Abstract
Cyanobacteria are often the dominant phototrophs in polar freshwater communities; yet, the phages that infect them remain unknown. Here, we present a genomic and morphological characterization of cyanophage S-EIV1 that was isolated from freshwaters on Ellesmere Island (Nunavut, High Arctic Canada), and which infects the polar Synechococcus sp., strain PCCC-A2c. S-EIV1 represents a newly discovered evolutionary lineage of bacteriophages whose representatives are widespread in aquatic systems. Among the 130 predicted open reading frames (ORFs) there is no recognizable similarity to genes that encode structural proteins other than the large terminase subunit and a distant viral morphogenesis protein, indicating that the genes encoding the structural proteins of S-EIV1 are distinct from other viruses. As well, only 19 predicted coding sequences on the 79 178 bp circularly permuted genome have homology with genes encoding proteins of known function. Although S-EIV1 is divergent from other sequenced phage isolates, it shares synteny with phage genes captured on a fosmid from the deep-chlorophyll maximum in the Mediterranean Sea, as well as with an incision element in the genome of Anabaena variabilis (ATCC 29413). Sequence recruitment of metagenomic data indicates that S-EIV1-like viruses are cosmopolitan and abundant in a wide range of aquatic systems, suggesting they have an important ecological role.
Introduction
Cyanobacteria are often the dominant phototrophs in polar and subpolar lakes (Vincent, 2000) and can account for >50% of the phytoplankton chlorophyll a in northern lakes (Bergeron and Vincent, 1997). In meromictic lakes in the High Arctic (Van Hove et al., 2008) and Antarctica (Rankin et al., 1997) planktonic cyanobacteria occur at abundances of up to 104 and 106 cells ml−1, respectively. Most planktonic polar cyanobacteria are related to Synechococcus spp. and fall within two groups that contain brackish and freshwater representatives from different latitudes (Van Hove et al., 2008). Arctic and Antarctic cyanobacteria may be mostly cosmopolitan, generalist taxa rather than endemic specialists, but this will require ongoing genomic analysis to fully resolve (Jungblut et al., 2010). Despite the ecological importance of cyanobacteria in polar freshwaters, and the long history of research on freshwater cyanophages (Safferman and Morris, 1963, 1964), there were no polar freshwater cyanophage systems in culture of which we were aware. This provided motivation to isolate a cyanophage-host system from high-arctic freshwaters.
In marine systems, titers of cyanophages infecting Synechococcus spp. can be in excess of 105 ml−1, and vary with temperature, salinity and host abundance (Waterbury and Valois, 1993; Suttle and Chan, 1993, 1994), and are estimated to remove up to a few percent of the Synechococcus population each day (Suttle and Chan, 1993; Suttle, 1994). Historically, cyanophages have been classified into the families Myoviridae, Siphoviridae and Podoviridae based largely on tail morphologies that are either contractile, non-contractile and flexible, or short and non-contractile, respectively (Suttle, 2000; Nelson, 2004; Lavigne et al., 2012). Representatives of all three families have been isolated from seawater (Wilson et al., 1993; Suttle and Chan, 1993; Waterbury and Valois, 1993; Sullivan et al., 2003) and freshwater (Safferman and Morris, 1963, 1964; Adolph and Haselkorn, 1971; Yoshida et al., 2006; Liu et al., 2007). Host-range studies have revealed that some cyanophages have broad host ranges and are able to infect strains that are distantly related (Suttle and Chan, 1993; Waterbury and Valois, 1993) or that even belong to different genera (Sullivan et al., 2003). Nonetheless, the mosaic architecture of phage genomes, including structural genes that result in similar morphology but that appear to have a different evolutionary origin (Sabehi et al., 2012), increasingly call into question the use of a Linnaean-based hierarchical classification, including morphology, as a basis for classifying phage (Lawrence et al., 2002; Nelson, 2004).
Many sequenced genomes exist for cyanophages that infect marine Synechococcus spp. ( Chen and Lu, 2002; Mann et al., 2005; Millard et al., 2009; Sullivan et al., 2010; Huang et al., 2012; Sabehi et al., 2012) and Prochlorococcus spp. (Sullivan et al., 2005, 2009; Sullivan et al., 2010; Labrie et al., 2013), as well as one from a myovirus that infects both genera (Weigele et al., 2007). Most cyanophage isolates are myoviruses with genome size ranges from 161 to 252 kb, and share core genes involved in virion structure, DNA replication and host-derived genes with T4-like phage (Mann et al., 2005; Millard et al., 2009; Sullivan et al., 2010) A number of sequenced marine cyanophages are also podoviruses, having genomes between 42 kb and 47 kb, and sharing similar genome architecture, as well as core genes with T7-like phages, including genes involved in virion structure, DNA replication and that are host-derived (Chen and Lu, 2002; Sullivan et al., 2005; Labrie et al., 2013). The few sequenced marine cyanophages that are siphoviruses have genome sizes between 30 and 108 kb (Sullivan et al., 2005; Huang et al., 2012; Ponsero et al., 2013), and, although divergent from other siphoviruses, they share several functional genes with lambda-like phages.
The comparatively limited data for freshwater cyanophages reveal that most are not closely related to their marine counterparts. Of the five sequenced cyanophages that infect Microcystis aeruginosa (Yoshida et al., 2008), Phormidium foveolarum (Liu et al., 2007, 2008), Planktothrix agardhii (Gao et al., 2012) and Synechococcus spp. (Dreher et al., 2011), only the myovirus S-CRM01 that infects the latter is related to myoviruses infecting marine Synechococcus spp. and Prochlorococcus spp. (Dreher et al., 2011).
Despite the widespread distribution and ecological importance of polar cyanobacteria (Vincent, 2000), little is known about viruses infecting these organisms. Here, we report on the isolation and genomic analysis of cyanophage S-EIV1 and its host, Synechococcus sp. strain PCCC-A2c, which were both isolated from polar freshwaters on northern Ellesmere Island, in the Canadian High Arctic. The virus bears little resemblance to previously characterized cyanophages and represents a new evolutionary lineage of viruses infecting cyanobacteria that serves as a new model system for investigating cyanophage–host interactions and a genomic template for exploring viral diversity.
Materials and Methods
Host cells
The phycoerythrin-rich picocyanobacterium Synechococcus sp. strain PCCC-A2c was isolated in July 2001 from the upper freshwater layer of Lake A, a meromictic lake at lat. 83°05'N, long. 75°30′W near the northern limit of the Canadian High Arctic (details in Van Hove et al. (2008)). The strain was isolated by sequential dilution from a sample taken immediately under the ice at 2 m depth, in the middle of the lake (Van Hove et al., 2008) in sterile BG-11 medium (Rippka et al., 1979) at 10 °C under continuous light (50 μmol photons m−2 s−1), and then transferred to batch cultures for maintenance at 8 °C under continuous low irradiance (33 μmol photons m−2 s−1). The isolate is maintained in the CEN Polar Cyanobacteria Culture Collection at Laval University as strain PCCC Number A2c.
Cyanophage isolation
Cyanophage S-EIV1 was isolated from a composite of virus concentrates collected from the surface waters of lakes and ponds on Ellesmere Island, Nunavut, Canada (Table 1). In brief, 20–40 l of water was filtered serially through 1.2-μm (GC50; Advantec MFS, Dublin CA) and 0.45-μm (HVLP; Millipore, Bedford, MA, USA) pore-size filters, and the remaining virus-size particles concentrated ∼100- to 200-fold using a 30-kDa-MWcutoff ultrafiltration cartridge (Prep/Scale-TFF-2; Millipore) (Suttle et al., 1991). Viral concentrates were stored at 4 °C in the dark until processed. S-EIV1 was isolated by pooling several subsamples from the virus concentrates (Table 1), adding the mix to an exponentially growing culture of Synechococcus sp. strain PCCC-A2c and incubating at 8 °C under continuous irradiance of 33 μmol photons m−2 s−1 for 14 to 17 days. Culture lysis was determined by a marked decrease in relative fluorescence (in vivo chlorophyll; Turner Designs TD-700 fluorometer, Sunnyvale, CA, USA) compared with control cultures. A clonal isolate of the virus was obtained by repetitive dilution to extinction in 96-well microtiter plates (Suttle and Chan, 1993) containing exponentially growing Synechococcus sp. strain PCCC-A2c.
Table 1. Site information for virus concentrates collected from freshwater systems on Ellesmere Island, Nunavut (Canada).
| Sample ID | Location | Latitude | Longitude | Date (August 2008) | Depth (m) |
|---|---|---|---|---|---|
| VC8 | Ward Hunt Lake | 83°05'N | 74°10'W | 14 | 0 |
| VC9 | Quttinirpaaq Lagoon | 83°05'N | 74°15'W | 17 | 0 |
| VC10 | Ward Hunt Ice Shelf | 83°01'N | 71°30'W | 19 | 0 |
| VC12 | Lake A-2m (oxic) | 83°00'N | 75°30'W | 20 | 2 |
| VC13 | Lake A-12 (oxicline) | 83°00'N | 75°30'W | 24 | 12 |
Amplification and purification of S-EIV1
The cyanophage was amplified by adding 0.1% (v/v) of the virus isolate to five 35 ml cultures of Synechococcus sp. strain PCCC-A2c and incubated until lysis. The lysates were pooled and filtered through a 0.45-μm pore-size filter (HVLP; Millipore) to remove cellular debris. The virus was then concentrated (∼50 × ) by ultrafiltration using Millipore Plus 70 Centricons. The concentrate was loaded onto a 20/30/40/50% (w/v in 50 mM Tris-HCl, pH 7.6) Optiprep (Sigma-Aldrich, St Louis, MO, USA) step gradient, and ultracentrifuged for 8 h at 86 711 g and 20 °C (SW40 rotor, Beckman Coulter, Indianapolis, IN, USA). After centrifugation, the single visible band was extracted from the gradient by puncturing the side of the tube with a sterile 1-ml syringe and dialyzed overnight in a 20,000 MWCO dialysis cassette (3 ml Slide-A-Lyzer; Thermo Scientific-Pierce, Rockford, IL, USA) against 500 ml of 200 mM Tris-HCl, pH 7.6, at 4 °C. These purified virus particles were used as the starting inoculum for subsequent experiments.
Transmission electron microscopy
S-EIV1 lysate (70 ml) was 0.45-μm filtered (HVLP; Millipore) and concentrated by ultracentrifugation for 6 h at 119 577 g and 8 °C in a 45Ti rotor (Beckman Coulter). The pelleted viruses were resuspended in 1 ml of supernatant. A portion of the virus suspension was fixed with glutaraldehyde (final 1% v/v) and adsorbed to the surface of formvar/carbon coated copper grids as previously described (Suttle and Chan, 1993). The grids were briefly stained with 2% phosphotungstic acid (pH 7), viewed and photographed on a FEI Tecnai G2 200 kV transmission electron microscope at the University of British Columbia Bioimaging Facility. Virus dimensions were estimated from electron micrographs of negatively stained particles.
Chloroform sensitivity
Sensitivity to chloroform was tested by adding 500 μl of 0.22-μm filtered lysate to an equal volume of chloroform and shaking by hand for 5 min. The chloroform was removed by centrifugation at 4100 g for 5 min at 10 °C (Allegra X-30, F2402 rotor, Beckman Coulter). The aqueous phase was transferred to a microfuge tube and incubated for 6 h at room temperature to evaporate any remaining chloroform. As a control, 500 μl of chloroform was added to 500 μl of BG-11 medium. Chloroform-treated virus, chloroform-treated medium and non-treated viruses were added to exponentially growing Synechoccoccus sp. strain PCCC-A2c cultures and relative fluorescence measured for 2 weeks.
Host range
Infectivity of S-EIV1 was tested against six replicate cultures of eight polar cyanobacterial strains (Supplementary Table 1) grown as described. Infectivity was determined by a decline in relative fluorescence compared with control cultures to which no viruses were added.
Structural proteins
To identify the structural proteins, purified S-EIV1 was diluted in SDS buffer (4:1, v/v) and heated at 95 °C for 5 min. The sample was then resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using a Mini-PROTEAN Tetra Cell (Bio-Rad Laboratories, Hercules, CA, USA). The 4–12% gel was run in a SDS running buffer (pH 8.3) at 100 V for 2 h using a Novex Sharp Protein Standard (Invitrogen, Carlsbad, CA, USA) for size calibration. The gel was stained overnight with Coomassie Blue and de-stained for 2 days in a solution of 20% methanol and 10% acetic acid.
DNA extraction, sequencing and assembly
Synechococcus sp. strain PCCC-A2c was grown at 8 °C in 800 ml of BG-11 medium (Rippka et al., 1979) in 1l flasks and 33 μmol photons m−2 s−1 continuous illumination. Exponentially growing cultures were infected with S-EIV1 and incubated as above for 14–17 days until lysis occurred. Sodium chloride (Sigma-Aldrich) was added to the lysate at a final concentration of 0.5 M at 4 °C, which after 1 h was filtered through 1.2-μm pore-size glass-fiber (GC50; Advantec MFS) and 0.22-μm pore-size membranes (GVWP; Millipore) to remove cellular debris. The filtered lysate was ultracentrifuged for 6 h at 119 577 g and 8 °C (Type 45Ti rotor, Beckman Coulter), the supernatant removed and the virus pellet resuspended in 200 μl of BG-11 medium.
The pellet was treated with DNase 1 and RNase A to remove free nucleic acids, and the nucleic acids extracted using a QiAamp MinElute Virus Spin Kit (Qiagen, Mississauga, ON, Canada). The DNA was sheared into ∼300 bp fragments using a Covaris M220 ultrasonicator (Covaris, Woburn, MA, USA) and purified using Agencourt AMPure XP beads (Beckham Coulter). The sequencing library was constructed using NxSeq DNA Sample Prep Kits (Lucigen, Middleton, WI, USA) and sequenced on an Illumina MiSeq at the Génome Québec Innovation Centre at McGill University (Montréal, QC, Canada). The adapters were trimmed from the reads using Trimmomatic-0.30 (Bolger et al., 2014) quality checked with Sickle (Joshi and Fass, 2011), and assembled using Ray with default parameters, and 23 as the k-value (Boisvert et al., 2012). The sequence data have been submitted to the GenBank databases under accession No. KJ410740.
Genome annotation and identification of regulatory elements and motifs
Open reading frames (ORFs) in S-EIV1 were predicted using GeneMark (Lukashin and Borodovsky, 1998) and GLIMMER (Delcher et al., 2007), where the predictions differed, the longer of the two was kept. The predicted ORFs were translated and assigned putative functions by using BLASTp to compare them with protein sequences in the GenBank (nr), Acclame and Procite databases. Sequences with e-values <10−3 were considered to be homologs. PSI-BLAST and HHpred were used to predict more distant homologs. The genome was also analyzed for regulatory elements and motifs such as tRNA genes, promoter motifs and transcriptional terminators. tRNA genes were identified using tRNAscan-SE (Lowe and Eddy, 1997) and Aragorn v1.1(Laslett and Canback, 2004). Putative promoter motifs were identified using PHIRE (Lavigne et al., 2004) with default parameters (20-mer sequences (S) with 4 base pair degeneracy (D=4)), and if they occurred in the 150 bp region immediately upstream of start codons of predicted protein-coding genes. Rho-independent terminators were identified using Softberry's FINDTERM, with the default energy threshold set to −16 kCal (Weigele et al., 2007). The genomic map was constructed using GCview (Grant and Stothard, 2008). The genome of S-EIV1 was compared with fosmid MEDDCM-OCT-S04-C348 (Ghai et al., 2010) by using a tBLASTx analysis (cutoff e-value>0.0001).
The prophage incision element, AvaD, in Anabaena variabilis ATC1495 was confirmed by translating the ORFs and using blastp to compare them with sequences in the GenBank (nr), Acclame and Procite databases, as outlined above.
Phylogenetic analysis
DNA polymerase A (DNApolA) and the large terminase subunit (terL) were compared phylogenetically with those from other cyanophages by aligning the inferred amino-acid sequences with ClustalX for DNApolA and Promals for terL (Pei and Grishin, 2007; Pei et al., 2007) using default parameters followed by manually refining the alignments with Geneious v4.7 (Drummond et al., 2011). Maximum likelihood trees were constructed with RAxML rapid bootstrapping and ML search (100 replicates) (Stamatakis et al., 2008) assuming the James-Taylor Thornton model of substitution using empirical base frequencies and estimating the proportion of invariable sites from the data.
Recruitments of reads to metagenomic data
To interrogate other aquatic systems for S-EIV1-like phages, reads were recruited from viral metagenomic data (Supplementary Table 2) onto the genome of S-EIV1 using tBLASTx with an e-value of 10−10. If a read was recruited more than once to the genome, the read was associated with the region that provided the lowest e-value. The reads were mapped on the S-EIV1 genome based on their percent amino acid identified using ggplot2 (Wickham, 2009). In addition, we constructed a database containing protein sequence from S-EIV1 and the NCBI viral database (n=181 331 proteins). Each read from the Pacific Ocean virome (Hurwitz and Sullivan, 2013) was used as a query in a BLASTx analysis against the protein database (cutoff e-value=10−3). Each read was assigned to a single best matching reference viral protein based on the e-value. Although the database contained all viral proteins, only reads with a best match to one of the 15 cyanophages that recruited the most reads are shown in Figure 6c. To reduce the effect of genome size, the read counts were normalized by the numbers of ORFs.
Results and Discussion
Cyanobacteria are major primary producers in freshwater ecosystems, and are often the most abundant phototrophs in polar lakes (Vincent, 2000); however, representative cyanophages from these waters have not been previously described. In the present study, we isolated and characterized cyanophage, S-EIV1, from the Canadian High Arctic. SEIV1 was isolated from a composite of virus concentrates collected from the surface waters of lakes and ponds on Ellesmere Island, Nunavut, Canada. This virus infects the freshwater polar cyanobacterium Synechococcus sp. strain PCCC-A2c. On the basis of morphology and genomic content cyanophage S-EIV1 represents a new evolutionary lineage of bacteriophages. The circularly permuted genome of 79 178 bp has little similarity to other sequenced phages; yet, interrogation of metagenomic data suggests that viruses related to S-EIV1 are widely distributed in aquatic systems.
General features
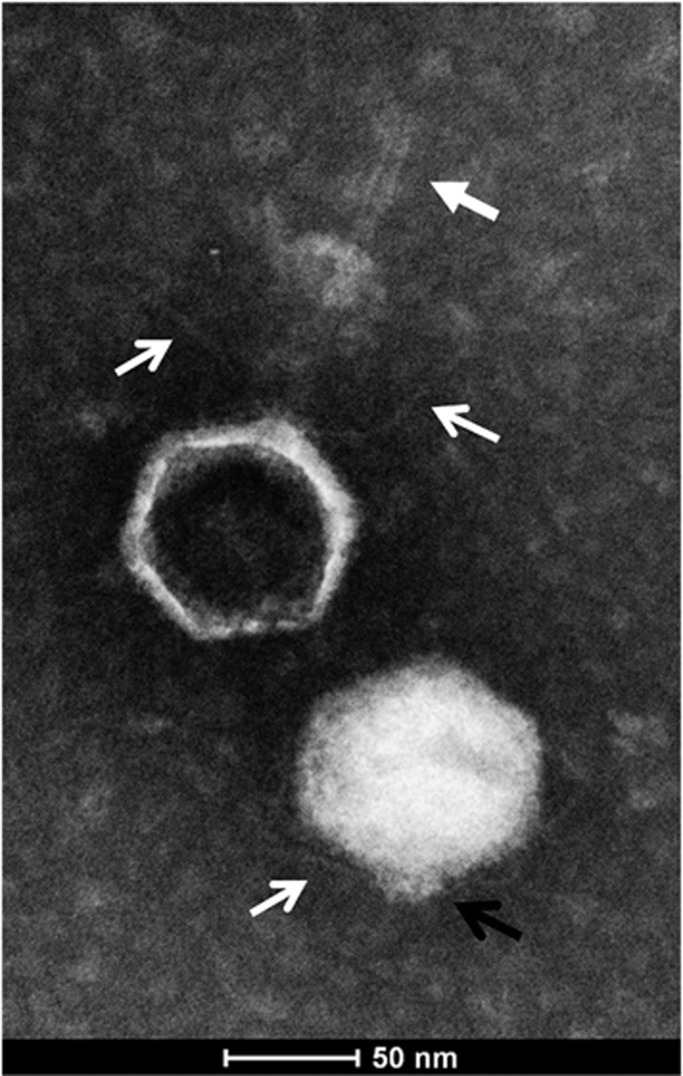
Electron micrographs of negatively stained cyanophage S-EIV1 particles revealed icosahedral capsids with an average diameter of ∼95 nm (n=33). Evidence of short spiky extensions and long, fine tail fibers (Figure 1) projecting from the base of the capsid were seen on both intact and empty capsids, but only capsids that were devoid of nucleic acids possessed a long tail-like structure that extended up to ∼125 nm in length (average=109 nm, n=5; Figure 1, closed white arrow) from the capsid. The observation that the extended tail was only associated with particles devoid of nucleic acids, while none of the particles containing nucleic acids had extended tails, suggests that ejection of the tail is associated with release of nucleic acids, as is seen in T4-like phages. The unusual morphology associated with ejection of a tail distinguishes S-EIV1 from other described phages. Other studies have shown that the release of nucleic acids can affect phage structure (Liu et al., 2011; Hu et al., 2013). Liu et al. (2011) demonstrated that the release of nucleic acids in a marine podovirus led to the tail fibers being extended horizontally rather than parallel to the capsid surface.
Figure 1.
General features of cyanophage S-EIV1. Transmission electron micrograph of S-EIV1 negatively stained with 2% phosphotungstic acid reveals icosahedral capsids with fine tail fibers (open white arrows) and short spiky extensions (black open arrow) that were present in filled and empty capsids, whereas delicate tail-like structures were only associated with empty capsids (closed white arrow).
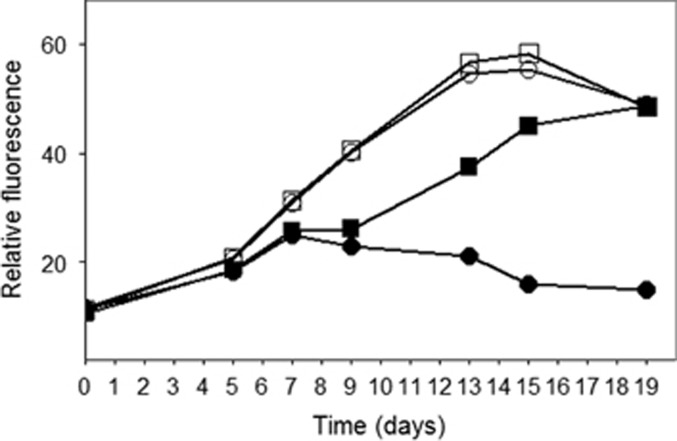
The infectivity of S-EIV1 is chloroform sensitive (Figure 2), similar to some tailed phages. Although sensitivity to chloroform can be associated with the presence of lipids in the phage, it is not necessarily the case (Ackermann, 2006). As well, S-EIV1 was unable to infect eight other polar cyanobacterial isolates from nearby freshwaters (Supplementary Table 1), suggesting that the virus has a limited host range. Hence, these data do not support the hypothesis of Säwström et al. (2008) that the high proportion of visibly infected cells observed in polar waters might be indicative of a broad host range.
Figure 2.
Effect of chloroform on the infectivity of S-EIV1. Relative fluorescence is shown for Synechococcus cultures grown in untreated (open circles) or chloroform-treated medium (open squares) or inoculated with chloroform-treated (black squares) or untreated viruses (black circles).
Genomic analysis
Despite the two distinct morphologies observed in the micrographs, the genomic data indicate that the particles are from a clonal isolate of a single phage. Both the TEM images and the purified nucleic acids for sequencing were scaled-up from the same cyanophage stock. The only phage-like sequences that were recovered belonged to the S-EIV1 genome; contaminating sequences were all bacterial.
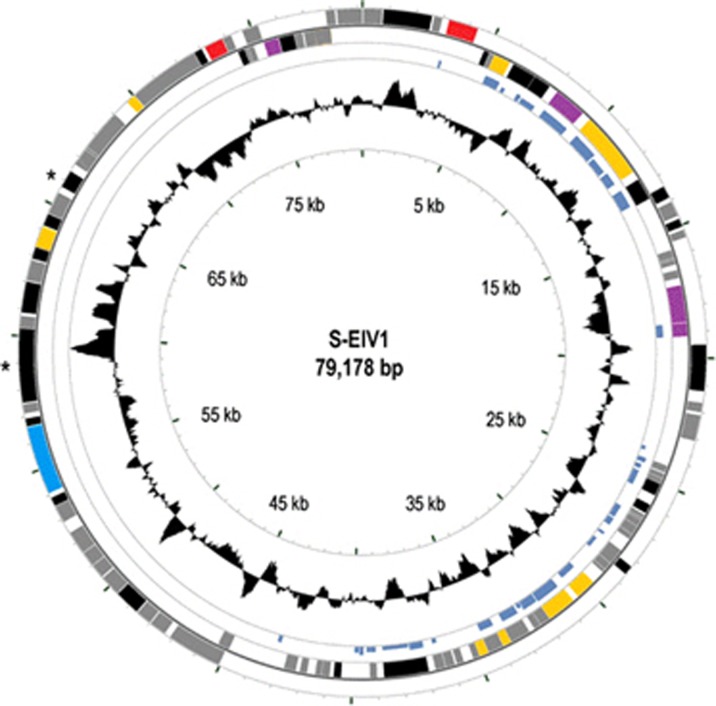
The 79 178 bp genome of S-EIV1 is circularly permuted (Figure 3), with a GC content of 46.2%. Most ORFs in S-EIV1 do not share significant similarity with genes of known function. Of the 130 predicted ORFs encoded on both strands, 42 have significant similarity to other sequences, although only 10 were to phage sequences, and only 15 were similar to genes of known function (BLASTp, e-value cutoff=10−3). Sequences that were predicted to encode proteins with known functions included those associated with DNA metabolism, replication and cell lysis (Table 2, Supplementary Table 3); no sequences coding for tRNAs were found. PSI-BLAST and HHpred were used to ascribe function to additional ORFs and resulted in the identification of putative coding sequences for a viral morphogenesis protein (ORF109), an exonuclease (ORF17), an o-methyltransferase (ORF19) and a restriction endonuclease (ORF33) (Table 3). This gives a total of 19 ORFs that show homology to genes of known function. Three transcriptional terminators were predicted by Findterm (Supplementary Table 4); two are downstream of ORFs with unknown function (ORF6 and ORF16), while one is downstream of a gene predicted to encode a peptidase (ORF106).
Figure 3.
Genomic map of S-EIV1. Circles from outmost to innermost correspond to (i) predicted ORFs (BLASTp, nr database, e-value>0.0001) on forward strand and (ii) reverse strand; (iii) tBLASTx hits (e-value>0.0001) against the fosmid MEDDCM-OCT-S04-C348 (the height of the bar is proportional to the e-value) and (iv) GC content plotted relative to the genomic mean of 46.2% G+C. Only ORFs >200 bp are shown and are colored as follows: red, lysis/lysogeny; gray, no homolog; black, hypothetical proteins; purple, host-derived genes; yellow, DNA metabolism and replication. *Indicates structural genes that were identified by SDS–PAGE.
Table 2. Predicted ORFs of cyanophage S-EIV1 with similarity to genes of known function.
| ORF | Length(bp) | Strand | Significant hit | Organism | e-Value | %Id (shared aa) |
|---|---|---|---|---|---|---|
| 5 | 1107 | + | Lysozyme | Synechococcus phage S-CBP3 | e−115 | 55% (205) |
| 8 | 888 | − | PurM | Synechococcus phage S-CAM1 | 9e−78 | 52% (114) |
| 13 | 1266 | − | Glycosyl transferase group 1 | Myxococcus fulvus HW-1 | e−46 | 30% (123) |
| 14 | 156 | − | S-adenosylmethionine decarboxylase proenzyme | Prochlorococcus marinus str. AS9601 | 2e−05 | 44%(22) |
| 15 | 2640 | − | P4 phage primase | Cyanothece sp. PCC7424 | 1e−78 | 37% (176) |
| 28 | 552 | − | DNA-binding ferritin-like protein | Opitutaceae bacterium TAV1 | 4e−04 | 30% (39) |
| 50 | 807 | − | ssDNA-binding protein | Thermodesulfobium narugense DSM | 5e−06 | 36%(47) |
| 52 | 1869 | − | DNA polymerase family A | Bilophila wadsworthia 316 | e−128 | 42% (259) |
| 55 | 627 | − | FAD dependent thymidylate synthase | Cyanophage S-TIM5 | 7e−57 | 51% (108) |
| 56 | 378 | − | Endodeoxyribonuclease | Pseudomonas sp. HPB0071] | 3e−05 | 47% (26) |
| 95 | 2538 | + | Putative terminase large subunit Ava_D0014 | Anabaena variabilis ATCC 29413 | 1e−67 | 43%(137) |
| 106 | 744 | + | Peptidase, M23 family | Acinetobacter sp. WC-743 | 3e−07 | 33%(44) |
| 115 | 360 | + | HNH nuclease | Synechococcus sp. CC9902 | 4e−16 | 42% (40) |
| 119 | 663 | + | Lysozyme | Acinetobacter sp. RUH2624 | 6e−18 | 37% (57) |
| 124 | 585 | − | Deoxycytidine triphosphate deaminase | Synechococcus sp. WH 7803 | 2e−53 | 53% (104) |
Table 3. Identification of distant homologs of S-EIV1 ORFs using HHpred analysis.
| ORF | Length(bp) | Strand | Predicted function | PfamA ID | e-Value |
|---|---|---|---|---|---|
| 17 | 849 | − | Exonuclease | PF12684 | 4.2e−17 |
| 19 | 588 | + | O-methyltransferase | PF01596 | 1.6e−20 |
| 33 | 621 | + | Restriction endonuclease | NA | 3.4e−11 |
| 109 | 327 | − | Morphogenesis protein | NA | 1.2e−39 |
Although S-EIV1 particles that are filled with DNA show morphological similarity with members of the Podoviridae, there are few genes shared between S-EIV1 and podoviruses, including no evident homology between genes encoding the core structural proteins, which indicates that S-EIV1 does not belong within this family. Similarly, S-EIV1 shares negligible genetic similarity with the large podoviruses that infect Cellulophaga baltica (Holmfeldt et al., 2013), including genes encoding the structural proteins. In fact, sequences encoding known phage structural proteins (that is, capsid, tail tube, portal or tail fiber) were not found within the S-EIV1 genome, with the exception of the terminase large subunit and a viral morphogenesis protein classified with HHpred, providing further evidence that S-EIV1 is distinct from podoviruses and represents a previously unknown phage lineage. However, SDS–PAGE analysis resolved six structural proteins of about 23, 32, 35, 39, 42 and 85 kDa (Supplementary Figure 1). Of these, the only structural proteins that could be matched with specific ORFs were the viral morphogenesis protein (ORF109) corresponding to the 23 kDa band, and ORF99 being the only putative coding sequence long enough to encode an 85 kDa protein. The detection of only six structural proteins for a phage of this size is undoubtedly an underestimation, especially given the complexity of the tail structure of S-EIV1. For example, SDS–PAGE analysis for cyanophage PaV-LD, which is about 80 nm in diameter and lacks a tail, resolved 13 structural proteins (Gao et al., 2012), while 14 structural proteins were resolved for Cyanophage Syn5 (Raytcheva et al., 2011). Ultimately, a mass spectrometry-based proteomics analysis would be a more sensitive approach for elucidating the structural proteins that make up the S-EIV1 virion.
Host-derived genes that have been found in other cyanophages, such as those encoding proteins involved in photosynthesis, carbon metabolism and phosphorus-related functions (Lindell et al., 2004; Millard et al., 2004; Mann et al., 2005; Sullivan et al., 2005; Weigele et al., 2007; Labrie et al., 2013), were not found in S-EIV1; however, genes for proteins involved in nucleotide metabolism and stress response were identified. First, S-EIV1 encodes a homolog of S-adenosylmethionine decarboxylase (SpeD), a key enzyme in the biosynthesis of spermidine and spermine, polyamines that are important in photoadaptation and photoinhibition in oxygenic phototrophs (Kotzabasis et al., 1999). For example, a mutant of Synechocystis sp. PCC6803 with reduced spermidine content exhibits reduced psbA2 transcript stability (Mulo et al., 1998). SpeD is also commonly found in marine T4-like cyanophage genomes (Clokie et al., 2010; Ignacio-Espinoza and Sullivan, 2012). Another sequence (ORF28) has distant homology (BLASTp, e-value=10−4) to genes encoding DNA binding protein from starved cells (DPS). These intracellular iron-binding proteins in the bacterioferritin/ferritin superfamily (Pen and Bullerjahn, 1995) act in iron storage, DNA binding and oxidative stress prevention. Prokaryotes have highly regulated enzymatic systems to protect DNA from oxidative damage due to reactive oxygen species such as hydroxyl radicals, superoxide and H2O2 (Storz et al., 1990). During starvation, when the ability to cope with environmental stress is compromised by the lack of nutrients, DPS efficiently and rapidly responds to oxidative and nutritional stresses by making cells more resistant to reactive oxygen (Storz et al., 1990; Farr and Kogoma, 1991; Martinez and Kolter, 1997). A gene encoding DPS might help cyanophages in polar lakes where oxygen tensions are high but DNA-repair rates are low because of cold temperatures, and low-nutrient availability, which constrains phytoplankton production (Vincent et al., 2008).
Other genes identified in the genome of S-EIV1 include a phosphoribosylaminoimidazole synthetase (purM) homolog that encodes an enzyme involved in purine ribonucleotide biosynthesis and a deoxycytidine triphosphate (dctp) homolog that encodes an enzyme required for pyrimidine metabolism. S-EIV1 also encodes a thymidylate synthase homolog, which may be involved in scavenging host nucleotides, and which may assist viruses with the synthesis of thymidylate from uridylate after host transcription has stopped (Thompson et al., 2011). As occurs in its marine counterparts, S-EIV1 uses the alternative form of thymidylate synthase (thyX) rather than the canonical form (thyA) (Ignacio-Espinoza and Sullivan, 2012).
S-EIV1: a new evolutionary lineage of cyanophage
S-EIV1 represents a new evolutionary lineage of cyanophages based on genome content and organization. In particular, with the exception of the terminase large subunit and a viral morphogenesis protein, we were unable to find any sequence similarity between the S-EIV1 genome and other phage structural proteins. The S-EIV1 genome has similarity to some of the core genes found in cyanopodoviruses including ssDNA-binding protein (ORF50), endonuclease (ORF 56), primase (ORF15), terminase large subunit (terL) (ORF95) and DNA polymerase family A (DNApol) (ORF52); many others are missing including core genes involved in DNA metabolism, assembly and capsid structure (Labrie et al., 2013). As well, cyanopodoviruses generally have a genomic architecture similar to coliphage T7, which encodes genes on a single strand arranged as follows: (1) transcription, (2) RNA polymerase, DNA metabolism and replication and (3) phage assembly and DNA maturation (Labrie et al., 2013). In contrast, S-EIV1 codes from both strands, and similar to Roseophage SIO1 (Rohwer et al., 2000) does not encode RNA polymerase, implying that host transcription machinery is used during infection; this has also been suggested for the siphovirus P-SS2 that infects Prochlorococcus sp. (MIT9313) (Sullivan et al., 2009).
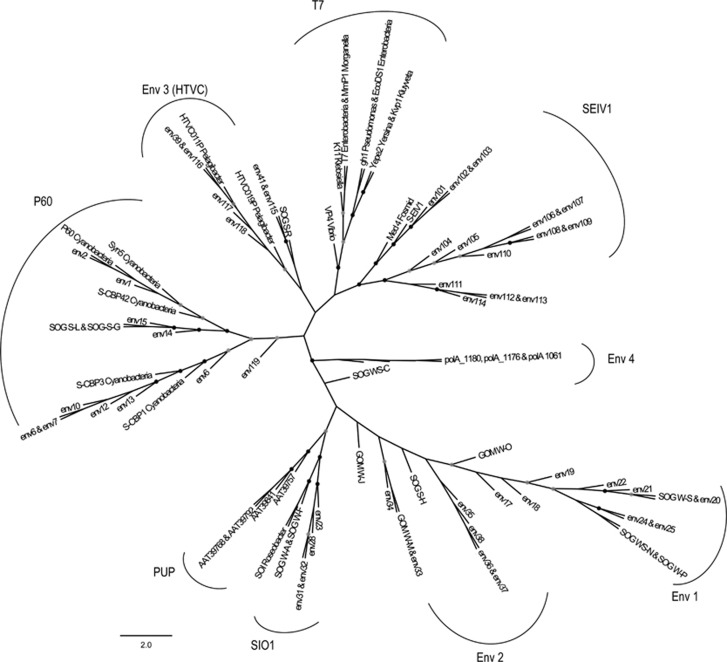
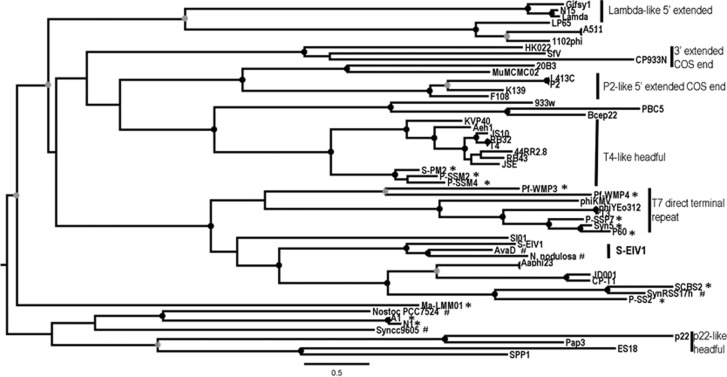
Phylogenetic analysis of DNApol and terL shows that S-EIV1 is evolutionarily distinct from other cyanophage isolates. Although DNApol is related to those found in podoviruses, it is evolutionarily distinct from other evolutionary groups including viruses infecting Pelagibacter ubique (HTCV-like) (Zhao et al., 2013) and Roseobacter sp. (SIO-like) (Rohwer et al., 2000) as well as the P60 group of cyanopodoviruses (Figure 4 and Supplementary Figure 2). The sequences were also very divergent from, and could not be reliably aligned with DNApol sequences from the freshwater cyanophages Pf-WMP3 and Pf-WMP4 (Liu et al., 2007, 2008). Further evidence of the evolutionary divergence of S-EIV1 from other viruses is provided by terL, which encodes a protein involved in DNA packaging. S-EIV1 clusters in a well-supported clade with terminases found in prophage elements in the filamentous cyanobacteria Anabaena variabilis (AvaD) and Nodosilinea nodulosa (Shih et al., 2013) (Figure 5). The phylogenetic divergence in terL between S-EIV1 and cyanopodoviruses is not surprising given that the genome of S-EIV1 is circularly permuted while in cyanopodoviruses it is linear with direct repeats, which likely involves different DNA packaging processes (Rao and Feiss, 2008).
Figure 4.
Unrooted maximum likelihood amino-tree of DNA polymerase A with sequences from amplicons (Labonté et al., 2009; Breitbart et al., 2004), as well as phage isolates and metagenomic sequences. Bootstrap values are indicated as black (90% to 100%) or gray (70% to 89%) dots at the nodes (100 replicates). The groups are labeled as follows: P-60, marine cyanopodoviruses; T7, Enterobacter podoviruses; SIO1-like, Roseobacter viruses. PUP clade was identified by Breitbart et al. (2004), Env clade 1–3 were identified by Labonté et al. (2009). Env-4 was identified by Schmidt et al. (2014). Env 5 is a new environmental clade that includes cyanophage S-EIV1. Scale bar represents amino-acid substitutions per site.
Figure 5.
Maximum likelihood amino-acid tree of the viral terminase large subunit (terL). Bootstrap values are indicated as black (90% to 100) or gray (75% to 89%) dots at the nodes (100 replicates). Cyanophage genomes are denoted by * and cyanobacterial host genomes by #. The clade containing S-EIV1 (S-EIV1-like) is in bold.
The isolation of S-EIV1 suggests that new evolutionary lineages of viruses are likely to be discovered if different host strains are screened. This has been shown recently with previously unknown groups of viruses isolated on Pelagibacter ubique (Zhao et al., 2013), a bacterium of SAR116 clade (Kang et al., 2013) and Cellulophaga baltica (Holmfeldt et al., 2013). Similarly, most cyanophages have been isolated using a few strains of Synechococcus spp.; however, a previously unknown lineage of myoviruses (S-TIM5) was isolated from the Red Sea using a different Synechococcus strain (Sabehi et al., 2012). Clearly, there is enormous potential to isolate representatives of previously unknown groups of viruses by screening untested taxa of host organisms.
S-EIV1-like viruses in nature
Although S-EIV1 represents a previously unknown phage lineage, it shares synteny with a sequence from an uncultured phage and an incision element in a filamentous cyanobacterium. During the genome annotation, the BLAST analysis demonstrated high similarity with putative proteins from an uncultured cyanophage (MEDDCM-OCT-S04-C348). A BLASTx analysis (e-value<0.0001) of the S-EIV1 genome against the sequence of MEDDCM-OCT-S04-C348, captured in a fosmid library from the deep-chlorophyll maximum in the Mediterranean Sea (Ghai et al., 2010), demonstrated synteny between a region of 40 kb from S-EIV1 and the fosmid (Figure 3, inner circle). A total of 26 ORFs are shared between S-EIV1 and MEDDCM-OCT-S04-C348. Of these 19 ORFs encode for hypothetical proteins and 7 encode for putative proteins with known function including lysozyme (ORF5), phosphoribosylaminoimidazole synthetase (ORF8), glycosyl transferase (ORF13), primase (ORF15), DNA-binding ferritin-like protein (ORF28), ssDNA-binding protein (ORF50) and DNApol (ORF52) (Supplementary Table 5). The similarity between S-EIV1 and MEDDCM-OCT-S04-C348, as well as the phylogenetic affiliation of DNApol indicates that MEDDCM-OCT-S04-C348 is from a relative of S-EIV1. Moreover, as S-EIV1 did not share significant sequence similarity with any other of the fosmids in the data base, it represents yet another evolutionary group of viruses within the deep-chlorophyll maximum of the Mediterranean (Mizuno et al., 2013).
Despite huge differences in temperature, salinity and wide geographic separation, High Arctic lakes and the Mediterranean Sea are oligotrophic regions where Synechococcus is a major primary producer. At the deep chlorophyll maximum in the Mediterranean, Synechococcus abundances range from 1.75 to 4 × 106 cells ml−1 (Agawin and Agustf, 1997), and in Lake A, picocyanobacterial populations reach up to 6 × 104 cells ml−1 (Van Hove et al., 2008). Moreover, Synechococcus sp. strain PCCC-A2c has high 16S rDNA gene sequence similarity to cyanobacteria isolated from freshwater, brackish and marine systems (Supplementary Table 6). This may also reflect the range of salinity conditions in meromictic Lake A, from freshwater at the surface where the strain was isolated to saline conditions at depth. Consequently, strains similar to Synechococcus sp. strain PCCC-A2c may occur in the Mediterranean Sea, as do closely related cyanophages such as MEDDCM-OCT-S04-C348.
Although S-EIV1 demonstrates a lytic lifestyle, it shares genes with prophage elements. A second 10 kb module on the positive strand of S-EIV1 shares synteny with, and has five ORFs with high similarity to a 37 kb incision element (AvaD) in the filamentous cyanobacterium Anabaena variabilis ATCC29413. Shared ORFs include putative genes encoding terL (Supplementary Figure 3) and a structural protein, suggesting an evolutionary relationship between S-EIV1 and AvaD. Annotation of AvaD revealed more phage-like genes including two integrases (AvaD0049 and AvaD0026), hnh nuclease (AvaD0046), endonuclease (AvaD0044), primase (AvaD0041), DNA polymerase (AvaD0033), RNA polymerase (AvaD0022) and DNA binding protein (AvaD0015), providing evidence that AvaD is a viral element.
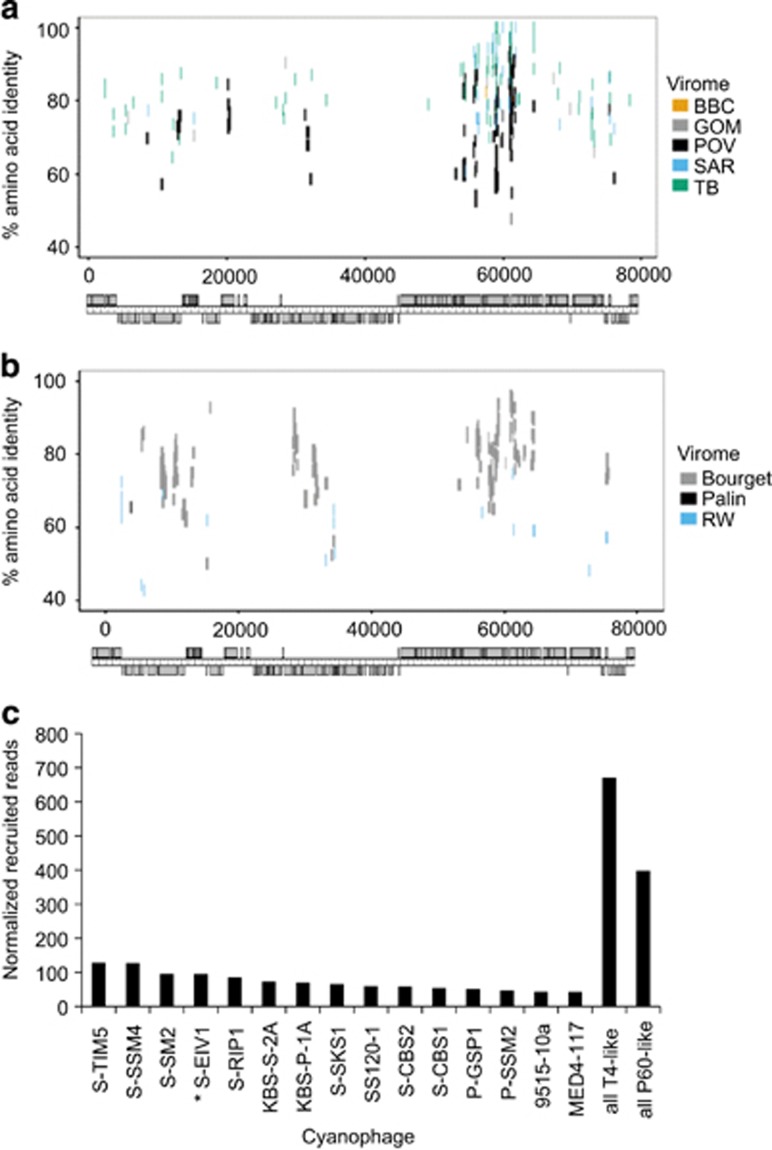
Phylogenetic analysis of DNApolA with known phages, metagenomic sequences and amplicons (Breitbart et al., 2004; Labonté et al., 2009; Schmidt et al., 2014) reveals that S-EIV1 along with environmental sequences forms a previously unrecognized evolutionary group of DNApolA sequences (Figure 4). Further evidence for S-EIV1-like phages in aquatic systems was obtained from the recruitment of reads from environmental viral metagenomic data (Figures 6a and b). Sequences from freshwater and marine viral environmental metagenomic databases were recruited onto the genome of S-EIV1 (Figures 6a and b), with most recruited sequences being similar to ORFs located between 55 and 65 kb into the genome. This region of the genome is believed to encode structural proteins, suggesting that phages with similar structural proteins are widespread in aquatic systems. In contrast, no reads were recruited to the region between 40 and 55 kb, suggesting that this is a variable region containing environment-specific genes or genes involved in host recognition.
Figure 6.
Prevalence of S-EIV1-like sequences in environmental viral metagenomic data. (a, b) Fragment recruitment of reads from environmental viral metagenomic data (Supplementary Table 2) onto the genome of S-EIV1. Each horizontal line represents a read recruited from one of the following publicly available metagenomic data sets: (a) marine viral metagenomes: Gulf of Mexico (GOM), Strait of Georgia (BBC), Sargasso Sea (SAR), Pacific Ocean (POV) and Tampa Bay (TB). (b) Freshwater viral metagenomes Lake Bourget (Bourget), Lake Pavin (Pavin) and Reclaimed water (RW). Reads were recruited against each of the assembled genomes using tBLASTx with an e-value of 10−10. The position of each line represents the percent similarity of the read to the genome. Only the hit with the highest e-value was used for each read. (c) Abundance of S-EIV1 relative to other cyanophages in the Pacific Ocean Virome. Only the 15 cyanophage isolates that recruited the most reads are shown in the bar chart. The number of reads was normalized to the number of ORFs in each genome.
Metagenomic databases retrieved from environments where Synechococcus is a major primary producer showed a higher number of recruited reads. For example, 249 reads were recruited from the Lake Bourget viral metagenomes while only 4 reads from the Lake Pavin viral metagenomes (Roux et al., 2012). Lake Bourget is an oligo-mesotrophic lake dominated by cyanobacteria (Zhong et al., 2013), whereas Lake Pavin is believed to be dominated by picoeukaryotes (Lefèvre et al., 2008). In addition, a BLASTx analysis of a database containing S-EIV1 and the NCBI viral proteins with the Pacific Ocean Virome (POV) metagenomes (Hurwitz and Sullivan, 2013) indicates that S-EIV1-like phages are prevalent in these samples (Figure 6c). Only sequences with higher similarity to the cyanophages S-SSM4, S-SM2 and S-TIM5 occurred more frequently in the POV metagenomes. This comparison overestimates the occurrence of S-EIV1 relative to other cyanophages, because each recruited read was only assigned to the phage with the most similar sequence. Hence, for phages with very similar sequences, such as the P60-like marine cyanophages P60, S-CBP3, P-SSP7 and P-RSP2, which share many core genes, only one phage genome would recruit each read. As there are several representative P-60 like cyanophages, but only one S-EIV1-like phage, the overall effect is to dilute the number of reads assigned to each P60-like cyanophage. Regardless, the data indicate that S-EIV1-like phages are widespread and relatively abundant in aquatic systems.
S-EIV1 infects a polar isolate of Synechococcus sp. and represents a previously unknown lineage of cyanophages. Metagenomic data indicate that related viruses are globally widespread in aquatic systems. Given the importance of picocyanobacteria for primary production in marine and fresh waters, this previously unknown evolutionary group of cyanophages lineage may have a major ecological role.
Acknowledgments
We are grateful to J.A. Gustavsen and C.E. Chow for help with library preparation and J.L. Clasen for helpful comments on the manuscript. We are also thankful to A.D. Jungblut, J. Veillette, S. Charvet and D. Sarrazin for field assistance and B. Ross for technical assistance. This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Institute for Advanced Research, the Network of Centres of Excellence ArcticNet, the FQRNT and UBC University Graduate Fellowship. We also thank Parks Canada, the Polar Continental Shelf Project (Natural Resources Canada) and the Northern Scientific Training Program (Aboriginal Affairs & Northern Development Canada) for logistical and infrastructure support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ackermann HW.2006Classification of bacteriophages The bacteriophagesCalendar R, (ed).Oxford University Press: New York, NY, USA; 8–16. [Google Scholar]
- Adolph KW, Haselkorn R. Isolation and characterization of a virus infecting the blue-green alga Nostoc muscorum. Virology. 1971;208:200–208. doi: 10.1016/0042-6822(71)90023-7. [DOI] [PubMed] [Google Scholar]
- Agawin NSR, Agustf S. Abundance, frequency of dividing cells and growth rates of Synechococcus sp. (Cyanobacteria) in the stratified Northwest Mediterranean Sea. J Plankton Res. 1997;19:1599–1615. [Google Scholar]
- Bergeron M, Vincent WF. Microbial food web responses to phosphorus and solar UV radiation in a subarctic lake. Aquat Microb Ecol. 1997;12:239–249. [Google Scholar]
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Miyake JH, Rohwer F. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett. 2004;236:249–256. doi: 10.1016/j.femsle.2004.05.042. [DOI] [PubMed] [Google Scholar]
- Chen F, Lu J. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl Environ Microbiol. 2002;68:2589–2594. doi: 10.1128/AEM.68.5.2589-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie MR, Millard AD, Mann NH. T4 genes in the marine ecosystem: studies of the T4-like cyanophages and their role in marine ecology. Virol J. 2010;7:291. doi: 10.1186/1743-422X-7-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Brown N, Bozarth CS, Schwartz AD, Riscoe E, Thrash C, et al. A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages. Environ Microbiol. 2011;13:1858–1874. doi: 10.1111/j.1462-2920.2011.02502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A.2011. Geneious v5.4. Available at http://www.geneious.com .
- Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Mol Biol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao E-B, Gui J-F, Zhang Q-Y. A novel cyanophage with cyanobacterial non-bleaching protein A gene in the genome. J Virol. 2012;86:236–245. doi: 10.1128/JVI.06282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Martin-Cuadrado A-B, Molto AG, Heredia IG, Cabrera R, Martin J, et al. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 2010;4:1154–1166. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, Verberkmoes NC. Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci USA. 2013;11031:12798–12803. doi: 10.1073/pnas.1305956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove P, Vincent WF, Galand PE, Wilmotte A. Abundance and diversity of picocyanobacteria in High Arctic lakes and fjords. Arch Hydrobiol Suppl Algol Stud. 2008;126:209–228. [Google Scholar]
- Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang K, Jiao N, Chen F. Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges. Environ Microbiol. 2012;14:540–558. doi: 10.1111/j.1462-2920.2011.02667.x. [DOI] [PubMed] [Google Scholar]
- Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One. 2013;8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignacio-Espinoza JC, Sullivan MB. Phylogenomics of T4 cyanophages: lateral gene transfer in the “core” and origins of host genes. Environ Microbiol. 2012;14:2113–2126. doi: 10.1111/j.1462-2920.2012.02704.x. [DOI] [PubMed] [Google Scholar]
- Joshi NA, Fass JN.2011. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. Available at http://github.com/najoshi/sickle .
- Jungblut AD, Lovejoy C, Vincent WF. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 2010;4:191–202. doi: 10.1038/ismej.2009.113. [DOI] [PubMed] [Google Scholar]
- Kang I, Oh H-M, Kang D, Cho J-C. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci USA. 2013;110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzabasis K, Strasser B, Navakoudis E, Senger H, Dornemann The regulatory role of polyamines in structure and functioning of the photosynthetic apparatus during photoadaptation. J Photochem Photobiol B Biol. 1999;50:45–52. [Google Scholar]
- Labonté JM, Reid KE, Suttle CA. Phylogenetic analysis indicates evolutionary diversity and environmental segregation of marine podovirus DNA polymerase gene sequences. Appl Environ Microbiol. 2009;75:3634–3640. doi: 10.1128/AEM.02317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Frois-Moniz K, Osburne MS, Kelly L, Roggensack S, Sullivan MB, et al. Genomes of marine cyanopodoviruses reveal multiple orgins of diversity. Environ Microbiol. 2013;15:1356–1376. doi: 10.1111/1462-2920.12053. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne R, Kropinski AM, Molineux IJ.2012Order Caudovirales Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of VirusesKing AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, (eds)Elsevier Academic Press: London [Google Scholar]
- Lavigne R, Sun WD, Volckaert G. PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics. 2004;20:629–635. doi: 10.1093/bioinformatics/btg456. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Hatfull GF, Roger W, Hendrix RW. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol. 2002;184:4891–4905. doi: 10.1128/JB.184.17.4891-4905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre E, Roussel B, Amblard C, Sime-Ngando T. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS One. 2008;3:e2324. doi: 10.1371/journal.pone.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kong S, Shi M, Fu L, Gao Y, An C. Genomic analysis of freshwater cyanophage Pf-WMP3 infecting cyanobacterium Phormidium foveolarum: the conserved elements for a phage. Microb Ecol. 2008;56:671–680. doi: 10.1007/s00248-008-9386-7. [DOI] [PubMed] [Google Scholar]
- Liu X, Shi M, Kong S, Gao Y, An C. Cyanophage Pf-WMP4, a T7-like phage infecting the freshwater cyanobacterium Phormidium foveolarum: complete genome sequence and DNA translocation. Virology. 2007;366:28–39. doi: 10.1016/j.virol.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Matthew B, Fu C, et al. Structural changes in a marine podovirus associated with the release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2011;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark. hmm: new solutions for gene finding. DNA Seq. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH, Clokie MRJ, Millard A, Cook A, Wilson WH, Wheatley PJ, et al. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J Bacteriol. 2005;187:3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard AD, Zwirglmaier K, Downey MJ, Mann NH, Scanlan DJ. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol. 2009;11:2370–2387. doi: 10.1111/j.1462-2920.2009.01966.x. [DOI] [PubMed] [Google Scholar]
- Mizuno CM, Rodriguez-Valera F, Kimes N, Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P, Laakso S, Mäenpää P, Aro EM. Stepwise photoinhibition of photosystem II. Studies with Synechocystis species PCC 6803 mutants with a modified D-E loop of the reaction center polypeptide D1. Plant Physiol. 1998;117:483–490. doi: 10.1104/pp.117.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. Phage taxonomy: We agree to disagree. J Bacteriol. 2004;186:7029–7031. doi: 10.1128/JB.186.21.7029-7031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Tang M, Grishin NV. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res. 2007;35:W649–W652. doi: 10.1093/nar/gkm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pen MMO, Bullerjahn GS. The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. J Biol Chem. 1995;270:22478–22482. doi: 10.1074/jbc.270.38.22478. [DOI] [PubMed] [Google Scholar]
- Ponsero A, Chen F, Lennon JT, Wilhelm SW. Complete genome sequence of cyanobacterial siphovirus KBS2A. Genome Announc. 2013;1:e00472–13. doi: 10.1128/genomeA.00472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin I, Franzmann PD, McMeekin TA, Burton HR. Antarctic Communities, Species, Structure and Survival. University of Cambridge Press; 1997. Seasonal distribution of picocyanobacteria in Ace Lake, a marine derived Antarctic Lake; pp. 178–184. [Google Scholar]
- Rao V, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Raytcheva DA, Haase-Pettingell C, Piret JM, King JA. Intracellular assembly of cyanophage Syn5 proceeds through a scaffold-containing procapsid. J Virol. 2011;85:2406–2415. doi: 10.1128/JVI.01601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Desruelles J, Waterbury JB, Herdman M, Stanier R. Generic assigments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Rohwer F, Segall A, Steward G, Seguritan V, Breitbart M, Wolven F, et al. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol Oceanogr. 2000;45:408–418. [Google Scholar]
- Roux S, Enault F, Robin A, Ravet V, Personnic S, Theil S, et al. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One. 2012;7:e33641. doi: 10.1371/journal.pone.0033641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, Lindell D. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc Natl Acad Sci USA. 2012;109:2037–2042. doi: 10.1073/pnas.1115467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safferman RS, Morris ME. Algal virus: isolation. Science. 1963;140:679–680. doi: 10.1126/science.140.3567.679. [DOI] [PubMed] [Google Scholar]
- Safferman RS, Morris ME. Growth characteristics of the blue-green algal virus LPP-1. J Bacteriol. 1964;88:771–775. doi: 10.1128/jb.88.3.771-775.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säwström C, Lisle J, Anesio AM, Priscu JC, Laybourn-Parry J. Bacteriophage in polar inland waters. Extremophiles. 2008;12:167–175. doi: 10.1007/s00792-007-0134-6. [DOI] [PubMed] [Google Scholar]
- Schmidt HF, Sakowski EG, Williamson SJ, Polson SW, Wommack KE. Shotgun metagenomics indicates novel family A DNA polymerases predominate within marine virioplankton. ISME J. 2014;8:103–114. doi: 10.1038/ismej.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2013;110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Storz G, Tartaglia LA, Farr SB, Ames BN. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. 2010;12:3035–3056. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D, et al. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial “ mobilome”. Environ Microbiol. 2009;11:2935–2951. doi: 10.1111/j.1462-2920.2009.02081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium. Prochlorococcus Nat. 2003;424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- Suttle CA.2000Cyanophages and their role in the ecology of cyanobacteria The Ecology of Cyanobacteria: Their diversity in time and spaceWhitton BA, Potts M, (eds)Kluwer Academic Publishers: Boston, MA, USA; 563–589. [Google Scholar]
- Suttle CA. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol. 1994;28:237–243. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- Suttle CA, Chan AM. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol. 1994;60:3167–3174. doi: 10.1128/aem.60.9.3167-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA, Chan AM. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar Ecol Prog Ser. 1993;92:99–109. [Google Scholar]
- Suttle CA, Chan AM, Cottrell MT. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl Environ Microbiol. 1991;57:721–726. doi: 10.1128/aem.57.3.721-726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108:E757–E764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent WF.2000Cyanobacterial dominance in the polar regions Ecology of Cyanobacteria Their Diversity in Time and SpaceWhitton BA, Potts M, (eds)Kluwer Academic Publishers: Boston, MA, USA; 321–340. [Google Scholar]
- Vincent WF, Hobbie JE, Laybourn-Parry J.2008Introduction to the limnology of high-latitude lake and river ecosystems Polar Lakes and Rivers Limnology of Arctic and Antarctic Aquatic EcosystemsVincent WF, Laybourn-Parry J, (eds)Oxford University Press: New York; 1–23. [Google Scholar]
- Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, et al. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer: New York, NY, USA; 2009. p. 2009. [Google Scholar]
- Wilson WH, Joint IR, Carr NG, Mann NH. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl Environ Microbiol. 1993;59:3736–3743. doi: 10.1128/aem.59.11.3736-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nagasaki K, Takashima Y, Shirai Y, Tomaru Y, Takao Y. Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J Bacteriol. 2008;190:1762–1772. doi: 10.1128/JB.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Takashima Y, Tomaru Y, Shirai Y, Takao Y, Hiroishi S, et al. Isolation and characterization of a cyanophage infecting the toxic cyanobacterium Microcystis aeruginosa. Appl Environ Microbiol. 2006;72:1239–1247. doi: 10.1128/AEM.72.2.1239-1247.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, et al. Abundant SAR11 viruses in the ocean. Nature. 2013;494:357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- Zhong X, Berdjeb L, Jacquet S. Temporal dynamics and structure of picocyanobacteria and cyanomyoviruses in two large and deep peri-alpine lakes. FEMS Microbiol Ecol. 2013;86:312–326. doi: 10.1111/1574-6941.12166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.