure S3), which is likely caused by primer bias or suboptimal PCR conditions. Nonetheless, the recovery of prevalent features in each metagenome using random shotgun metagenomic sequencing provides a basis for comparing the three metagenomes.
Table 3. The six most abundant bacterial and archaeal operational taxonomic units (OTUs) in enrichment cultures, determined by performing PCR amplification, sequencing and phylogenetic analysis of 16S rRNA genes.
| Phylum | Taxonomic affiliationa |
Percentage of reads in each taxon |
||
|---|---|---|---|---|
| NAPDC | SCADC | TOLDC | ||
| Bacterial OTUs | ||||
| Firmicutes | Anaerobacter | 9.5 | 3.0 | 30.2 |
| Firmicutes | Lachnospiraceae | 0.0 | 0.0 | 23.8 |
| Firmicutes | Peptococcaceae | 19.5 | 0.2 | 0.0 |
| Firmicutes | Desulfotomaculum | 15.3 | 0.0 | 0.0 |
| Firmicutes | Sedimentibacter | 0.0 | 0.0 | 11.1 |
| Firmicutes | Desulfosporosinus | 4.0 | 0.0 | 3.0 |
| Spirochaetes | Spirochaetaceae | 0.6 | 6.5 | 2.2 |
| Spirochaetes | Spirochaetaceae | 2.7 | 14.3 | 0.2 |
| Deltaproteobacteria | Smithella | 0.6 | 11.6 | 0.0 |
| Deltaproteobacteria | Syntrophus | 0.1 | 6.8 | 0.0 |
| Deltaproteobacteria | Geobacter | 2.8 | 2.6 | 0.1 |
| Deltaproteobacteria | Desulfovibrio | 0.6 | 4.2 | 0.0 |
| Deltaproteobacteria | Desulfobacterium | 0.1 | 3.6 | 0.0 |
| Synergistetes | Thermanaerovibrio | 4.2 | 2.5 | 0.0 |
| Chloroflexi | Anaerolineaceae | 0.0 | 0.0 | 5.4 |
| Candidate division | Candidate division OP11 | 2.4 | 0.0 | 0.0 |
| Candidate division | Candidate division OP8 | 0.0 | 0.0 | 3.3 |
| Archaeal OTUs | ||||
| Euryarchaeota | Methanosaeta | 51.5 | 55.9 | 50.6 |
| Euryarchaeota | Methanosaeta | 5.4 | 5.4 | 2.2 |
| Euryarchaeota | Methanoculleus | 32.1 | 21.3 | 15.1 |
| Euryarchaeota | Methanoculleus | 3.6 | 2.4 | 0.4 |
| Euryarchaeota | Methanolinea | 0.4 | 1.3 | 27.3 |
| Euryarchaeota | Candidatus Methanoregula | 0.6 | 4.8 | 0.6 |
| Euryarchaeota | Methanomicrobia | 1.4 | 1.4 | 0.0 |
| Euryarchaeota | WCHA1 | 0.2 | 0.3 | 3.0 |
Abbreviations: NAPDC, naphtha-degrading culture; SCADC, short chain alkane-degrading culture; TOLDC, toluene-degrading culture.
Each OTU was assigned a taxonomic affiliation using RDP classifier. Only the lowest identified taxon is shown.
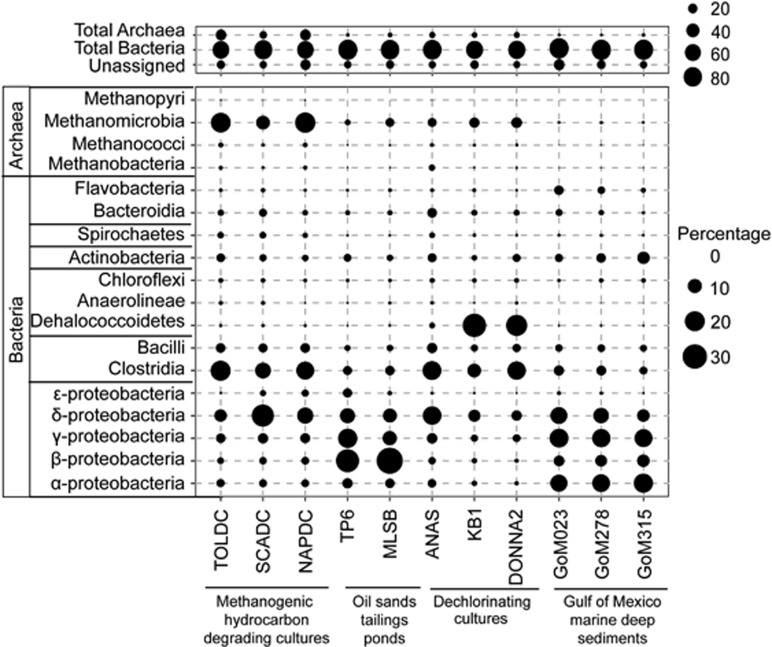
Figure 1.
Relative abundance of microbial classes detected in metagenomes from NAPDC, TOLDC, SCADC and eight relevant environments (Supplementary Table S1), based on MG-RAST assignment of unassembled 454 metagenomic reads and expressed as the proportion of total reads in each metagenome.
The archaeal reads in all three metagenomes, determined using 16S rRNA amplicon pyrosequencing, were dominated by Methanomicrobia, represented by Methanosarcinales (two OTUs affiliated with Methanosaeta; total 53–61% of archaea) and Methanomicrobiales (two Methanoculleus OTUs; total 16–36%), plus one Methanolinea OTU that was abundant (27%) only in TOLDC (Table 3). These methanogens are associated with acetotrophic and hydrogenotrophic methanogenesis, suggesting that both pathways contribute to methane production in the cultures.
The bacterial communities in NAPDC and TOLDC were dominated by members of the phylum Firmicutes, followed by Proteobacteria, Bacteroidetes, Actinobacteria and Spirochaetes (Figure 1; Table 3; Supplementary Figure S3). Despite similarities in bacterial community structure at the phylum level, there are noteworthy distinctions between the dominant OTUs affiliated with the Firmicutes as determined by pyrotag sequencing of amplicons (Table 3). The dominant Firmicutes-related OTUs in NAPDC belonged to the Peptococcaceae (Desulfotomaculum and Desulfosporosinus), whereas other Clostridiales (Anaerobacter and Lachnospiraceae) were most abundant in TOLDC (Table 3). This is in contrast to the recent finding that Desulfosporosinus sp. is a key toluene activating organism in TOLDC (Fowler et al., 2014). Binning of metagenomic contigs from TOLDC (Fowler, 2014) suggests that there is an overrepresentation of Anaerobacter and Lachnospiraceae, and an underrepresentation of Desulfosporosinus sp. and specific members of the Deltaproteobacteria in the TOLDC 16S rRNA pyrotag data set. This is likely due to PCR bias, as Desulfosporosinus and Deltaproteobacteria were well represented in the TOLDC metagenome (Fowler, 2014). Another possibility is that the overrepresented organisms contain multiple rRNA operons; for example, Clostridium acetobutylicum (closely related to Anaerobacter sp.) harbors eleven 16S rRNA gene copies (Acinas et al., 2004). Further sequencing and genome reconstruction of TOLDC could reveal why this bias impacted TOLDC but not other cultures. The bacterial community in SCADC, as determined using metagenomic reads (Figure 1), mainly comprised Deltaproteobacteria and Clostridia, although the latter were not abundant in amplified sequences (Table 3) likely due to PCR bias. Not surprisingly, SCADC and NAPDC shared more OTUs with each other than with TOLDC (for example, Deltaproteobacteria, Table 3), consistent with their common environmental origin and substrate pressure (for example, the presence of alkanes in the enrichment substrates; Table 1).
The high abundance of Firmicutes in TOLDC and NAPDC is consistent with recent reports of anaerobic monoaromatic hydrocarbon-degrading cultures that implicate Firmicutes (for example, Peptococcaceae) as primary hydrocarbon degraders (Abu Laban et al., 2009; Winderl et al., 2010; Sun and Cupples, 2012; Fowler et al., 2014) or playing key roles alongside Deltaproteobacteria (Ficker et al., 1999; Ulrich and Edwards, 2003; Sakai et al., 2009). In contrast, members of the Deltaproteobacteria, particularly Syntrophus/Smithella sp., have been implicated in methanogenic alkane degradation, particularly of longer-chain alkanes (Zengler et al., 1999; Gray et al., 2011; Cheng et al., 2013). Indeed, all of the known strictly anaerobic (for example, sulfate-reducing/syntrophic) alkane degraders are Deltaproteobacteria (Mbadinga et al., 2011; Agrawal and Gieg, 2013; Callaghan, 2013). However, the implication of a Peptococcaceae in degradation of low molecular weight n-alkanes (for example, propane) under sulfate-reducing conditions (Kniemeyer et al., 2007) and iso-alkanes under methanogenic conditions (Abu Laban et al., in press; Tan et al., 2014B) supports a role for Firmicutes in anaerobic alkane degradation. Based on the dominant OTUs in these cultures and the phylogenetic distribution of genes involved in fumarate addition, the primary hydrocarbon degraders in NAPDC, SCADC and TOLDC appeared to consist of different species from a few bacterial taxa (Table 3; Supplementary Figure S3). Indeed, the genomes of novel putative hydrocarbon degraders including Smithella spp. and Peptococcaceae capable of fumarate addition have been obtained from SCADC using single-cell amplified genomes (Tan et al., 2014B, 2014C). Novel species capable of anaerobic hydrocarbon degradation are likely also present in NAPDC and TOLDC, as metagenomic reads from these cultures did not map well to reference genomes of known hydrocarbon degraders or their close relatives (Supplementary Table S2).
The high diversity (for example, 150–320 OTUs) present in NAPDC, SCADC and TOLDC after multiple culture transfers (particularly TOLDC, which has been passaged on toluene for >10 years) is likely due to the presence of secondary fermenters and syntrophs that are essential to the growth of the community. These organisms (for example, Clostridiales, Spirochaetes, Anaerolinea, Desulfovibrio; Figure 1, Table 3) may function in the transformation of hydrocarbon degradation products (such as fatty acids) to methanogenic substrates, maintaining a low redox potential and hydrogen partial pressure, scavenging or recycling of waste products and dead biomass within the enrichments, and/or providing essential nutrients such as vitamin B12 and amino acids for other community members (Ahring et al., 1991; Beller and Edwards, 2000; Gray et al., 2011; Walker et al., 2012). The stability and persistence of diverse microorganisms in methanogenic hydrocarbon-degrading environments after long-term incubation suggests that these organisms are important for syntrophic growth of the communities.
Anaerobic hydrocarbon activation genes
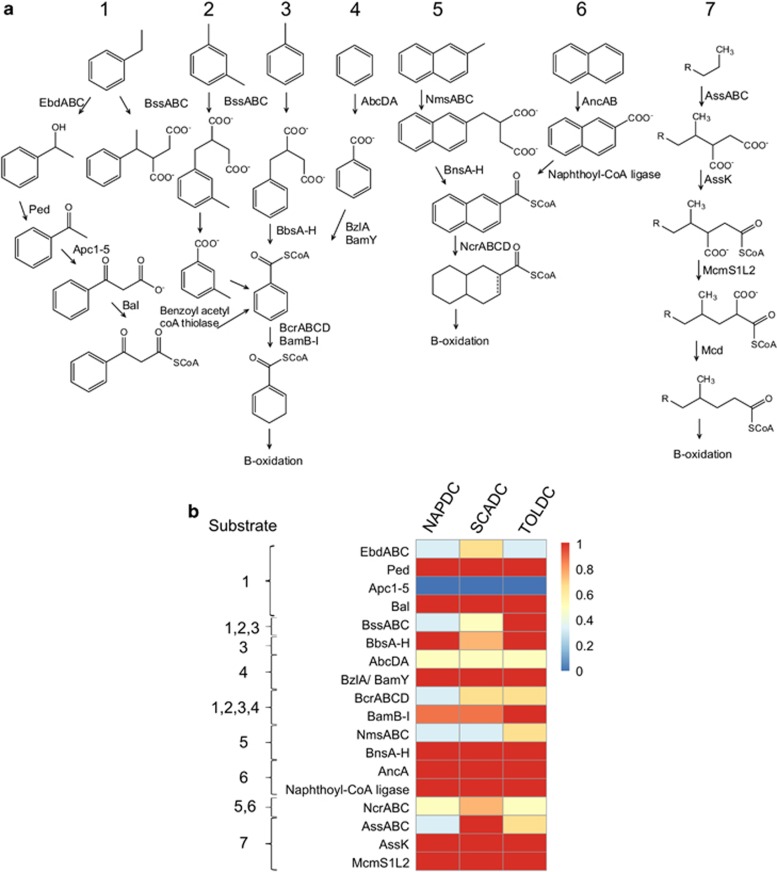
Figure 2a shows the pathways and genes/enzymes known to be involved in the anaerobic metabolism of various hydrocarbons (for example, reviewed by Foght, 2008; Callaghan, 2013). Characterized hydrocarbon activation mechanisms (Figure 2) include addition to fumarate (pathways 1, 2, 3, 5 and 7), hydroxylation (pathway 1) and carboxylation (pathways 4 and 6). Thus, the three enrichment cultures were examined for the presence of genes associated with these pathways by using HMMs and BLAST to infer their complete hydrocarbon substrate spectrum (Figure 2; Supplementary Table S3).
Figure 2.
Enzymes involved in anaerobic hydrocarbon degradation and the corresponding genes detected in the metagenomes of NAPDC, SCADC and TOLDC. (a) Known pathways (substrates) for anaerobic hydrocarbon metabolism and associated enzymes. (b) Heatmap showing the presence of various genes and putative genes involved in anaerobic hydrocarbon metabolism in the metagenomes. The color scale indicates the inferred enzyme completeness, in the case of enzymes with multiple subunits. Enzyme abbreviations: EbdABC, ethylbenzene dehydrogenase; Ped, (S)-1-phenylethanol dehydrogenase; Apc1 to Apc5, acetophenone carboxylase; Bal, benzoylacetate-CoA ligase; BssABC, benzylsuccinate synthase trimer; BbsEF, succinyl-CoA:(R)-benzylsuccinate CoA transferase; BbsG, (R)-benzylsuccinyl-CoA dehydrogenase; BbsH, phenylitaconyl-CoA hydratase; BbsCD, 2-[hydroxyl(phenyl)methyl]- succinyl-CoA dehydrogenase; BbsAB, benzoylsuccinyl-CoA thiolase; AbcDA, anaerobic benzene carboxylase; BzlA/BamY, benzoate-CoA ligase; BcrABCD, benzoyl-CoA reductase (ATP-dependent); BamB to BamI, benzoyl-CoA reductase (ATP-independent); NmsABC, naphthyl-2-methyl-succinate synthase; BnsEF, naphthyl-2-methyl-succinate CoA transferase; BnsG, naphthyl-2-methyl-succinate dehydrogenase; BnsH, naphthyl-2-methylene-succinyl CoA hydratase; BnsCD, naphthyl-2-hydroxymethyl-succinyl CoA dehydrogenase; BnsAB, naphthyl-2-oxomethyl-succinyl CoA thiolase; AncA, anaerobic naphthalene carboxylase; NcrABC, 2-naphthoyl CoA reductase; AssABC, alkylsuccinate synthase; AssK, AMP-dependent synthetase and ligase; Mcm, methylmalonyl CoA mutase; Mcd, methylmalonyl-CoA decarboxylase. Based on the models used, Mcd and benzoyl-CoA thiolase were abundant in all metagenomes. It was not known whether any of the genes identified were associated with hydrocarbon-degrading pathways or more general decarboxylase and thiolase functions, thus they have been omitted from the heatmap to curtail over-interpretation of the data.
Fumarate addition reactions
Examination of the culture metagenomes revealed several full-length and/or partial genes associated with fumarate addition (bssA for methylated monoaromatics, assA for alkanes, nmsA for methylnaphthalene) showing that all cultures have the potential to activate multiple substrates by fumarate addition.
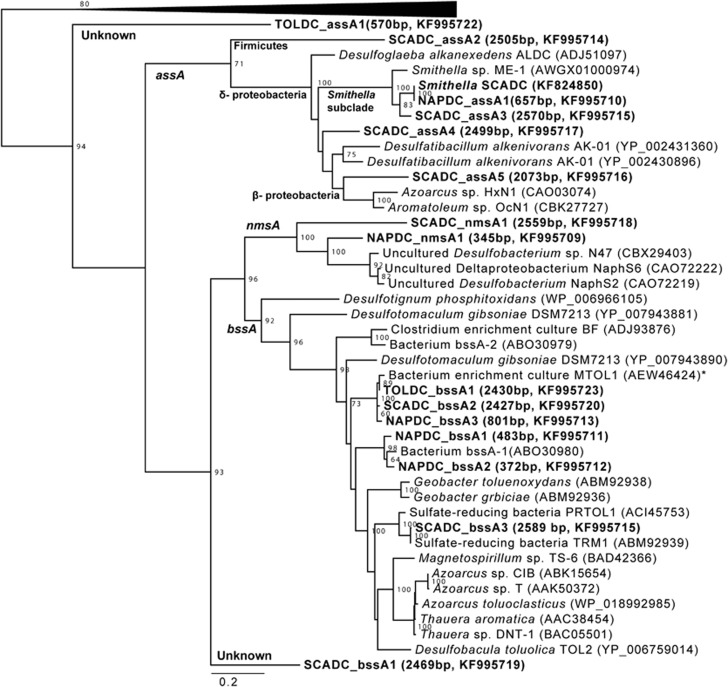
Putative assA genes were identified in NAPDC and SCADC, three of which (Smithella_SCADC, NAPDC_assA1 and SCADC_assA3) shared high homology with each other and a putative assA gene identified in Smithella sp. ME-1 (Embree et al., 2014; Tan et al., 2014A) (Figure 3). Five distinct putative assA genes were identified in SCADC (Figure 3), reflecting the diversity of alkane substrates degraded by this culture (Tan et al., 2013). A partial putative assA was also detected in TOLDC, but it was not closely affiliated with assA reference sequences nor SCADC and NAPDC assA genes (Figure 3). Instead, the amino-acid sequence of TOLDC_assA1 gene had a best BLAST hit to assA in Desulfoglaeba alkanexedens ALDC (ADJ51097, similarity 44.5%). Apparently this gene has persisted in TOLDC despite long-term enrichment on toluene, and either belongs to a bacterium with unique taxonomic affiliations, encodes an enzyme having a substrate spectrum yet to be identified, and/or may even be involved in toluene activation (Rabus et al., 2011).
Figure 3.
Maximum likelihood tree of translated full-length and partial assA, bssA, and nmsA homologs recovered from TOLDC, SCADC and NAPDC metagenomes (bold font). Bootstrap support ⩾60% is indicated. Full-length translated pyruvate formate lyase (PFL) sequences were used as an outgroup (collapsed in figure). Sequence length of genes recovered from TOLDC, SCADC and NAPDC, and their corresponding GenBank accession numbers are indicated in parentheses. A tree with the same overall topology was obtained when including only full-length sequences with gaps (not shown).
Full-length bssA genes having high sequence homology to one another were also detected (SCADC_bssA2, TOLDC_bssA1 and NAPDC_bssA3; Figure 3), implying similar substrate spectra and/or phylogeny (for example, Firmicutes). Detection of bssA in the TOLDC metagenome is consistent with a previous report of benzylsuccinate formation from toluene by this culture (Fowler et al., 2012). The NAPDC and SCADC metagenomes contained additional bssA genes most closely related to those recovered from a methanogenic toluene-degrading culture (ABO30980) and sulfate-reducing environments (ACI45753, ABM92939). SCADC also contained a deeply branching bssA gene (SCADC_bssA1) with best a BLAST hit to bssA from Desulfobacula toluolica (YP_006759359, similarity 55%) rather than pyruvate formate lyase or assA genes (Figure 3), despite enrichment on aliphatic hydrocarbons. The detection of deeply branching putative assA and bssA genes is especially interesting because recent evidence suggests that an archaeal species (Archaeoglobus fulgidus strain VC-16) activates long chain alkanes by fumarate addition using a homolog of pyruvate formate lyase that is phylogenetically distant from canonical assA and bssA (Khelifi et al., 2014). Although genes matching the HMM for bssA were detected in all cultures, bssB and bssC genes encoding the other two subunits of benzylsuccinate synthase were not detected in NAPDC (Figure 2, Supplementary Table S3), underlining the importance of screening for all enzyme subunits to indicate the presence of functions (as performed here), especially for metagenomes suffering from low sequencing coverage.
Whereas all three metagenomes contained assA and bssA, only NAPDC and SCADC contained nmsA (pathway 5, Figures 2 and 3). Recent evidence suggests that some fumarate addition genes that cluster with nmsA may function in the activation of toluene or other monoaromatics rather than polycyclic aromatic hydrocarbons (Acosta-González et al., 2013; von Netzer et al., 2013); thus, the physiological role of nms-like genes in SCADC and NAPDC is unclear as polycyclic aromatic hydrocarbon degradation has not been tested with these cultures. Previous studies noted that bssA and nmsA exhibit a high degree of homology (for example, Selesi et al., 2010), thus enzymes involved in fumarate addition might potentially co-activate several structurally diverse substrates. For example, Azoarcus sp. HxN1, which actives n-alkanes by using Ass (Mas), can co-activate cyclo-alkanes in the presence of n-alkanes (Wilkes et al., 2003). Similarly, other Ass enzymes can activate toluene in the presence of n-alkanes (Rabus et al., 2011), whereas Bss can catalyze fumarate addition to both toluene and xylenes (Beller and Spormann, 1997).
A point of interest in comparing the fumarate addition genes in these cultures is the relatively high abundance and sequence divergence of alpha gene subunits in NAPDC and SCADC compared with TOLDC (Figures 2 and 3). This may reflect the degree of enrichment of the cultures, with NAPDC and SCADC being established relatively recently, vs TOLDC that was established >10 years ago. Another contributing factor may be the diversity of hydrocarbon substrates during enrichment: NAPDC was established on a mixed alkane and monoaromatic hydrocarbon substrate (naphtha) and SCADC was enriched on a mixture of alkanes, whereas TOLDC was only exposed to toluene. The initial environments may also have dictated the genetic diversity (oil sands tailings ponds vs contaminated aquifer). However, other enrichments derived from the same aquifer sediments as TOLDC were shown to degrade a broad range of hydrocarbons such as other monoaromatics, n- and cyclo-alkanes, and polycyclic aromatic hydrocarbons (Gieg et al., 1999; Townsend et al., 2003 and 2004; Berdugo-Clavijo et al., 2012), thus we speculate that long-term enrichment on a sole hydrocarbon is more likely the reason for lower gene diversity in TOLDC. The presence of multiple assA/bssA/nmsA genotypes in these cultures is of interest because hydrocarbon contaminants often exist as diverse, structurally complex hydrocarbon mixtures. Thus, a microbial community that can degrade multiple substrates may be useful for in situ bioremediation and biostimulation. However, the presence of enzyme subunits does not necessarily denote functionality, and some enzymes may activate multiple substrates as discussed above. Further physiological studies are needed to define the complete range of substrates that these cultures can activate by fumarate addition but these results suggest that all three cultures harbor the genetic ability to degrade a range of hydrocarbon substrates.
Anaerobic pathways downstream of fumarate addition
Following toluene activation by fumarate addition, benzylsuccinate is converted to benzoyl-CoA by a multi-subunit enzyme encoded by the bbsABCDEFGH genes (pathway 3, Figure 2). Analogous genes (bns) have also been reported in the degradation of 2-methylnaphthalene (Figure 2, pathway 5, Selesi et al., 2010). We found matches to all HMMs for bbs genes in TOLDC and for the majority of bbs genes in NAPDC and SCADC (Figure 2), showing that these genes are present in all cultures regardless of the hydrocarbon substrate used for enrichment. BLAST matches were found for all of the bns genes in all three cultures (Figure 2, Supplementary Table S3), although it is unknown whether these genes are involved in the degradation of 2-methylnaphthalene by the cultures. Anaerobic degradation of monoaromatic compounds converges at the central intermediate benzoyl-CoA (Figure 2) that is then further metabolized by ring reduction. Reductive dearomatization can occur via an ATP-dependent benzoyl-CoA reductase found in facultative organisms (bcrABCD), or via an ATP-independent reductase found in strict anaerobes (bamBCDEFGHI; Carmona et al., 2009). We found matches to the alpha and delta subunits of the ATP-dependent ring reductases in all three cultures, but beta and gamma subunits were detected only in SCADC and TOLDC (Figure 2, Supplementary Table S3). Virtually all genes encoding the strictly anaerobic ring reductase (bamB-I) were also found in the three cultures, an expected result as the bacterial communities consist mainly of strict anaerobes, rather than facultative organisms (Table 3).
Following alkane activation (pathway 7; Figure 2), alkylsuccinates are activated to a CoA derivative, followed by carbon-skeleton rearrangement and decarboxylation (Wilkes et al., 2002). In D. alkenivorans AK-01, these reactions are presumably catalyzed by AssK, a putative methylmalonyl-CoA mutase, and a putative methylmalonyl-CoA carboxyl transferase before β-oxidation (Callaghan et al., 2012). We found BLAST or Pfam matches to these genes in all three metagenomes (Figure 2,Supplementary Table S3). However, because CoA-synthetases, along with methylmalonyl-CoA mutase and carboxylase (involved in the methylmalonyl-CoA pathway) are widespread in microbes, their presence cannot be solely associated with n-alkane degradation. It has been proposed that, before carbon skeleton rearrangement, the diastereomers produced from fumarate addition undergo epimerization catalyzed by alpha-methylacyl-CoA racemase to produce one intermediate for carbon skeleton rearrangement (Jarling et al., 2012). In a partial ass operon found in the genome of Peptococcaceae from SCADC (Tan et al., 2014B), putative genes involved in fumarate addition and downstream pathways clustered together, including alpha-methylacyl-CoA racemase, methyl malonyl-CoA mutase and β-oxidation, suggesting that alpha-methylacyl-CoA racemase may indeed be involved in n-alkane degradation. The putative alpha-methylacyl-CoA racemase gene in Peptococcaceae SCADC was used as a probe in tBLASTn searches to screen assembled contigs from TOLDC and NAPDC and to reveal homologs that have identity (~80%) but low sequence coverage (<50%). Unfortunately, the operon structure of the genes involved in n-alkane degradation in TOLDC and NAPDC is cryptic due to the short length of the associated contigs, so similarity to operons from known alkane degraders cannot be confirmed yet.
Alternate anaerobic hydrocarbon activation mechanisms
Hydroxylation and carboxylation are alternate mechanisms of anaerobic hydrocarbon activation. Genes for ethylbenzene hydroxylation (pathway 1, Figure 2) have been described (Ebd, encoded by ebdAB) (Rabus and Widdel, 1995; Kniemeyer and Heider, 2001) and it was recently proposed that Desulfococcus oleovorans Hxd3 activates alkanes by hydroxylation at C2 using an enzyme homologous to Ebd to produce a secondary alcohol (Callaghan, 2013). BLAST searches of TOLDC, SCADC and NAPDC assembled contigs using Ebd sequences from D. oleovorans Hxd3 revealed that ebd homologs are not abundant in these metagenomes. All genes identified had higher similarity to dehydrogenases with other functions (for example, DMSO dehydrogenase and nitrate reductase) than to Ebd, suggesting that these genes are not involved in ethylbenzene and alkane hydroxylation in the three cultures.
Putative genes encoding a benzene carboxylase (abcDA) (pathway 4, Figure 2) were recently identified under iron- (Abu Laban et al., 2010) and nitrate-reducing conditions (Luo et al., 2014) along with analogous genes encoding a putative naphthalene carboxylase (ancA) and naphthoyl-CoA ligase under sulfate-reducing conditions (pathway 6, Figure 2) (Bergmann et al., 2011). Several genes with homology to abcA were found in the three metagenomes, however, no delta subunit (abcD) genes were identified (Figure 2, Supplementary Table S3). The ancA gene was also detected in all three metagenomes (Figure 2) and matches to the CoA-ligases of benzene and naphthalene carboxylation were also identified (pathway 4, Figure 2), but none were detected on contigs adjacent to putative benzene carboxylase genes, as previously observed (Abu Laban et al., 2010; Luo et al., 2014). Overall, metagenomic analysis of the three cultures showed the genetic potential for carboxylation as a hydrocarbon activation strategy for non-substituted aromatic hydrocarbons but this activity remains to be tested in the cultures.
Other metabolic pathways
The metagenomes were interrogated for metabolic pathways including methanogenesis, fumarate regeneration, β-oxidation, carbon fixation, the glyoxylate cycle and alternate electron-accepting pathways (Supplementary Figures S4 and S5). Notably, genes were found for acetotrophic and hydrogenotrophic pathways of methane production in the cultures, but not the methylotrophic pathway (Supplementary Figure S5). These findings along with other pathways analyzed in this study are discussed further in the Supplementary Information.
Comparison of function among TOLDC, SCADC and NAPDC metagenomes
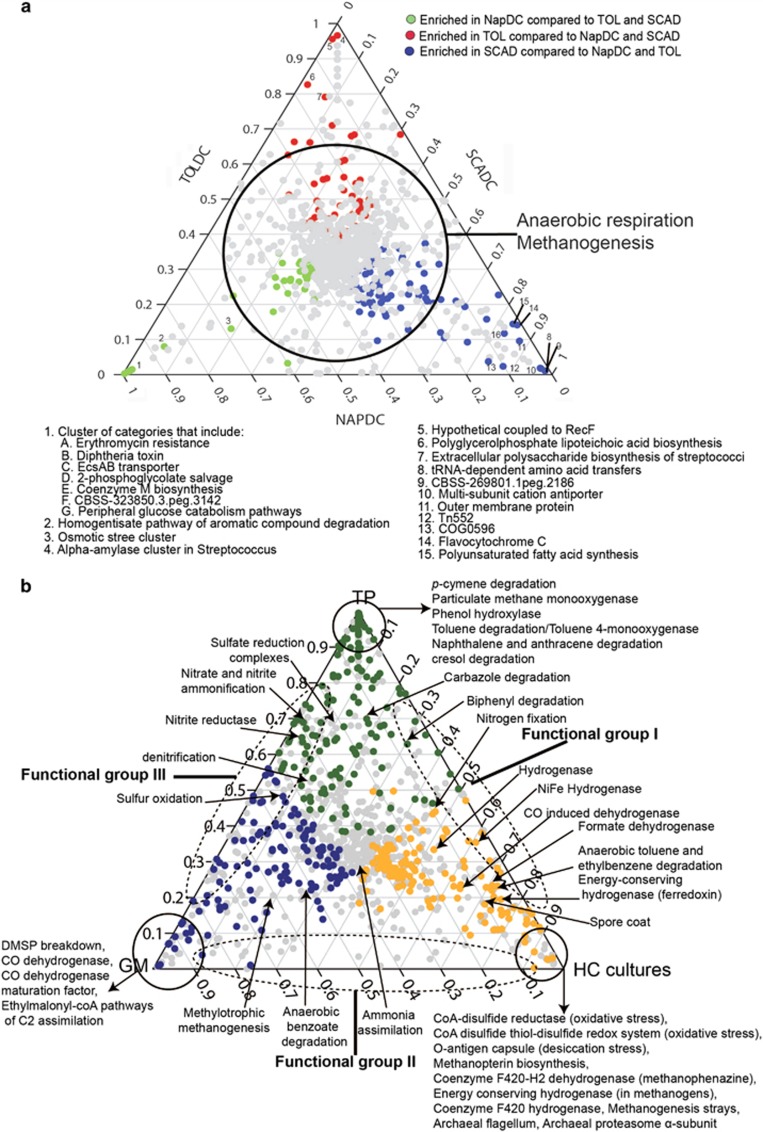
To examine the individual contributions of NAPDC, SCADC and TOLDC to their aggregate functional profile, we performed a three-way comparison of the relative abundance of SEED functional categories according to the method of Hug et al. (2012) (Figure 4a). Key functions related to methanogenesis, anaerobic hydrocarbon metabolism, redox condition regulation and hydrogen and formate transfer (important for syntrophy) were shared among the three cultures (Figure 4a). Features that were highly enriched in any single metagenome were related to non-specific functions associated with specific taxa present in each of the cultures (Supplementary Table S4). The observation that NAPDC, SCADC and TOLDC share similar genetic potentials despite being enriched from two distinct environments on different hydrocarbon substrates for different incubation periods (~2 years vs>10 years) indicates that the cultures comprise communities having flexible taxonomic composition and high functional redundancy. A similar conclusion was reached recently for a very different anoxic environment (termite hindgut) wherein, despite the presence of diverse taxa in different termite species, common functionality was observed (He et al., 2013).
Figure 4.
Ternary plots showing three-way comparisons of functional categories in SEED subsystems level 3 shared among individual metagenomes or groups of metagenomes. (a) Comparison of individual NAPDC, SCADC and TOLDC metagenomes. (b) Comparison of metagenome groups from hydrocarbon-impacted communities: HC (TOLDC, SCADC and NAPDC); GM (three metagenomes from Gulf of Mexico deep marine sediments); and TP (two metagenomes from oil sands tailings ponds). Each point on the ternary plot represents a subsystem category in the three metagenomes or groups of metagenomes, with the proportion of each SEED being normalized to a value of 1. Data points are colored according to the source of each metagenome or group; gray dots represent functional categories present at low, statistically non-significant (P<0.01) abundance. Points located near the vertices are enriched within the metagenome or group associated with that vertex, points along each axis are shared only between the two vertices associated with that axis (labelled as Functional Groups I, II and III in b), and points located near the center of the plot have similar proportions in all three metagenomes or groups (that is, show no specific enrichment; Hug et al., 2012).
Taxonomic and functional comparison of enrichment cultures with other environments and cultures
To investigate the effects of environmental selection on functional potential in methanogenic microbial communities, the three metagenomes from the hydrocarbon-degrading cultures were compared with 41 metagenomes available through MG-RAST including two oil sands tailings ponds (MLSB and TP6), three Gulf of Mexico (GoM) marine sediment samples collected from the Deepwater Horizon oil spill area (Kimes et al., 2013), three dechlorinating enrichment cultures (the only other anaerobic pollutant-degrading enrichment culture metagenomes available on MG-RAST at time of analysis), and 33 metagenomes from a range of environments with varying physiochemical properties (Supplementary Table S1). A comparison of this nature assumes that random shotgun sequencing has recovered the prevalent genetic composition of the microbial community in any given environmental sample, validated by the taxonomic composition expected in their respective niches (for example, Proteobacteria in GoM and Dehalococcoidetes in dechlorinating cultures). Nonetheless, comparisons using functional categories (COG, KEGG, SEED) may effectively remove discovery of novel functions which otherwise are annotated as hypothetical proteins, and are not examinable due to unassigned functions. This study investigated the overall functional properties and dissimilarities among the 41 environmental metagenomes, and may not provide insights into functions of the rare biosphere due to unequal sequence coverage in the metagenomes.
Taxonomic analysis of metagenomes from hydrocarbon-impacted environments including the methanogenic cultures, tailings ponds, GoM sediments, and dechlorinating cultures revealed that all of these cultures contained methanogenic archaea (albeit in different proportions; Figure 1) indicating that they are anoxic environments or contain anoxic microenvironments. However, the bacterial communities supporting methanogenesis differed considerably in diversity and relative abundance (Figure 1).
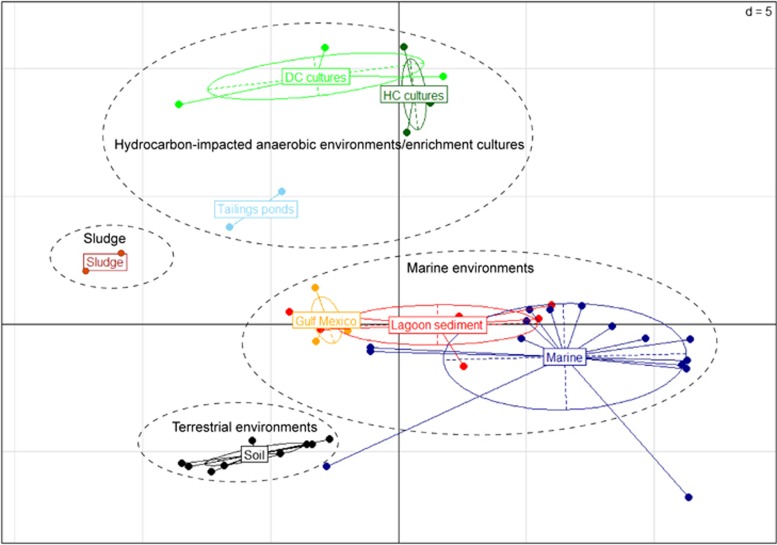
Using principal component analysis based on the abundance of SEED functional categories, the metagenomes from each type of environment or enrichment culture were found to be more functionally related to each other than to metagenomes from other environments (Figure 5), presumably due to selective pressures imposed by diverse environmental conditions. The 44 metagenomes clustered into four distinct groups: (1) terrestrial environments including different soil types; (2) sludge; (3) marine environments including lagoon and marine sediments and the GoM deep sediment samples; and (4) methanogenic hydrocarbon-degrading enrichments, tailings ponds and dechlorinating enrichment cultures (Figure 5). NAPDC, SCADC and TOLDC were more functionally similar to each other than to any other environments, including the tailings pond (MLSB) from which NAPDC and SCADC were enriched (Figure 5). The cultures also grouped closely with methanogenic dechlorinating cultures enriched from environments that were co-contaminated with chlorinated compounds and hydrocarbons (Hug et al., 2012). This grouping presumably reflects the process of enrichment compared with unenriched samples from tailings and other environmental samples. All three GoM metagenomes grouped more closely with marine samples and lagoon sediments than with hydrocarbon-impacted environments, despite the recent exposure of at least two of the GoM sediments to hydrocarbons during the Deepwater Horizon oil spill (Kimes et al., 2013).
Figure 5.
Principal component analysis of NAPDC, SCADC and TOLDC plus 41 additional relevant metagenomes available in MG-RAST (Supplementary Table S1); symbols are colored according to their environment of origin. All counts were normalized against total annotated sequences of each metagenome. Metagenomes from related environments are enclosed with broken lines.
To further interrogate the functions enriched in TOLDC, NAPDC and SCADC vs hydrocarbon-impacted environments, we performed a three-way comparison of pooled metagenomes from the cultures, tailings ponds and the GoM marine sediments (Figure 4b). Features specifically enriched in the three cultures included those associated with hydrogen and formate transfer, methanogenesis and regulation of redox conditions (for example, CoA-disulfide redox systems) (HC vertex, Figure 4b), processes that are important for syntrophy and methanogenesis. GoM metagenomes shared little functional potential with the methanogenic hydrocarbon-degrading enrichments, as shown by the paucity of data points along the bottom axis (Functional Group II; Figure 4b). This is consistent with the taxonomic profiles of GoM metagenomes that were characterized by an abundance of Alpha- and Gammaproteobacteria, distinctly different from the methanogenic enrichments and oil sands tailings ponds (Figure 1). The GoM metagenomes did share some similarities in functional potential with tailings ponds (Functional Group III; Figure 4b), particularly related to sulfur cycling (likely due to the abundance and types of sulfur compounds present in both environments) and denitrification processes. Despite the exposure of two of the GoM samples to hydrocarbons following the Deepwater Horizon oil spill, there was a notable absence of enriched functions specifically related to hydrocarbon metabolism (Figure 4b).
Both MLSB and TP6 are located in the same geographic region and contain residual naphtha, components of which can serve as substrates for methanogenic communities (Siddique et al., 2006, 2007). On the basis of principal component analysis (Figure 5), the genetic potentials of the two tailings ponds were more similar to each other than to SCADC and NAPDC that were enriched from MLSB. Both tailings ponds harbor microbes genetically capable of anaerobic alkane and aromatic hydrocarbon metabolism by fumarate addition (An et al., 2013). In addition, three-way comparison revealed that, in agreement with the recent report by An et al. (2013), the metagenomes of the tailings ponds are also enriched in functions associated with aerobic metabolism of pollutants and hydrocarbons (Figure 4b), likely due to the presence of oxic surface water layers in the ponds and sedimentation of aerobic or facultative microbes from the water into the underlying tailings. Indeed, the ponds harbor a high abundance of Beta- and Gammaproteobacteria including Burkholderia, Pseudomonas and Rhodococcus (Figure 1), genera associated with aerobic hydrocarbon metabolism (Das and Chandran, 2011). In deeper strata of TP6, the microbial communities consist mainly of syntrophs, sulfate reducers and methanogens (Ramos-Padrón et al., 2011). The addition of gypsum (CaSO4•2H2O) to this pond stimulates sulfate reduction (Ramos-Padrón et al., 2011), a function that was enriched in tailings ponds relative to the methanogenic cultures (Figure 4b). The oil sands tailings ponds were found to share some functions with the methanogenic hydrocarbon-degrading cultures, particularly those associated with anaerobic or methanogenic metabolism (Functional Group I, Figure 4b). For example, previous analyses revealed the presence of genes associated with fumarate addition in TP6 (An et al., 2013) and in clone libraries from GoM sediments (Kimes et al., 2013), suggesting that this is a widespread mechanism of hydrocarbon activation in anoxic environments.
Conclusions
Metagenomic analysis of three distinct methanogenic hydrocarbon-degrading enrichment cultures (NAPDC, SCADC and TOLDC) revealed that despite differences in enrichment pressures and putative hydrocarbon-degrading taxa, all harbored multiple functional genes for different mechanisms of anaerobic hydrocarbon activation (fumarate addition and carboxylation) and subsequent conversions. The diversity and abundance of fumarate addition genes increased in the cultures enriched with a mixture of hydrocarbon substrates. However, it remains difficult to assess the effect of any given substrate on the enrichment of a particular fumarate addition gene as the cognate enzymes may exhibit relaxed substrate specificity (Rabus et al., 2011). Comparative metagenomic analyses showed that the functional profiles of the methanogenic hydrocarbon-degrading cultures were similar to each other but were distinct from hydrocarbon-impacted environments including the tailings pond from which two of the cultures were derived. Specialized functions related to methanogenesis, anaerobic hydrocarbon metabolism, regulation of redox conditions and assorted hydrogenases and formate dehydrogenases were shared among the culture metagenomes and enriched compared with other metagenomes suggesting that these are hallmark features of methanogenic hydrocarbon degradation. Because the methanogenic hydrocarbon-degrading cultures were functionally similar, the study of such cultures can provide fundamental information about environmentally important syntrophic processes in hydrocarbon-associated methanogenic environments.
Acknowledgments
This research was supported by a Genome Canada grant (JF, LMG, CWS, http://www.hydrocarbonmetagenomics.com/). SJF was supported by NSERC Alexander Graham Bell and Alberta Innovates Technology Futures Graduate Scholarships. We are grateful to Camilla Nesbø (University of Alberta, University of Oslo) for bioinformatics support and to Payal Sipahimalani and Steven Hallam (University of British Columbia) for genomic DNA extraction from NAPDC, SCADC and TOLDC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abu Laban N, Selesi D, Jobelius C, Meckenstock RU. Anaerobic benzene degradation by Gram-positive sulfate-reducing bacteria. FEMS Microbiol Ecol. 2009;68:300–311. doi: 10.1111/j.1574-6941.2009.00672.x. [DOI] [PubMed] [Google Scholar]
- Abu Laban N, Selesi D, Rattei T, Tischler P, Meckenstock RU. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ Microbiol. 2010;12:2783–2796. doi: 10.1111/j.1462-2920.2010.02248.x. [DOI] [PubMed] [Google Scholar]
- Abu Laban N, Dao A, Semple K, Foght J.(in press). Biodegradation of C7 and C8iso-alkanes under methanogenic conditions Environ Microbioldoi: 10.1111/1462-2920.12643 [DOI] [PubMed]
- Acinas S, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-González A, Rosselló-Móra R, Marqués S. Diversity of benzylsuccinate synthase-like (bssA genes in hydrocarbon-polluted marine sediments suggests substrate-dependent clustering. Appl Environ Microbiol. 2013;79:3667–3676. doi: 10.1128/AEM.03934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Gieg LM. In situ detection of anaerobic alkane metabolites in subsurface environments. Front Microbiol. 2013;4:140. doi: 10.3389/fmicb.2013.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahring BK, Westermann P, Mah RA. Hydrogen inhibition of acetate metabolism and kinetics of hydrogen consumption by Methanosarcina thermophila Tm-1. Arch Microbiol. 1991;157:38–42. [Google Scholar]
- Aitken CM, Jones DM, Maguire MJ, Gray ND, Sherry A, Bowler BFJ, et al. Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions. Geochim Cosmochim Acta. 2013;109:162–174. [Google Scholar]
- An DS, Brown D, Chatterjee N, Dong X, Ramos-Padrón E, Wilson S, et al. Microbial community and potential functional gene diversity involved in anaerobic hydrocarbon degradation and methanogenesis in an oil sands tailings pond. Genome. 2013;56:612–618. doi: 10.1139/gen-2013-0083. [DOI] [PubMed] [Google Scholar]
- Beller HR, Spormann AM. Anaerobic activation of toluene and o-xylene by addition to fumarate in denitrifying strain T. J Bacteriol. 1997;179:670–676. doi: 10.1128/jb.179.3.670-676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller HR, Edwards EA. Anaerobic toluene activation by benzylsuccinate synthase in a highly enriched methanogenic culture. Appl Environ Microbiol. 2000;66:5503–5505. doi: 10.1128/aem.66.12.5503-5505.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdugo-Clavijo C, Dong X, Soh J, Sensen CW, Gieg LM. Methanogenic biodegradation of 2-ringed polycyclic aromatic hydrocarbons. FEMS Microbiol Ecol. 2012;81:124–133. doi: 10.1111/j.1574-6941.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- Bergmann FD, Selesi D, Meckenstock RU. Identification of new enzymes potentially involved in anaerobic naphthalene degradation by the sulfate-reducing enrichment culture N47. Arch Microbiol. 2011;193:241–250. doi: 10.1007/s00203-010-0667-4. [DOI] [PubMed] [Google Scholar]
- Callaghan AV, Morris BEL, Pereira IAC, McInerney MJ, Austin RN, Groves JT, et al. The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation. Environ Microbiol. 2012;14:101–113. doi: 10.1111/j.1462-2920.2011.02516.x. [DOI] [PubMed] [Google Scholar]
- Callaghan AV. Enzymes involved in the anaerobic oxidation of n-alkanes: from methane to long-chain paraffins. Front Microbiol. 2013;4:89. doi: 10.3389/fmicb.2013.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona M, Zamarro MT, Blazquez B, Durante-Rodriguez G, Juarez JF, Valderrama JA, et al. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev. 2009;73:71–133. doi: 10.1128/MMBR.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Um Y, Hoffman B, Holoman T. Molecular characterization of anaerobic microbial communities from benzene-degrading sediments under methanogenic conditions. Biotechnol Prog. 2005;21:1789–1794. doi: 10.1021/bp050250p. [DOI] [PubMed] [Google Scholar]
- Cheng L, Ding C, Li Q, He Q, Dai LR, Zhang H. DNA-SIP reveals that Syntrophaceae play an important role in methanogenic hexadecane degradation. PLoS One. 2013;8:e66784. doi: 10.1371/journal.pone.0066784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N, Chandran P. Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int. 2011;2011:941810. doi: 10.4061/2011/941810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embree M, Nagarajan H, Movahedi N, Chitsaz H, Zengler K. Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J. 2014;8:757–767. doi: 10.1038/ismej.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Essaid HI, Bekins BA, Herkelrath WN, Delin GN. Crude oil at the Bemidji site: 25 years of monitoring, modeling, and understanding. Ground Water. 2011;49:706–726. doi: 10.1111/j.1745-6584.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Ficker M, Krastel K, Orlicky S, Edwards E. Molecular characterization of a toluene-degrading methanogenic consortium. Appl Environ Microbiol. 1999;65:5576–5585. doi: 10.1128/aem.65.12.5576-5585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foght J, Aislabie J, Turner S, Brown CE, Ryburn J, Saul DJ, et al. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb Ecol. 2004;47:329–340. doi: 10.1007/s00248-003-1036-5. [DOI] [PubMed] [Google Scholar]
- Foght J. Anaerobic biodegradation of aromatic hydrocarbons: Pathways and prospects. J Mol Microb Biotech. 2008;15:93–120. doi: 10.1159/000121324. [DOI] [PubMed] [Google Scholar]
- Fowler SJ, Dong XL, Sensen CW, Suflita JM, Gieg LM. Methanogenic toluene metabolism: community structure and intermediates. Environ Microbiol. 2012;14:754–764. doi: 10.1111/j.1462-2920.2011.02631.x. [DOI] [PubMed] [Google Scholar]
- Fowler SJ, Gutierrez-Zamora M-L, Manefield M, Gieg LM. Identification of toluene degraders in a methanogenic enrichment culture. FEMS Microbiol Ecol. 2014;89:625–636. doi: 10.1111/1574-6941.12364. [DOI] [PubMed] [Google Scholar]
- Fowler SJ. Syntrophic hydrocarbon metabolism under methanogenic conditions. PhD Thesis: University of Calgary; 2014. [Google Scholar]
- Gieg LM, Kolhatkar RV, McInerney MJ, Tanner RS, Harris SH, Sublette KL, Suflita JM. Evidence for intrinsic bioremediation in a gas condensate-contaminated aquifer. Environ Sci Technol. 1999;33:2550–2560. [Google Scholar]
- Gieg LM, Fowler SJ, Berdugo-Clavijo C. Syntrophic biodegradation of hydrocarbon contaminants. Curr Opin Biotechnol. 2014;27:21–29. doi: 10.1016/j.copbio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Gray ND, Sherry A, Grant RJ, Rowan AK, Hubert CRJ, Callbeck CM, et al. The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol. 2011;13:2957–2975. doi: 10.1111/j.1462-2920.2011.02570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haque MM, Ghosh TS, Komanduri D, Mande SS. SOrt-ITEMS: Sequence orthology based approach for improved taxonomic estimation of metagenomic sequences. Bioinformatics. 2009;25:1722–1730. doi: 10.1093/bioinformatics/btp317. [DOI] [PubMed] [Google Scholar]
- He S, Ivanova N, Kirton E, Allgaier M, Bergin C, Scheffrahn RH, et al. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One. 2013;8:361126. doi: 10.1371/journal.pone.0061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Beiko RG, Rowe AR, Richardson RE, Edwards EA. Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: the role of the non-dechlorinating community. BMC Genomics. 2012;13:327. doi: 10.1186/1471-2164-13-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarling R, Sadeghi M, Drozdowska M, Lahme S, Buckel W, Rabus R, et al. Stereochemical investigations reveal the mechanism of the bacterial activation of n-alkanes without oxygen. Angew Chem Int Ed. 2012;51:1334–1338. doi: 10.1002/anie.201106055. [DOI] [PubMed] [Google Scholar]
- Jones DM, Head IM, Gray ND, Adams JJ, Rowan AK, Aitken CM, et al. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature. 2008;451:176–180. doi: 10.1038/nature06484. [DOI] [PubMed] [Google Scholar]
- Kasai Y, Takahata Y, Hoaki T, Watanabe K. Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ Microbiol. 2005;7:806–818. doi: 10.1111/j.1462-2920.2005.00754.x. [DOI] [PubMed] [Google Scholar]
- Kato S, Kosaka T, Watanabe K. Substrate-dependent transcriptomic shifts in Pelotomaculum thermopropionicum grown in syntrophic co-culture with Methanothermobacter thermautotrophicus. Microb Biotechnol. 2009;2:575–584. doi: 10.1111/j.1751-7915.2009.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi N, Amin Ali O, Roche P, Grossi V, Brochier-Armanet C, Valette O, et al. Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. ISME J. 2014;8:2153–2166. doi: 10.1038/ismej.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes NE, Callaghan AV, Aktas DF, Smith WL, Sunner J, Golding B, et al. Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front Microbiol. 2013;4:50. doi: 10.3389/fmicb.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinsteuber S, Schleinitz KM, Vogt C. Key players and team play: anaerobic microbial communities in hydrocarbon-contaminated aquifers. Appl Microbiol Biotech. 2012;94:851–873. doi: 10.1007/s00253-012-4025-0. [DOI] [PubMed] [Google Scholar]
- Kniemeyer O, Heider J. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J Biol Chem. 2001;276:21381–21386. doi: 10.1074/jbc.M101679200. [DOI] [PubMed] [Google Scholar]
- Kniemeyer O, Musat F, Sievert SM, Knittel K, Wilkes H, Blumenberg M, et al. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature. 2007;449:898–901. doi: 10.1038/nature06200. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Luo F, Gitiafroz R, Devine CE, Gong YC, Hug LA, Raskin L, et al. Metatranscriptome of an anaerobic benzene-degrading, nitrate-reducing enrichment culture reveals involvement of carboxylation in benzene ring activation. Appl Environ Microbiol. 2014;80:4095–4107. doi: 10.1128/AEM.00717-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykidis A, Chen CL, Tringe SG, McHardy AC, Copeland A, Kyrpides NC, et al. Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J. 2011;5:122–130. doi: 10.1038/ismej.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles CN, Le HM, Beasley WH, McInerney MJ, Suflita JM. Anaerobic hydrocarbon and fatty acid metabolism by syntrophic bacteria and their impact on carbon steel corrosion. Front Microbiol. 2014;5:114. doi: 10.3389/fmicb.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbadinga SM, Wang LY, Zhou L, Liu JF, Gu JD, Mu BZ. Microbial communities involved in anaerobic degradation of alkanes. Int Biodeter Biodegr. 2011;65:1–13. [Google Scholar]
- McInerney MJ, Bryant MP, Pfennig N. Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens. Arch Microbiol. 1979;122:129–135. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Rabus R, Widdel F. Anaerobic degradation of ethylbenzene and other aromatic-hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103. doi: 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- Rabus R, Jarling R, Lahme S, Kühner S, Heider J, Widdel F, et al. Co-metabolic conversion of toluene in anaerobic n-alkane-degrading bacteria. Environ Microbiol. 2011;13:2576–2586. doi: 10.1111/j.1462-2920.2011.02529.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A.2012. FigTree v1.4.Institute of Evolutionary Biology, University of Edinburgh. Link http://tree.bio.ed.ac.uk/software/figtree/ .
- Ramos-Padrón E, Bordenave S, Lin S, Bhaskar IM, Dong X, Sensen CW, et al. Carbon and sulfur cycling by microbial communities in a gypsum-treated oil sands tailings pond. Environ Sci Technol. 2011;45:439–446. doi: 10.1021/es1028487. [DOI] [PubMed] [Google Scholar]
- Sakai N, Kurisu F, Yagi O, Nakajima F, Yamamoto K. Identification of putative benzene-degrading bacteria in methanogenic enrichment cultures. J Biosci Bioeng. 2009;108:501–507. doi: 10.1016/j.jbiosc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Selesi D, Jehmlich N, von Bergen M, Schmidt F, Rattei T, Tischler P, et al. Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47. J Bacteriol. 2010;192:295–306. doi: 10.1128/JB.00874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique T, Fedorak PM, Foght JM. Biodegradation of short-shain n-alkanes in oil sands tailings under methanogenic conditions. Environ Sci Technol. 2006;40:5459–5464. doi: 10.1021/es060993m. [DOI] [PubMed] [Google Scholar]
- Siddique T, Fedorak PM, MacKinnon MD, Foght JM. Metabolism of BTEX and naphtha compounds to methane in oil sands tailings. Environ Sci Technol. 2007;41:2350–2356. doi: 10.1021/es062852q. [DOI] [PubMed] [Google Scholar]
- Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Ann Rev Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- Soh J, Dong XL, Caffrey SM, Voordouw G, Sensen CW. Phoenix 2: a locally installable large-scale 16S rRNA gene sequence analysis pipeline with web interface. J Biotechnol. 2013;167:393–403. doi: 10.1016/j.jbiotec.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Strapoc D, Mastalerz M, Dawson K, Macaladay JL, Callaghan AV, Wawrik B, et al. Biogeochemistry of microbial coal-bed methane. Annu Rev Earth Planet Sci. 2011;39:617–656. [Google Scholar]
- Sun WM, Cupples AM. Diversity of five anaerobic toluene-degrading microbial communities investigated using stable isotope probing. Appl Environ Microbiol. 2012;78:972–980. doi: 10.1128/AEM.06770-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Dong X, Sensen CW, Foght J. Metagenomic analysis of an anaerobic alkane-degrading microbial culture: potential hydrocarbon-activating pathways and inferred roles of community members. Genome. 2013;56:599–611. doi: 10.1139/gen-2013-0069. [DOI] [PubMed] [Google Scholar]
- Tan B, Nesbø C, Foght J. Re-analysis of omics data indicates Smithella may degrade alkanes by addition to fumarate under methanogenic conditions. ISME J. 2014;8:2353–2356. doi: 10.1038/ismej.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Charchuk R, Li C, Nesbø C, Abu Laban N, Foght J. Draft genome sequence of uncultivated Firmicutes (Peptococcaceae SCADC) single cells sorted from methanogenic alkane-degrading cultures. Genome Announc. 2014;2:e00909–e00914. doi: 10.1128/genomeA.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, de Araújo e Silva R, Rozycki T, Nesbø C, Foght T. Draft genome sequences of three Smithella spp. obtained from a methanogenic alkane-degrading culture and oil field produced water. Genome Announc. 2014;2:e0185–e0214. doi: 10.1128/genomeA.01085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend GT, Prince RC, Suflita JM. Anaerobic oxidation of crude oil hydrocarbons by the resident microorganisms of a contaminated aquifer. Environ Sci Technol. 2003;37:5213–5218. doi: 10.1021/es0264495. [DOI] [PubMed] [Google Scholar]
- Townsend GT, Prince RC, Suflita JM. Anaerobic biodegradation of alicyclic constituents of gasoline and natural gas condensate by bacteria from an anoxic aquifer. FEMS Microbiol Ecol. 2004;49:129–135. doi: 10.1016/j.femsec.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Ulrich AC, Edwards EA. Physiological and molecular characterization of anaerobic benzene-degrading mixed cultures. Environ Microbiol. 2003;5:92–102. doi: 10.1046/j.1462-2920.2003.00390.x. [DOI] [PubMed] [Google Scholar]
- von Netzer F, Pilloni G, Kleindienst S, Krüger M, Knittel K, Grundger F, et al. Enhanced gene detection assays for fumarate-adding enzymes allow uncovering of anaerobic hydrocarbon degraders in terrestrial and marine systems. Appl Environ Microbiol. 2013;79:543–552. doi: 10.1128/AEM.02362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, He ZL, Yang ZK, Ringbauer JA, He Q, Zhou JH, et al. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol. 2009;191:5793–5801. doi: 10.1128/JB.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, Redding-Johanson AM, Baidoo EE, Rajeev L, He ZL, Hendrickson EL, et al. Functional responses of methanogenic archaea to syntrophic growth. ISME J. 2012;6:2045–2055. doi: 10.1038/ismej.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washer CE, Edwards EA. Identification and expression of benzylsuccinate synthase genes in a toluene-degrading methanogenic consortium. Appl Environ Microbiol. 2007;73:1367–1369. doi: 10.1128/AEM.01904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrik B, Mendivelso M, Parisi VA, Suflita JM, Davidova IA, Marks CR, et al. Field and laboratory studies on the bioconversion of coal to methane in the San Juan Basin. FEMS Microbiol Ecol. 2012;81:26–42. doi: 10.1111/j.1574-6941.2011.01272.x. [DOI] [PubMed] [Google Scholar]
- Widdel F, Bak F.1992Gram-negative mesophilic sulfate-reducing bacteriaIn: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds)The Prokaryotes Springer-Verlag: New York, USA; 3352–3378. [Google Scholar]
- Wilkes H, Rabus R, Fischer T, Armstroff A, Behrends A, Widdel F. Anaerobic degradation of n-hexane in a denitrifying bacterium: further degradation of the initial intermediate (1-methylpentyl)succinate via C-skeleton rearrangement. Arch Microbiol. 2002;177:235–243. doi: 10.1007/s00203-001-0381-3. [DOI] [PubMed] [Google Scholar]
- Wilkes H, Kuhner S, Bolm C, Fischer T, Classen A, Widdel F, et al. Formation of n-alkane- and cycloalkane-derived organic acids during anaerobic growth of a denitrifying bacterium with crude oil. Org Geochem. 2003;34:1313–1323. [Google Scholar]
- Winderl C, Penning H, von Netzer F, Meckenstock RU, Lueders T. DNA-SIP identifies sulfate-reducing Clostridia as important toluene degraders in tar-oil-contaminated aquifer sediment. ISME J. 2010;4:1314–1325. doi: 10.1038/ismej.2010.54. [DOI] [PubMed] [Google Scholar]
- Wright JJ, Lee S, Zaikova E, Walsh DA, Hallam SJ. DNA extraction from 0.22 μM sterivex filters and cesium chloride density gradient centrifugation. J Vis Exp. 2009;31:e1352. doi: 10.3791/1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C, Banning E, Mincer TJ, Drenzek NJ, Coolen MJL. Microbial diversity and methanogenic activity of Antrim Shale formation waters from recently fractured wells. Front Microbiol. 2013;4:367. doi: 10.3389/fmicb.2013.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K, Richnow HH, Rossello-Mora R, Michaelis W, Widdel F. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature. 1999;401:266–269. doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.