Abstract
Loss-of-function mutations in PALB2 are associated with an increased risk of breast cancer, with recent data showing that female breast cancer risks for PALB2 mutation carriers are comparable in magnitude to those for BRCA2 mutation carriers. This study applied targeted massively parallel sequencing to characterize the mutation spectrum of PALB2 in probands attending breast cancer genetics clinics in the USA. The coding regions and proximal intron–exon junctions of PALB2 were screened in probands not known to carry a mutation in BRCA1 or BCRA2 from 1,250 families enrolled through familial cancer clinics by the Breast Cancer Family Registry. Mutation screening was performed using Hi-Plex, an amplicon-based targeted massively parallel sequencing platform. Screening of PALB2 was successful in 1,240/1,250 probands and identified nine women with protein-truncating mutations (three nonsense mutations and five frameshift mutations). Four of the 33 missense variants were predicted to be deleterious to protein function by in silico analysis using two different programs. Analysis of tumors from carriers of truncating mutations revealed that the majority were high histological grade, invasive ductal carcinomas. Young onset was apparent in most families, with 19 breast cancers under 50 years of age, including eight under the age of 40 years. Our data demonstrate the utility of Hi-Plex in the context of high-throughput testing for rare genetic mutations and provide additional timely information about the nature and prevalence of PALB2 mutations, to enhance risk assessment and risk management of women at high risk of cancer attending clinical genetic services.
Keywords: Breast cancer, PALB2, Mutation screening, Massively parallel sequencing, Hi-Plex, Genetic variant
Introduction
Partner and localiser of BRCA2 (PALB2) encodes a protein whose interaction with BRCA1 and BRCA2 is critical for homologous recombination repair of double-stranded DNA breaks and for checkpoint control functions. Although PALB2 mutations were initially suggested to be associated with moderate breast cancer risk (2 to 3-fold) [1], accumulation of more data recently has supported to higher risk estimates. In the largest study to date, involving 154 PALB2 mutation-carrying families, Antoniou et al. reported that breast cancer risks for PALB2 mutation carriers are comparable to those of pathogenic BRCA2 mutation carriers, with a risk higher among those younger than 40 years of age (8 to 9-fold) and slight decrements in risk with age (approximately fivefold in those older than 60 years) [2]. Heterozygous loss-of-function germline mutations in this gene account for ~2.4 % of the familial aggregation of breast cancer [3–5]. Germline mutations in PALB2 have also been identified in individuals with pancreatic and ovarian cancers, both with and without family history of breast cancer [5–9].
In this study, we used massively parallel sequencing to screen the PALB2 gene for germline mutations in 1,250 probands of families recruited from genetic clinics into the Breast Cancer Family Registry (BCFR). We applied Hi-Plex, a platform for library preparation that has previously been demonstrated to facilitate accurate, cost-effective, and rapid high-throughput mutation screening [10].
Methods
Subjects
Participants were probands (defined as the first enrolled family member, who may or may not have had a personal history of breast cancer) from families seen in clinical settings and recruited by the New York (n = 825) [11–15], Utah (n = 67) and Philadelphia (n = 358) sites of the BCFR [16]. All subjects had been tested previously and found negative for mutations in BRCA1 and BRCA2 [17]. Study recruitment was approved by the Institutional Review Board (IRB) of the University of Melbourne and the local IRBs of the BCFR centers involved in this study. Written informed consent was obtained from each participant for general research using their data and biospecimens. This study was approved by the University of Melbourne Human Research Ethics Committee.
Where relevant and available, pathology data such as estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor-2 (HER2) status of the PALB2 mutation-associated tumors were obtained from the relevant BCFR.
Mutation screening
We conducted mutation screening of the coding exons and proximal splice junction regions of PALB2 (LRG_308; NM_024675.3) by targeted massively parallel sequencing using Hi-Plex, an amplicon-based approach for library building [18] [Nguyen-Dumont et al., accepted]. Hi-Plex gene-specific primers were designed to target the protein coding and flanking intronic and untranslated regions of PALB2 as described in Nguyen-Dumont et al. [10]. All oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA, USA). We applied the improved Hi-Plex protocol reported in [Nguyen-Dumont et al. accepted] to 25 ng genomic DNA obtained from the BCFR sites involved in this study. Sequencing on the MiSeq (Illumina, San Diego, CA, USA) and mapping were performed as described in [10]. For a given DNA sample, successful sequencing was defined as ≥95 % of all amplicons covered by a minimum of ten read pairs (depth = 20X). For the minority of DNA samples falling below this threshold, fresh libraries were prepared and another sequencing run was performed. Sequencing data from the initial and the repeat run were merged, and sequencing statistics were assessed again. Variant calling was performed using ROVER [19] with the settings previously published [10]. Details of sequencing statistics calculations reported in this paper (on-target and coverage) are also described in Nguyen-Dumont et al. [10]. Variant confirmation was performed by Sanger Sequencing using the primers and conditions described in [3].
In silico analysis
Variant annotation was performed using Annovar [20]. The probability that missense substitutions observed during our mutation screening of PALB2 were deleterious to protein function was assessed with Align-GVGD [21] using the curated alignment reported by Tischkowitz et al. [22], and with PolyPhen2 [23] using its precompiled alignments. Align-GVGD scores provide a seven-tiered classifier: C0, C15, C25, C35, C45, C55, and C65 where C0 refers to the category of variants least likely to be deleterious and C65 describes the category of variants most likely to be deleterious to protein function. Genetic variants assessed with PolyPhen2 are categorized as probably damaging (score ≥0.957), possibly damaging (score ≥0.453), or benign (score ≤0.452) [24].
Results and discussion
Sequencing data
Of the 1,250 BCFR specimens, 5.9 % (74 specimens: 69 from the New York BCFR, two from the Utah BCFR, and three from the Philadelphia BCFR) failed sequencing in the first instance and were repeated as described in “Methods” section. Following repeated sequencing, ten (nine from the New York BCFR and one from the Utah BCFR) were excluded from further analysis because they did not reach our criteria for successful sequencing. Thus, sequencing success rate with Hi-Plex was 99.2 %. Across the 1,240 successfully sequenced specimens, the median depth of coverage was 1110X, with 98.8 and 99.2 % of all amplicons represented within 20- and 25-fold of the median coverage, respectively. The median on-target rate was 98.2 %.
Results from the mutation screening
PALB2 mutation screening identified 55 different genetic variants in total. We observed three nonsense mutations—c.1984A>T, p.(K662*); c.2108T>G, p.(L703*); and c.3113G>A, p.(W1038*) (rs180177132)—and five frameshift mutations resulting in predicted premature termination codons—c.172_175del, p.(Q60fs); c.1546delA, p.(R516fs); c.2120delC, p.(P707fs); c.2325dupA, p.(F776fs); and c.3426dupA, p.(L1143fs) (Table 1). All these variants were identified in one proband each, except for c.3426dupA, which was observed in two probands in the New York BCFR.
Table 1.
Nonsense and frameshift PALB2 variants identified in 1,240 probands participating in the clinic-based resource of the BCFR
| Nucleotide changea | Protein change | rs numberb | LOVDc | Frequency (n = 1,240) | % | |
|---|---|---|---|---|---|---|
| Nonsense | PALB2:c.1984A>T | p.K662* | – | No | 1 | 0.08 |
| PALB2:c.2108T>G | p.L703* | – | No | 1 | 0.08 | |
| PALB2:c.3113G>A | p.W1038* | rs180177132 | Yes | 1 | 0.08 | |
| Frameshift | PALB2:c.172_175del | p.Q60fs | – | Yes | 1 | 0.08 |
| PALB2:c.1546delA | p.R516fs | – | Yes | 1 | 0.08 | |
| PALB2:c.2120delC | p.P707fs | – | No | 1 | 0.08 | |
| PALB2:c.2325dupA | p.F776fs | – | No | 1 | 0.08 | |
| PALB2:c.3426dupA | p.L1143fs | – | No | 2 | 0.16 |
Number based on transcript sequence LRG_308; NM_024675.3, +1 as A of ATG start codon
rs number from dbSNP v.137
Present (yes) or absent (no) from the LOVD v.2.0 Build 36, PALB2 version 140217 [29]
The asterisk describes the stop codon at protein level according to the Human Genome Variation Society (HGVS) recommendations v.2
Of the 33 missense variants identified in the mutation screening, ten were predicted to be possibly or probably damaging by an in silico analysis using PolyPhen2: c.53A>G, p.(K18R) (11 carriers); c.2087C>T, p.(T696 M), c.2674G>A, p.(E892 K) (three carriers); c.2816T>G, p.(L939 W) (seven carriers); c.2897T>C, p.(I966T); c.3054G>C, p.(E1018D); c.3061G>Q, p.(G1021R); c.3278T>C; p.(I1093T); c.3356T>C, p.(L1119P); and c.3513G>C p.(L1171F).
Of those, four were also predicted to be deleterious to protein function using Align-GVGD: c.2816T>G, c.3061G>Q,c.3278T>C,and c.3356T>C,withAlign-GVGD scores of C55, C65, C25, and C65, respectively (Table 2). The remaining variants were graded as C0 by Align-GVGD.
Table 2.
PALB2 missense variants predicted to affect protein function by Align-GVGD and Polyphen2 identified in 1,240 probands participating in the clinic-based resource of the BCFR
| Nucleotide changea | Protein change | rs numberb | LOVDc | PolyPhen2d | Align-GVGD | Frequency (n = 1,240) | % |
|---|---|---|---|---|---|---|---|
| PALB2:c.53A>G | p.K18R | rs138789658 | Yes | D, 1.000 | C0 | 11 | 0.89 |
| PALB2:c.2087C>T | p.T696M | – | No | P, 0.954 | C0 | 1 | 0.08 |
| PALB2:c.2674G>A | p.E892K | rs45476495 | Yes | D, 1.000 | C0 | 3 | 0.24 |
| PALB2:c.2816T>G | p.L939W | rs45478192 | Yes | D, 1.000 | C55 | 7 | 0.56 |
| PALB2:c.2897T>C | p.I966T | – | No | D, 1.000 | C0 | 1 | 0.08 |
| PALB2:c.3054G>C | p.E1018D | rs183489969 | Yes | D, 0.998 | C0 | 1 | 0.08 |
| PALB2:c.3061G>A | p.G1021R | rs143808171 | Yes | D, 1.000 | C65 | 1 | 0.08 |
| PALB2:c.3278T>C | p.I1093T | rs45616636 | Yes | D, 0.999 | C25 | 1 | 0.08 |
| PALB2:c.3356T>C | p.L1119P | – | Yes | D, 1.000 | C65 | 1 | 0.08 |
| PALB2:c.3513G>C | p.L1171F | – | No | D, 1.000 | C0 | 1 | 0.08 |
Number based on transcript sequence LRG_308; NM_024675.3, +1 as A of ATG start codon
rs number from dbSNP v.137
Present (yes) or absent (no) from the LOVD v.2.0 Build 36, PALB2 version 140217 [29]
PolyPhen2 prediction (D probably damaging, P possibly damaging), PolyPhen2 score [23]
PALB:c.3278T>C and PALB2:c.3356T>C are not reported in the exome variant server (EVS) [25]. PALB2:c.2816T>G and PALB2:c.3061G>A are both present in the EVS at a minor allele frequency in the European ancestry American population of 0.24 and 0.02 %, respectively. PALB2:c.3061G>A has not been previously reported in the literature. PALB2:c.2816T>G has been previously identified in studies assessing the role of PALB2 in multiple-case breast cancer families but not reported to be associated with risk of disease [1, 26]. Overall, there has not been strong evidence so far that rare missense variants in PALB2 are associated with an increased risk of breast cancer [22, 27]. However, the first functional characterization study of missense variants in PALB2, focusing on variants occurring in the protein domain involved in PALB2/BRCA2 interactions, has recently showed that L939 W mutant proteins display a decreased capacity for DNA double-strand break-induced homologous recombination and an increased cellular sensitivity to ionizing radiation [28]. These results suggest that functional characterization of other missense variants of PALB2 could contribute to inform the pathogenicity conferred by such variants.
The remaining variants were synonymous variants. No variant affecting consensus splice sites has been detected. Of the 18 PALB2 truncating variants or missense variants predicted to be deleterious to protein function by either in silico programs, ten are present in the PALB2 LOVD database [29] or have been previously reported elsewhere, and eight have not been reported previously (Tables 1, 2).
We sought confirmation of the truncating and predicted damaging missense variants by Sanger sequencing of the 37 mutation-carrying probands. We found that sequencing of Hi-Plex libraries yielded 100 % specificity, confirming the high accuracy achievable with this high-throughput, cost- and time-efficient approach for targeted massively parallel sequencing [10].
Pathology reviews and family information
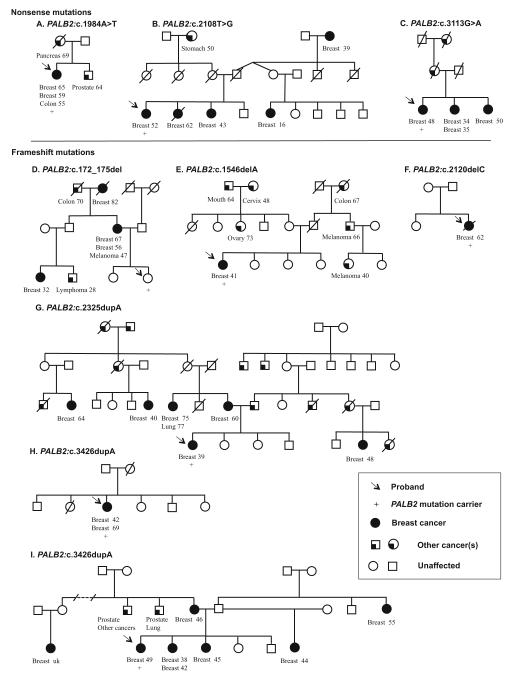
The details of available pathology information and family history of the probands identified to carry nonsense or frameshift mutations in PALB2 are shown in Table 3 and in the pedigrees of Fig. 1.
Table 3.
Histopathology features of PALB2-associated tumors from probands who carry truncating mutations
| Mutation | Pedigree | Breast cancer diagnoses (age at diagnosis) |
Grade | Histological type | ER | PR | HER2 |
|---|---|---|---|---|---|---|---|
| PALB2:c.1984A>T | A | Proband (59) | |||||
| Proband (65) | 3 | IDC | + | + | + | ||
| PALB2:c.3113G>A | C | Proband (48) | 3 | IDC | + | − | Eq |
| PALB2:c.2120delC | F | 3 | IDC | + | + | ||
| PALB2:c.2325dupA | G | Proband (39) | 3 | IDC | + | + | |
| PALB2:c.3426dupA | H | Proband (42) | |||||
| Proband (69) | 3 | IDC | + | + | + |
Histopathology information was presented where available
ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor-2, IDC invasive ductal carcinoma, + positive, − negative, uk unknown, Eq equivocal, tested to be 2+ by immunohistochemistry but not confirmed by fluorescence in situ hybridisation
Fig. 1.
Family pedigrees of probands found to carry PALB2 nonsensec (a–c) and frameshift (d–i) mutations. When known, cancer diagnoses and age of onset are indicated for affected family members
In total, there were 11 breast cancers recorded in the three families with the nonsense mutations PALB2:c.1984A>T, PALB2:c.2108T>G, and PALB2:c.3113G>A.
Additional cancers in the kindred with PALB2:c.1984A>T were colon cancer (proband, dx 55 years), prostate cancer (brother, dx 64 years), and pancreatic cancer (mother, dx 69 years) (Fig. 1a). Stomach cancer (maternal grandmother, dx 50 years) was seen in the family carrying PALB2:c.2108T>G (Fig. 1b). The family harboring PALB2:c.3113G>A had five affected women, with high-risk features including histological grade three, invasive ductal carcinoma bilateral disease, and young onset of cancer (Fig. 1c).
In the six families with frameshift mutations, 22 breast cancers were recorded.
PALB2:c.172_175del was observed in one proband who was cancer free at last contact, in the context of a cancer dense family with bilateral breast cancer and reported breast cancer aged 32 years. Other cancers included melanoma (dx 47 years), lymphoma (dx 28 years), and colon cancer (dx 70 years) (Fig. 1d).
PALB2:c.1546delA was identified in a proband affected by breast cancer (dx 41 years) and melanoma (dx 33 years), with paternal relatives with colon cancer (grandmother, dx 67 years) and melanoma (uncle, dx 66 years; first cousin, dx 40 years). On the maternal side, cancers included cervical cancer (maternal grandmother, dx 48 years), ovarian cancer (aunt, dx 73 years), and cancer of the mouth (grandfather, dx 64 years) (Fig. 1e).
PALB2:c.2120delC was observed in a woman diagnosed at the age of 62 years with histological grade three, ER+/PR+, invasive ductal carcinoma. HER2 status was not tested (Table 3). No other cancers were known in this limited pedigree (Fig. 1f).
PALB2:c.2325dupA was observed in one proband (dx 39) who had breast cancer, with four maternal and one paternal family members with breast cancer, as well as multiple unspecified reported cancers, five on her maternal side and six on her paternal side, including her father (Fig. 1g).
PALB2:c.3426dupA was observed in two probands. One carrier had bilateral breast cancers (left breast, dx 42 years; right breast, dx 69 years). The second breast cancer was an invasive ductal carcinoma (grade three), ER+/PR+/HER2+ (immunochemistry) (Fig. 1h; Table 3). The second carrier had breast cancer diagnosed at the age of 49 years. Breast cancers were reported in six family members, comprising young onset and bilateral disease. Prostate cancer, lung cancer, and a hematologic cancer were also reported (Fig. 1i).
Although information on ER/PR/HER2 status was available for a limited numbers of tumors in the present study (Table 3), our findings are consistent with those from Teo et al. and Antoniou et al. who have found that the majority of the breast tumors arising in PALB2 loss-of-function mutation carriers were positive for ER and PR expression (11/19–58 % and 95/129–74 %, respectively) [2, 30]. A study investigating the Finnish founder mutation PALB2:c.1592delT reported that carriers with a family history of breast cancer were more likely to be diagnosed with triple-negative breast cancer [31]. The phenotype of PALB2 mutation-associated breast cancers is thus potentially variable and could be influenced by mutation type.
Conclusions
We screened probands of multiple-case breast cancer families enrolled in the BCFR for genetic variants in the breast cancer susceptibility gene PALB2 and identified truncating mutations in 9/1,240 individuals (0.73 %), a frequency consistent with similar reports of PALB2 mutation frequency in multiple-case breast cancer families [1, 26, 27]. Ten missense variants were predicted to be damaging to protein function by PolyPhen2, four of which were also predicted to be damaging by Align-GVGD. Eight of the 18 truncating or possibly pathogenic variants identified in this study were not reported in the PALB2 LOVD database.
When available, examination of breast tumor pathology from subjects carrying a truncating mutation showed that the tumors were histological high grade, invasive ductal carcinomas and, in majority, positive for ER and PR expression. Bilateral breast cancer was reported in two of the probands and three of the family members. Young onset was apparent in most families, with 19 cancers under the age 50 years of age, including eight under the age of 40 years.
Our data demonstrate the utility of Hi-Plex to test for rare genetic mutations in the context of breast cancer predisposition in an efficient, accurate, cost-effective, and rapid manner, as previously reported [10]. This work also provides additional timely information about the nature and prevalence of PALB2 mutations in high-risk women attending clinical genetic services, as PALB2 is becoming commonly included in panel testing for breast cancer susceptibility.
Large-scale studies are now required to further elucidate mutation prevalence, family history, tumor morphology, and to refine breast cancer risk estimates (penetrance) associated with mutations in PALB2. With testing of PALB2 ready to enter clinical practice, additional families with mutations will be identified and contribute to a better understanding of the breast cancer risk conferred by PALB2 mutations.
Acknowledgments
TN-D is a Susan G. Komen for the Cure Postdoctoral Fellow. ZLT was supported by Postgraduate Scholarships provided by the Faculty of Medicine, Dentistry and Health Sciences, The University of Melbourne and the National Health and Medical Research Council (NHMRC, Australia). RL is supported by UROP, a program of Biomedical Research Victoria, by the Victorian Life Sciences Computation Initiative (VLSCI) and by the Department of Pathology, The University of Melbourne. MCS is an NHMRC Senior Research Fellow. The Utah, New York, and Philadelphia sites of the Breast Cancer Family Registry were supported by Grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), or does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. This work was supported by the Australian National Health and Medical Research Council (NHMRC) (APP1025879 and APP1029974), the Victorian Breast Cancer Research Consortium and by a VLSCI Grant (number VR0182) on its Peak Computing Facility, an initiative of the Victorian Government.
List of abbreviations
- BCFR
Breast Cancer Family Registry
- ER
Estrogen receptor
- HER2
Human epithelial growth factor-2
- LCL
Lymphoblastoid cell line
- LOVD
Leiden open variant database
- PR
Progesterone receptor
Footnotes
Conflict of interests The authors declare that they have no competing interests.
Contributor Information
Tú Nguyen-Dumont, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia.
Fleur Hammet, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia.
Maryam Mahmoodi, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia.
Helen Tsimiklis, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia.
Zhi L. Teo, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia; Peter MacCallum Cancer Centre, St Andrews Place, East Melbourne, VIC 3002, Australia
Roger Li, Victorian Life Sciences Computation Initiative, 187 Grattan Street, Carlton, Melbourne, VIC 3010, Australia.
Bernard J. Pope, Victorian Life Sciences Computation Initiative, 187 Grattan Street, Carlton, Melbourne, VIC 3010, Australia; Department of Computing and Information Systems, The University of Melbourne, Melbourne, VIC 3010, Australia
Mary Beth Terry, Columbia University Mailman School of Public Health, New York, NY, USA; Herbert Irving Comprehensive Cancer Center, New York, NY, USA.
Saundra S. Buys, Huntsman Cancer Institute, Salt Lake City, UT, USA
Mary Daly, Fox Chase Cancer Center, Philadelphia, PA, USA.
John L. Hopper, Centre for Epidemiology and Statistics, The University of Melbourne, Carlton, VIC 3010, Australia
Ingrid Winship, Department of Medicine, The University of Melbourne Health, Parkville, VIC 3010, Australia; The Royal Melbourne Hospital, Parkville, VIC 3050, Australia.
David E. Goldgar, Huntsman Cancer Institute, Salt Lake City, UT, USA
Daniel J. Park, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia
Melissa C. Southey, Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, VIC 3010, Australia
References
- 1.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, Lee A, Subramanian D, De Leeneer K, Fostira F, Tomiak E, Neuhausen SL, Teo ZL, Khan S, Aittomaki K, Moilanen JS, Turnbull C, Seal S, Mannermaa A, Kallioniemi A, Lindeman GJ, Buys SS, Andrulis IL, Radice P, Tondini C, Manoukian S, Toland AE, Miron P, Weitzel JN, Domchek SM, Poppe B, Claes KB, Yannoukakos D, Concannon P, Bernstein JL, James PA, Easton DF, Goldgar DE, Hopper JL, Rahman N, Peterlongo P, Nevanlinna H, King MC, Couch FJ, Southey MC, Winqvist R, Foulkes WD, Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(6):497–506. doi: 10.1056/NEJMoa1400382. doi:10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Southey MC, Teo ZL, Dowty JG, Odefrey FA, Park DJ, Tischkowitz M, Sabbaghian N, Apicella C, Byrnes GB, Winship I, Baglietto L, Giles GG, Goldgar DE, Foulkes WD, Hopper JL, kConFab for the Beast Cancer Family R A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12(6):R109. doi: 10.1186/bcr2796. doi:10.1186/bcr2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erkko H, Dowty JG, Nikkila J, Syrjakoski K, Mannermaa A, Pylkas K, Southey MC, Holli K, Kallioniemi A, Jukkola-Vuorinen A, Kataja V, Kosma VM, Xia B, Livingston DM, Winqvist R, Hopper JL. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clinical Cancer Res. 2008;14(14):4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. doi:10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- 5.Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, Kallioniemi A, Pylkas K, Karppinen SM, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma VM, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446(7133):316–319. doi: 10.1038/nature05609. doi:10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 6.Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, Taherian N, Srivastava A, Holter S, Rothenmund H, Ghadirian P, Foulkes WD, Gallinger S. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137(3):1183–1186. doi: 10.1053/j.gastro.2009.06.055. doi:10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterlongo P, Catucci I, Pasquini G, Verderio P, Peissel B, Barile M, Varesco L, Riboni M, Fortuzzi S, Manoukian S, Radice P. PALB2 germline mutations in familial breast cancer cases with personal and family history of pancreatic cancer. Breast Cancer Res Treat. 2011;126(3):825–828. doi: 10.1007/s10549-010-1305-1. doi:10.1007/s10549-010-1305-1. [DOI] [PubMed] [Google Scholar]
- 8.Dansonka-Mieszkowska A, Kluska A, Moes J, Dabrowska M, Nowakowska D, Niwinska A, Derlatka P, Cendrowski K, Kupryjanczyk J. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20. doi: 10.1186/1471-2350-11-20. doi:10.1186/1471-2350-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southey MC, Teo ZL, Winship I. PALB2 and breast cancer: ready for clinical translation. The Appl Clinical Genet. 2013;6:43–52. doi: 10.2147/TACG.S34116. doi:10.2147/TACG.S34116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen-Dumont T, Teo ZL, Pope BJ, Hammet F, Mahmoodi M, Tsimiklis H, Sabbaghian N, Tischkowitz M, Foulkes WD, Giles GG, Hopper JL, Southey MC, Park DJ. Hi-Plex for high-throughput mutation screening: application to the breast cancer susceptibility gene PALB2. BMC Med Genomics. 2013;6(1):48. doi: 10.1186/1755-8794-6-48. doi:10.1186/1755-8794-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strahm B, Malkin D. Hereditary cancer predisposition in children: genetic basis and clinical implications. Int J Cancer. 2006;119(9):2001–2006. doi: 10.1002/ijc.21962. doi:10.1002/ijc.21962. [DOI] [PubMed] [Google Scholar]
- 12.Wu HC, Delgado-Cruzata L, Machella N, Wang Q, Santella RM, Terry MB. DNA double-strand break repair genotype and phenotype and breast cancer risk within sisters from the New York site of the Breast Cancer Family Registry (BCFR) Cancer Causes Control. 2013;24(12):2157–2168. doi: 10.1007/s10552-013-0292-z. doi:10.1007/s10552-013-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, Stavrou T, Scherer SW, Dura WT, Wainwright B, Squire JA, Rutka JT, Hogg D. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31(3):306–310. doi: 10.1038/ng916. doi:10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 14.Delgado-Cruzata L, Wu HC, Perrin M, Liao Y, Kappil MA, Ferris JS, Flom JD, Yazici H, Santella RM, Terry MB. Global DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Epigenetics. 2012;7(8):868–874. doi: 10.4161/epi.20830. doi:10.4161/epi.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu HC, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, Ferris JS, Santella RM, Terry MB. Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis. 2012;33(10):1946–1952. doi: 10.1093/carcin/bgs201. doi:10.1093/carcin/bgs201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, Ziogas A, Andrulis IL, Anton-Culver H, Boyd N, Buys SS, Daly MB, O’Malley FP, Santella RM, Southey MC, Venne VL, Venter DJ, West DW, Whittemore AS, Seminara D, Breast Cancer Family R The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast cancer Res. 2004;6(4):R375–R389. doi: 10.1186/bcr801. doi:10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuhausen SL, Ozcelik H, Southey MC, John EM, Godwin AK, Chung W, Iriondo-Perez J, Miron A, Santella RM, Whittemore A, Andrulis IL, Buys SS, Daly MB, Hopper JL, Seminara D, Senie RT, Terry MB, Breast Cancer Family R BRCA1 and BRCA2 mutation carriers in the Breast Cancer Family Registry: an open resource for collaborative research. Breast Cancer Res Treat. 2009;116(2):379–386. doi: 10.1007/s10549-008-0153-8. doi:10.1007/s10549-008-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen-Dumont T, Pope BJ, Hammet F, Southey MC, Park DJ. A high-plex PCR approach for massively parallel sequencing. Biotechniques. 2013;55(2):69–74. doi: 10.2144/000114052. doi:10.2144/000114052. [DOI] [PubMed] [Google Scholar]
- 19.Pope BJ, Nguyen-Dumont T, Hammet F, Park DJ. ROVER variant caller: read-pair overlap considerate variant-calling software applied to PCR-based massively parallel sequencing datasets. Source Code Biol Med. 2014;9(1):3. doi: 10.1186/1751-0473-9-3. doi:10.1186/1751-0473-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. doi:10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum Mutat. 2008;29(11):1342–1354. doi: 10.1002/humu.20896. doi:10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tischkowitz M, Capanu M, Sabbaghian N, Li L, Liang X, Vallee MP, Tavtigian SV, Concannon P, Foulkes WD, Bernstein L, Group WSC. Bernstein JL, Begg CB. Rare germline mutations in PALB2 and breast cancer risk: a population-based study. Hum Mutat. 2012;33(4):674–680. doi: 10.1002/humu.22022. doi:10.1002/humu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. doi:10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7:Unit7.20. doi:10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Exome Variant Server [Accessed 2 Oct 2014]; http://evs.gs.washington.edu/EVS/
- 26.Hellebrand H, Sutter C, Honisch E, Gross E, Wappenschmidt B, Schem C, Deissler H, Ditsch N, Gress V, Kiechle M, Bartram CR, Schmutzler RK, Niederacher D, Arnold N, Meindl A. Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum Mutat. 2011;32(6):E2176–E2188. doi: 10.1002/humu.21478. doi:10.1002/humu.21478. [DOI] [PubMed] [Google Scholar]
- 27.Teo ZL, Park DJ, Provenzano E, Chatfield CA, Odefrey FA, Nguyen-Dumont T, kConFab. Dowty JG, Hopper JL, Winship I, Goldgar DE, Southey MC. Prevalence of PALB2 mutations in Australasian multiple-case breast cancer families. Breast cancer Res. 2013;15(1):R17. doi: 10.1186/bcr3392. doi:10.1186/bcr3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JY, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, Meetei AR, Andreassen PR. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014;33(40):4803–4812. doi: 10.1038/onc.2013.421. doi:10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LOVD P PALB2 LOVD [Accessed 6 Oct 2014]; https://grenada.lumc.nl/LOVD2/shared1/home.php?select_db=PALB2.
- 30.Teo ZL, Provenzano E, Dite GS, Park DJ, Apicella C, Sawyer SD, James PA, Mitchell G, Trainer AH, Lindeman GJ, Shackleton K, Cicciarelli L, kConFab. Buys SS, Andrulis IL, Mulligan AM, Glendon G, John EM, Terry MB, Daly M, Odefrey FA, Nguyen-Dumont T, Giles GG, Dowty JG, Winship I, Goldgar DE, Hopper JL, Southey MC. Tumour morphology predicts PALB2 germline mutation status. Br J Cancer. 2013;109(1):154–163. doi: 10.1038/bjc.2013.295. doi:10.1038/bjc.2013.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heikkinen T, Karkkainen H, Aaltonen K, Milne RL, Heikkila P, Aittomaki K, Blomqvist C, Nevanlinna H. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clinical Cancer Res. 2009;15(9):3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. doi:10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]