Recent advances in medical research have revealed how humans rely on their microbiome for diverse traits and functions. Similarly, microbiomes of other higher organisms play key roles in disease, health, growth and development of their host. Exploring microbiome functions across kingdoms holds enormous potential to understand common mechanisms and concepts underlying microbiome assembly and microbial processes that sustain life of Eukaryotes. The gut and plant rhizosphere are both open systems with large surface areas overpopulated with microbes. Despite distinct differences in microbiome composition, these two ecosystems share striking similarities in microbiome functions related to nutrient acquisition, immune system modulation and protection against infections. We also discuss how humans and plants exchange microbes, for better or for worse. We propose that adopting ecological theory, combined with modeling and synthetic microbial ecosystems, provides a promising strategy to identify host traits and cues involved in microbiome assembly on and in Eukaryotes.
A staggering number of microbes lives on and in cell tissues of eukaryotic organisms, thereby influencing the functioning and evolution of their host. The microbiome concept, that is, the collective communities of microorganisms, their genomes and interactions, was first used in the context of microorganisms that inhabit the human body. Since then, numerous studies have adopted this term to describe microbial communities associated with other mammals, insects, fish or plants. For humans, the microbiome significantly contributes to metabolism and provides functions that humans did not need to evolve on their own (Gill et al., 2006). Hence, the genes present in the human microbiome are considered the secondary genome. Plants also rely on their microbiome for specific traits and functions, including nutrient acquisition and tolerance to (a)biotic stress factors (Mendes et al., 2013). In the human microbiome, the highest density of microbes is found in the gastrointestinal tract, where ‘they synthesize essential amino acids and vitamins, and process components of otherwise indigestible contributions to our diet such as plant polysaccharides' (Backhed et al., 2005). Similarly, microbes overpopulate the plant rhizosphere, that is, the narrow zone of soil surrounding and influenced by plant roots via the exudation of nutrients and other chemically diverse compounds. In this context, the rhizosphere can be seen as ‘the gut inside out' (Ramírez-Puebla et al., 2013).
Technological advances, including next-generation sequencing, bioinformatics and metabolomics, have enabled more detailed analyses of the microbiome structures and functions during their association with the host. In mammalians, the microbiome structure is correlated with host phylogeny, diet, infection and medical interventions. Various studies have now shown that also the microbiome structure of plants is determined by host genotype, soil characteristics, fertilization, pathogen infection and pesticide application. Although in humans the diet is the main source of gut microbes, microbes on plant roots are primarily selected from the soil in which seeds germinate and plants grow. Furthermore, for both mammalians and plants some members of the microbiome may be inherited from the mother.
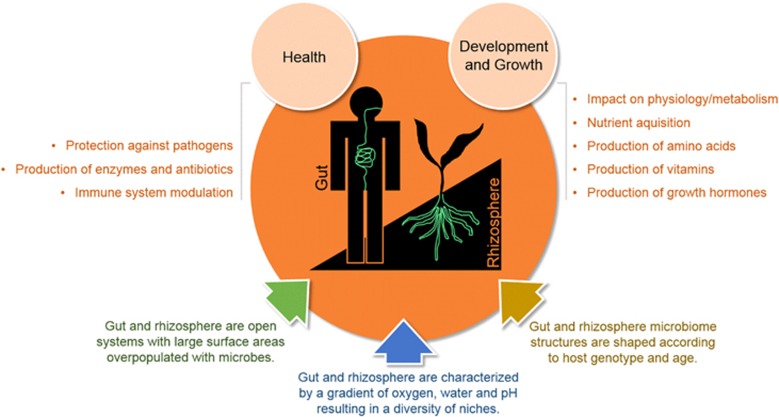
Comparisons between bacterial communities associated with the rhizosphere and the mammals' gut show that both ecosystems are dominated by the same four bacterial phyla (Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria), albeit in different relative abundances. Taxonomic classification at the lower hierarchy levels shows a distinct microbiome composition, which is correlated with, among others, differences in oxygen availability in the gut and in the rhizosphere. In both ecosystems, the stability of the microbiome structure is key for the functioning and well-being of their hosts (Figure 1). Perturbations to the microbiome structure of healthy individuals, also referred to as dysbiosis (Petersen and Round, 2014), can lead to changes in specific functions. For example, changes in the ratio of Firmicutes to Bacteroidetes in the gut of mammals affects the capacity for energy harvest and obesity (Turnbaugh et al., 2006), whereas a reduction in the diversity was associated with inflammatory bowel diseases (Khoruts et al., 2010). In the rhizosphere, the relative abundances of Proteobacteria and Actinobacteria are correlated with the protection against infections by a fungal pathogen (Mendes et al., 2011). Hence, in both human and plant microbiome research, there is a strong interest to define and characterize ‘healthy microbiomes'. This fundamental knowledge will enable therapies toward ‘rebiosis', that is, re-establishment of a healthy, complex microbiome after dysbiosis. It is striking to note that fecal transplantations used to re-establish a health microbiome in the gut have a similar counterpart in rhizosphere research. Similar to curing human diarrhea by fecal bacteriotherapy (Khoruts et al., 2010), transplantation of the so-called disease suppressive soils to nonsuppressive (conducive) soil can restore the protection of plants against soil-borne fungal and bacterial infections (Mendes et al., 2011). Hence, fecal bacteriotherapy and soil transplantation share the same rationale, which is to reinstate a balanced, ‘healthy' microbiome.
Figure 1.
Commonalities in the life support functions of the microbiomes of the human gut and the plant rhizosphere.
Meta-analysis of the fecal bacterial community from 60 mammal species revealed that bacterial diversity was higher in herbivores, followed by omnivores and carnivores; humans clustered with other omnivores (Ley et al., 2008). Considering that plant tissues, including the roots, are colonized by a large and diverse number of microbes, herbivory provides a link between microbiomes of animal and plant kingdoms, that is, when animals feed on plants, microbes are transferred from plant tissues and establish in the gut system. Here specific attention should be given to the opportunistic human pathogenic bacteria present in or on plant tissues, which can cause diseases in debilitated consumers of contaminated plant materials. Comparing the rhizosphere microbiomes of plants grown in soils that are suppressive or conducive to a specific fungal plant pathogen showed a higher richness and abundance of putative opportunistic human pathogenic bacteria in the conducive soil than in the suppressive soil (Mendes et al., 2013). This finding suggests that perturbations in the healthy microbiomes of plants may contribute to the proliferation, transfer and invasion of opportunistic pathogens to humans through diet. Also rhizosphere bacteria with beneficial effects on plant growth and plant health can represent a threat when introduced in humans, considering that these isolates sometimes are closely related or genetically indistinguishable from the opportunistic human pathogenic strains.
Although we focused our discussion on microbiomes of two seemingly different ecosystems (humans, plants), also insects, nematodes, fish and birds are interacting with and depend on their microbiomes for specific traits. How to explore the increasing number of microbiome data obtained across diverse ecosystems to design strategies for selection, reconstruction or augmentation of microbial consortia that benefit their host? Are there cross-kingdom similarities in the mechanisms underlying microbiome assembly? Scheuring and Yu (2012) proposed a model on how beneficial microbes are assembled in/on their hosts. They assumed that a large initial abundance of beneficial microbes is required to establish a healthy core microbiome, which can be achieved by vertical transmission, as in the cuticular microbiome of the attine ants, or by higher immigration rates, as in disease suppressive soils (Scheuring and Yu, 2012). Adopting ecological theory in microbiome research, combined with modeling and synthetic microbial ecosystems, is a promising complementary strategy to unravel the host traits and cues involved in the assembly as well as evolution of human and plant microbiomes, with the ultimate goal to improve agricultural sustainability and human health.
Acknowledgments
We thank Dr Tim Mauchline for the critical reading of the manuscript.
The authors declare no conflict of interest.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Puebla ST, Servín-Garcidueñas LE, Jiménez-Marín B, Bolaños LM, Rosenblueth M, Martínez J, et al. Gut and root microbiota commonalities. Appl Environ Microbiol. 2013;79:2–9. doi: 10.1128/AEM.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring I, Yu DW. How to assemble a beneficial microbiome in three easy steps. Ecol Lett. 2012;15:1300–1307. doi: 10.1111/j.1461-0248.2012.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]