Abstract
Background
Avian hepatitis E virus (HEV) is the main causative agent of big liver and spleen disease, as well as hepatitis-splenomegaly syndrome in chickens. To date, conventional reverse transcriptase polymerase chain reaction (RT-PCR) and nested RT-PCR methods have been used for the diagnosis of avian HEV infection in chickens. However, these assays are time consuming, inconvenient, and cannot detect the virus quantitatively. In this study, a rapid and sensitive SYBR Green real-time RT-PCR assay was developed to detect avian HEV RNA quantitatively in serum, liver, spleen, and fecal samples from chickens.
Results
Based on the sequence of the most conserved HEV gene, ORF3, the primers for the assay were designed, and the standard plasmid was constructed. The detection limit of the assay was shown to be 10 copies/μl of standard plasmid/reaction, with a corresponding cycle-threshold value of 29.3. The standard curve exhibited a dynamic linear range across at least 7 log units of DNA copy number. The specificity and reproducibility of this assay was high, showing that the assay detected avian HEV RNA specifically and with little variability. Compared to conventional RT-PCR, the current assay is more sensitive for detecting avian HEV in serum, liver, spleen, and fecal samples from chickens.
Conclusions
A rapid, specific, and reproducible SYBR Green real-time RT-PCR assay was developed for the diagnosis of avian HEV infection in chickens. This assay can accurately detect avian HEV RNA in serum, liver, spleen, and fecal samples with more sensitivity than conventional RT-PCR.
Keywords: Avian hepatitis E virus, Diagnosis, SYBR Green real-time RT-PCR
Background
Avian hepatitis E virus (HEV) is the main causative agent of big liver and spleen disease, as well as hepatitis-splenomegaly syndrome in chickens [1–3]. Both of these diseases result in a decrease in egg production, an increase in mortality, and an enlargement of liver and spleen in broiler breeders and egg laying hens [4–7]. In addition, the antibodies and viral RNA of avian HEV were universally detected in healthy chicken flocks, indicating that the virus can cause subclinical infections in chickens [8–11].
Avian HEV is a non-enveloped, positive-sense, single-stranded RNA virus, belonging to the family Hepeviridae along with human and swine HEVs [12]. The complete genome of avian HEV is approximately 6.6 kb and shares roughly 50 % nucleotide sequence identity with human and swine HEVs [12–15]. Similar to human and swine HEVs, the genome of avian HEV contains two noncoding regions and three open reading frames (ORFs). ORF1 encodes a non-structural protein, ORF2 encodes a capsid protein, and ORF3 (overlapping partially with ORF2), encodes a small phosphoprotein [12, 13]. Based on phylogenetic analysis, avian HEV forms a separate genus from mammalian HEV and is divided into at least three different genotypes [15, 16]. The genomes of the different genotypes share approximately 80 % nucleotide sequence identity, while there is over 90 % nucleotide sequence identity within a single genotype [14, 15].
Currently there is not an efficient cell culture system for propagating avian HEV. Avian HEV infection is generally diagnosed using ELISA to test for the presence of antibodies, and RT-PCR to detect viral RNA [8, 16–19]. To date, some conventional RT-PCR and nested RT-PCR methods have been developed for the diagnosis of avian HEV infection [8, 17]. Among these methods, the RT-PCR assays developed by Huang et al. were widely used for detection avian HEV infection in the flocks from the different countries [17]. However, these assays are time consuming, cumbersome, and prone to falsely positive results due to cross-contamination. Compared to conventional RT-PCR, real-time RT-PCR (using SYBR Green and TaqMan) has many advantages, including simplicity, shorter detection times, and lower contamination rates, as well as higher sensitivity and specificity rates. This assay is also high throughput and easily standardized. One TaqMan real-time RT-PCR assay has been previously described for the detection and quantification of avian HEV in clinical samples [20]. In comparison with a TaqMan assay, SYBR Green real-time RT-PCR is preferable due to the relatively low cost and simplified primer design. However, there is currently no report validating the use of the SYBR Green real-time RT-PCR for the detection and quantification of avian HEV RNA.
The aim of the present study was to develop a rapid, sensitive, and specific SYBR Green real time RT-PCR assay for the identification and quantification of avian HEV RNA in chicken samples and to compare its sensitivity and specificity with the currently prevailing RT-PCR methods which were developed by Huang et al. and widely used for detection avian HEV RNA in clinical samples from the chickens in different countries.
Results
Preparation of the standard plasmid
The 264 base pair (bp) ORF3 gene from a Chinese avian HEV strain (CaHEV) was amplified by RT-PCR and cloned into the commercial clone vector pMD 18-T (TaKaRa Biotech Corporation, Dalian, China). Then, the recombinant plasmid was prepared for the standard control plasmid. The inserted fragment was sequenced and confirmed for 100 % nucleotide identity with the expected ORF3 sequence of CaHEV. The concentration of the recombinant plasmid was 42.5 ng/μl, and the copy number was 1.30 × 1010 copies/μl.
Detection limit and standard curve of the assay
The detection limit of the SYBR Green real-time PCR assay was determined by testing 10-fold serial dilutions of the standard plasmids (107 to 101 copies/μl) in duplicate. The minimum limit of detection was shown to be 10 copies/μl of standard plasmids/reaction, with a corresponding cycle-threshold (Ct) value of 29.3 (Fig. 1a). By plotting the Ct values against the copy numbers of the diluted standard plasmid DNA, the standard curve exhibited a dynamic linear range across at least 7 log units of DNA copy number (107 to 101 copies/μl). Linear regression analysis revealed that the efficiency of the assay was 0.895 and the correlation coefficient (R2) was 1, with a slope value of 3.60 (Fig. 1b).
Fig. 1.
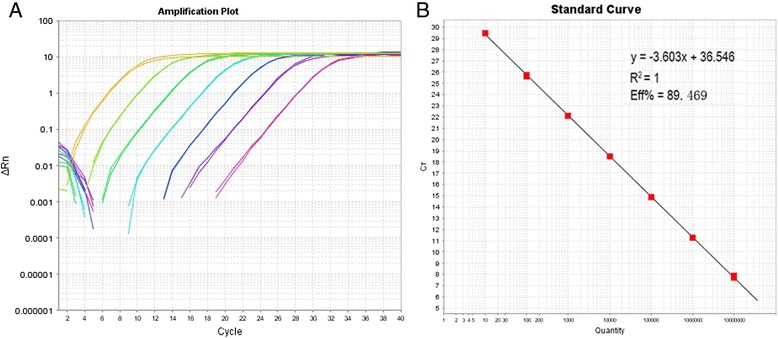
Detection of 10-fold serial dilutions (107 to 101 copies/μl) of standard plasmid using the SYBR Green real-time RT-PCR. a: Avian HEV SYBR Green real-time RT-PCR amplification plot. b: Standard curve where each dot represents the cycle threshold value for the amplified standard plasmid at a given copy number
Specificity and reproducibility of the assay
The specificity of the SYBR Green real-time RT-PCR assay was determined by testing the following 6 avian viruses in triplicate: Newcastle disease virus (NDV), avian influenza virus (H9N2), infectious bronchitis virus (IBV), infectious bursal disease virus (IBDV), Marek’s disease virus (MDV), and J group of avian leucosis virus (ALV-J). In addition, the CaHEV stock and ultrapure water were used as positive and negative controls, respectively. In the assay only CaHEV was detected as a single melt peak and the other six viruses and negative control were not found (Fig. 2). The mean Ct value of CaHEV was 26.8 and no Ct values were measured for the other viruses or the ultrapure water.
Fig. 2.
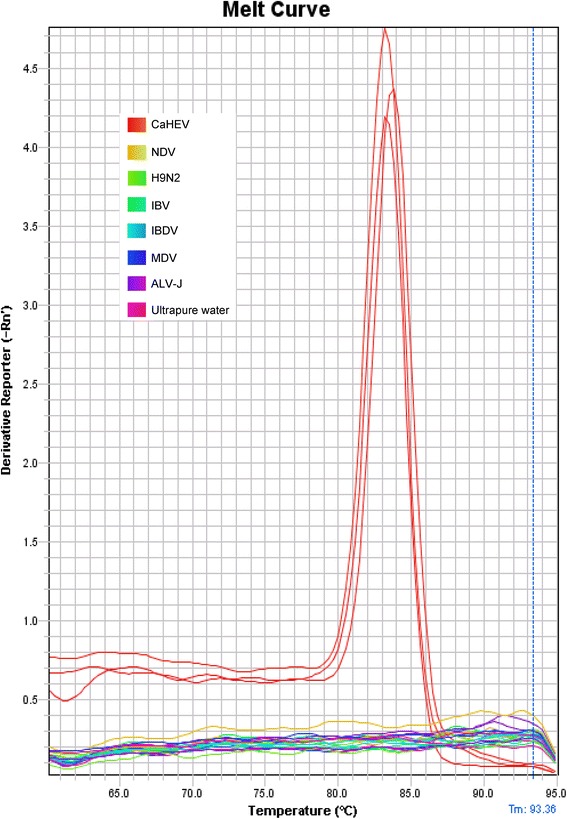
Specificity of the SYBR Green real-time RT-PCR assay. DNA or RNA from CaHEV and six other avian viruses were tested in triplicate using the SYBR Green real-time RT-PCR assay
The reproducibility of the assay was determined by testing five 10-fold serial dilutions of standard plasmids (10−1 to 10−5) in triplicate using inter- and intra-assay comparisons. For the standard plasmids, using dilutions from 10−1 to 10−5, the values of the intra-assay standard deviation (SD) and coefficient of variation (CV) ranged from 0.050 to 0.33 and 0.23 % to 1.6 %, respectively, and the values of the inter-assay SD and CV ranged from 0.070 to 1.3 and 0.32 % to 5.4 %, respectively (Table 1). These data indicated that the assay was repeatable and reproducible with low variation.
Table 1.
Reproducibility of intra- and inter-assay with different dilution standard plasmids by SYBR Green real-time RT-PCR
| Variation | Different dilutions | Ct values for the different dilution of standard plasmids | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Mean Ct ± SD | CV (%) | ||
| Inter-assay | 10−5 | 27.91 | 28.23 | 28.07 | 28.07 ± 0.16 | 0.58 |
| 10−4 | 23.24 | 25.67 | 25.38 | 24.76 ± 1.33 | 5.37 | |
| 10−3 | 21.89 | 21.97 | 22.03 | 21.96 ± 0.07 | 0.32 | |
| 10−2 | 17.25 | 18.12 | 18.23 | 17.87 ± 0.54 | 3.02 | |
| 10−1 | 14.10 | 14.02 | 14.97 | 14.36 ± 0.53 | 3.69 | |
| Intra-assay | 10−5 | 27.38 | 27.54 | 28.01 | 27.64 ± 0.33 | 1.19 |
| 10−4 | 25.10 | 25.23 | 24.98 | 25.10 ± 0.13 | 0.52 | |
| 10−3 | 22.07 | 22.16 | 22.13 | 22.12 ± 0.05 | 0.23 | |
| 10−2 | 17.98 | 18.12 | 18.05 | 18.05 ± 0.07 | 0.39 | |
| 10−1 | 14.54 | 14.78 | 15.01 | 14.78 ± 0.24 | 1.62 | |
Detection of viral RNA in sequential serum samples from CaHEV challenged chickens
Chickens were challenged with CaHEV, and sequential serum samples were collected at 0, 3, 7, 10, 14, 21, 28, 35, and 42 days post-inoculation (dpi) and analyzed for the presence of viral RNA using SYBR Green real-time RT-PCR and conventional RT-PCR. Four of the five challenged chickens were observed with viremia. Using SYBR Green real-time RT-PCR, viral RNA was detected in the infected chickens starting at 7 dpi and ending at 28 dpi. Conventional RT-PCR detected viral RNA in samples from infected chickens starting at 10 dpi and ending at 21 dpi (Fig. 3). The highest amount of viral RNA detected was in the sera of chicken No. 3 at 10 dpi, with a mean titre of 3601 copies/ml. Chickens No. 1, 4, and 5 showcased their viral RNA peak at 14 dpi, with titres ranging from 1120–3121 copies/ml (Fig. 3). No viral RNA was detected in the five chickens from the control group or in chicken No. 2 from the challenged group (Fig. 3).
Fig. 3.
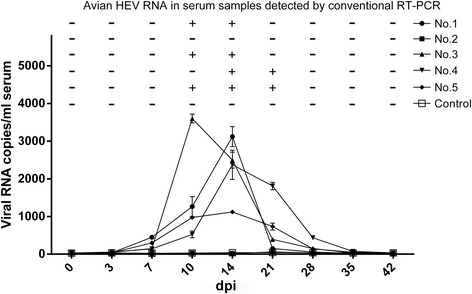
SYBR Green real-time RT-PCR and conventional RT-PCR detection of HEV viral RNA in serum samples from chickens challenged with CaHEV. In the linear graph (bottom), viral RNA in serum samples was quantitatively detected using the SYBR Green real-time RT-PCR. Each data point represents the mean value (±SD) generated from three replicates. The line graph plotted above depicts qualitative detection of avian HEV RNA by conventional RT-PCR. A “-” represents a negative sample and a “+” represents a positive sample
Comparison of SYBR Green real-time RT-PCR and conventional RT-PCR for sensitivity in detecting positive clinical samples
SYBR Green real-time RT-PCR and conventional RT-PCR were performed simultaneously to test 90 clinical samples (32 livers, 16 spleens, and 42 fecal samples), which were collected at different time intervals from birds showing a decrease in egg production in the Shaanxi province of China. Of the 90 samples collected, 22 samples (7 livers, 4 spleens, and 11 fecal samples) were found to be HEV positive using conventional RT-PCR (Table 2). However, 11 additional samples (4 livers and 7 fecal samples) were identified to be positive by SYBR Green real-time RT-PCR (Table 2). These additional samples were confirmed to be positive using sequence analysis (Data not shown). These data indicated that SYBR Green real-time RT-PCR can detect avian HEV RNA in clinical samples with more sensitivity than conventional RT-PCR.
Table 2.
Detection of the clinical samples from the chickens with a decrease in egg production by SYBR Green real-time RT-PCR and conventional RT-PCR
| Types of samples | Total number | Positive number for conventional RT-PCR | Positive number for SYBR Green real-time RT-PCR |
|---|---|---|---|
| Liver | 32 | 7 | 11 |
| Spleen | 16 | 4 | 4 |
| Faeces | 42 | 11 | 18 |
| Total | 90 | 22 | 33 |
Discussion
Over the last two decades remarkable progress has been made in the application of RT-PCR for the diagnosis of avian HEV infection in chickens [8, 17, 18]. However there are currently no efficient cell culture lines available for the propagation of HEV. Several conventional RT-PCR systems have been used for the detection of partial ORF2 and ORF1 genes from avian HEV in various biological samples such as fecal matter, serum, bile, and liver [9, 16–18]. However, these conventional RT-PCR assays are time consuming and labor-intensive with a high risk of contamination. Compared with conventional RT-PCR, the real-time RT-PCR assay is a more rapid and sensitive test and is less likely to produce false positive by contamination during sample preparation. Recently, a more rapid and sensitive TaqMan real-time RT-PCR assay has been reported and are currently under extensive evaluation with clinical chicken samples including livers, fecal and serum samples [20]. However, since the TaqMan real-time RT-PCR assay uses the 5’-3’ nuclease oligoprobe, it is not economical to perform routine testing. As an alternative, the SYBR Green real-time RT-PCR assay can be applied directly to any gene without the need to design and synthesize fluorescently labeled target-specific probes. In addition, the SYBR Green fluorogenic molecules is less expensive, simple in primer design and its universal RT-PCR protocols are suitable for multiple target sequences than the 5’-nuclease TaqMan assay. In this study, a more rapid, simple to operate, and sensitive SYBR Green real-time RT-PCR assay was developed and evaluated for its accuracy and sensitivity in detecting the avian HEV RNA in chicken fecal, liver, spleen, and serum samples.
The RT-PCR assays which have been established in the past use almost exclusively primers designed to target the ORF1 and ORF2 genes of avian HEV. However, sequence alignment showed that the ORF3 gene is actually the most conserved region of the avian HEV genome. Consequently, the SYBR Green real-time PCR assay developed in this study used primers targeting ORF3 gene. In addition, based on sequence alignment, the 3’ terminal nucleotide sequences used to create these primers are the same in different genotypes of avian HEV strains. Based on this information this SYBR Green real-time RT-PCR assay may be used for the universal detection of avian HEV.
To compare the SYBR Green real-time RT-PCR with conventional RT-PCR assays, sequential serum samples were obtained from chickens challenged with CaHEV, and clinical samples were obtained from chickens with a decrease in egg production. These samples were tested simultaneously with the two assays. For the sequential serum samples, viral RNA was detected as a sign of infection. The SYBR Green real-time RT-PCR assay proved to be more sensitive than conventional RT-PCR, as viral RNA was detected over a larger range of time during the progression of infection compared to conventional RT-PCR (Fig. 3). For the 90 clinical samples, only 22 samples were found to be positive using conventional RT-PCR, while 33 were identified as positive using SYBR Green real-time RT-PCR (Table 2). These present data indicated that this newly validated assay is more sensitive than the conventional RT-PCR for early detection and surveillance of avian HEV infection in chickens. This development will be economically important for the poultry industry in the prevention of losses due to HEV infection.
Conclusions
A SYBR Green real-time RT-PCR assay for the detection and quantification of avian HEV RNA was developed. This assay was proven to be efficient, specific, reproducible, and more sensitive than conventional RT-PCR in the detection of avian HEV RNA from serum, liver, spleen, and fecal samples. Effective disease management relies on diagnostic kits to confirm viral infection early and accurately. In this regard, the commercial availability of this assay may be useful for the diagnosis and management of avian HEV infection in poultry farms.
Methods
Virus strain
The CaHEV stock used in this study was prepared from bile suspensions collected from a chicken with hepatitis-splenomegaly syndrome in the Shandong province of China (GenBank accession number: GU954430) [15].
Chickens and samples collection
Five 8-week-old specific-pathogen-free (SPF) chickens (Beijing Merial Vital Laboratory Animal Technology Co., Beijing, China) were housed in an isolation facility and were inoculated intravenously with 0.8 ml of CaHEV stock (104 genomic amount/ml). A group of uninfected chickens (n = 5) were kept separately as negative controls. Sequential serum samples (200 μl) were collected from these chickens at 0, 3, 7, 10, 14, 21, 28, 35, and 42 dpi. In addition, a set of clinical samples from liver (n = 32), spleen (n = 16), and feces (n = 42), were collected from birds showing a decrease in egg production in the Shaanxi province of China. Each sample was homogenized in 10 % sterile 0.1 M phosphate-buffered saline (PBS, pH = 7.4) and clarified by centrifugation at 3000 rpm for 15 min at 4 °C. Total RNA was extracted from the supernatant. Animal experiments were conducted under the guidelines of the Institutional Animal Care and Use Committee of Northwest A&F University and were approved by the Institutional Animal Care and Use Committee of Northwest A&F University with the approval license number of NWAFU (Shaan) 20120910/12.
Primer design
The complete genome of CaHEV was aligned with four other avian HEV genomes from GenBank (GenBank accession numbers AM943647, AM943646, AY535004, and EF206691) using the Clustal W method of Lasergene 7.1 MegAlign program (DNASTAR Inc., Madison, WI, USA). As documented in a previous study, the ORF3 gene of avian HEV was the most conserved (Data not shown). The region selected for primer design was based on the nucleotide sequences of the CaHEV ORF3 gene. The F1 primer 5’-CGTGACAACTCAGCCCAGTG-3’ (covering nucleotides 4801–4821), and R1 primer 5’-GCGGTGACAACGTCGGTA-3’ (complementary to nucleotides 4866–4883), were used for real-time amplification of avian HEV. The F2 primer 5’-ATGTATCTTAGTTGCCAGTTCTGG-3’ (covering nucleotides 4654–4682), and R2 primer 5’-CTACATCTGGTACCGTG-3’ (complementary to nucleotides 4900–4917), were designed for the construction of the standard plasmid.
Viral RNA extraction and reverse transcription
Total RNA was extracted from the CaHEV stock, serum, liver, spleen, and fecal samples using RNAiso Plus reagents according to the manufacturer’s instructions (TaKaRa Biotech Corporation, Dalian, China). RNA samples were then dissolved in 16 μl of DNase-, RNase-, and Proteinase-free water. The first strand of cDNA was synthesized using the Reverse Transcriptase M-MLV (RNase H−) Kit (TaKaRa Biotech Corporation, Dalian, China). The reaction volume was as follows: 2 μl 5×M-MLV buffer, 0.5 μl dNTP mix (10 mmol/L), 1 μl Random 6 mers (50 μM), 0.25 μl RNase Inhibitor (40 U/μl), 0.25 μl M-MLV (200 U/μl), and 6 μl of RNA. The reaction was performed at 42 °C for 30 min, and finished at 80 °C for 15 s.
Standard plasmid DNA
A fragment containing the complete sequence of the CaHEV ORF3 gene was amplified using the F2 and R2 primers to prepare the standard control plasmid. The RT-PCR products were then cloned into the pMD 18-T vector according to the manufacturer’s instructions (TaKaRa Biotech Corporation, Dalian, China). The standard plasmid DNA was extracted from transformed Escherichia coli DH5α competent cells using the EasyPure Plasmid MiniPrep Kit (Transgen Biotech Corporation, Beijing, China) and confirmed with DNA sequencing (Genscript Biotech Corporation, Nanjing, China). DNA concentration was determined by spectrophotometry (Epoch) at 260 nm, and the average concentration was determined using five independent measurements. The copy number of the recombinant plasmid was calculated using the following formula: (DNA concentration in ng/μl×10−9×6.0233×1023 copies/mol)/[DNAsize (bp)×660] [21].
SYBR Green real-time RT-PCR assay
The real-time RT-PCR assay was carried out using the FastStart Universal SYBR Green Master (ROX) Kit (Roche Diagnostics GmbH, Mannheim, Germany). Optimization of primer concentration was achieved using dilutions of RNA (0.1 μM to 0.7 μM) from CaHEV stock. A concentration of 0.5 μM of each primer was found to yield the highest sensitivity. Real-time RT-PCR was performed with a final volume of 10 μl containing 5 μl of 2 × SYBR Green Master Mix, 0.25 μl each of 0.5 μM F1 forward and R1 reverse primers, 2.5 μl cDNA or standard plasmid DNA, and 2 μl of ultrapure water. The optimized thermal cycling conditions were as follows: 95 °C for 5 min, 95 °C for 15 s, and 60 °C for 30 s, for 40 cycles; 95 °C for 30 s, 60 °C for 20 s, and 95 °C for 30 s, for 1 cycle. For each run, an ultrapure water negative control was included. All real-time RT-PCR reactions were conducted using the Step One Plus Real-Time PCR System (Applied Biosystems®, California, USA). The fluorescence was measured at the end of each cycle.
Detection limit and standard curves
To determine the detection limit of the assay, 10-fold serial dilutions of the standard plasmid, (107 to 101 copies/μl in ultrapure water), were run in duplicate through the SYBR Green real-time RT-PCR assay. The obtained Ct values were plotted against the DNA copy number to construct a standard curve.
Specificity and reproducibility
To determine the specificity of the assay, RNA or DNA extracted from six other avian viruses (NDV, H9N2, IBV, IBDV, MDV and ALV-J) were tested in triplicate. Avian HEV positive samples and an ultrapure water negative control were included in each run.
To determine the reproducibility of the real-time PCR assay, intra-assay and inter-assay tests were performed using five different 10-fold dilutions (10−1 to 10−5) of standard plasmid. The intra-assay test was performed in triplicate within the same run, and the inter-assay test was conducted independently as three different runs on different days. The mean, SD, and CV for both the intra-assay and inter-assay tests were calculated separately for each standard DNA dilution based on their Ct values using Microsoft Excel software.
Conventional RT-PCR
The conventional RT-PCR assays for amplification of avian HEV RNA from serum and clinical samples were performed using a 2720 thermal cycler (Applied Biosystems®, California, USA) as described previously with some optimized modifications [17]. Briefly, the PCR mixture contained 12.5 μl of 2 × Premix Taq (loading mix) (TaKaRa Biotech Corporation, Dalian, China), 1 μl each of forward and reverse primers, 5 μl of the cDNA, and 5.5 μl of ultrapure water. The PCR parameters included a denaturation at 95 °C for 10 min followed by 39 cycles of denaturation for 50 s at 94 °C, annealing for 50 s at 50 °C, and extension for 50 s at 72 °C, with a final incubation at 72 °C for 10 min. The PCR yielded a 372 bp product, which was analyzed by electrophoresis on a 1.0 % agarose gel stained with ethidium bromide.
Acknowledgements
This study was supported by grants from Natural Science Foundation of China (No. 31372464) to EMZ and (No. 31402233) to QZ, the Open Fund of Key Laboratory of Veterinary Etiologic Biology (SKLVEB2013KFKT008) and the Specialized Fund for the Basic Research Operating Expenses Program of Central College (Z109021307) to QZ.
Abbreviations
- HEV
Hepatitis E virus
- ORF
Open reading frame
- ELISA
Enzyme-linked immunosorbent assay
- RT-PCR
Reverse transcriptase polymerase chain reaction
- Ct
Cycle-threshold
- SD
Standard deviation
- CV
Coefficient of variation
- SPF
Specific-pathogen-free
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
QZ and SX performed the experiment, arranged the data and drafted the manuscript. YNS, JMG and HYL analyzed the data. XJW, BYL, YYC and LZW participated in animal experiment and clinical samples collection. SFS and GPZ analyzed the data and revised the manuscript. EMZ designed the study, analyzed the data and revised the manuscript. All of the authors read and approved the final manuscript.
Contributor Information
Qin Zhao, Email: qinzhao_2004@sina.com.
Sha Xie, Email: 602267802@qq.com.
Yani Sun, Email: yanisun_2005@sina.com.
Yiyang Chen, Email: 414165606@qq.com.
Jiming Gao, Email: jimingao2006@163.com.
Huiya Li, Email: lihui.xia@163.com.
Xinjie Wang, Email: wang_xin_jie@126.com.
Shahid Faraz Syed, Email: farazvet@yahoo.com.
Baoyuan Liu, Email: lby358891054@126.com.
Lizhen Wang, Email: 917110906@qq.com.
Gaiping Zhang, Email: Zhanggaiping2003@163.com.
En-Min Zhou, Email: zhouem@nwsuaf.edu.cn.
References
- 1.Payne CJ, Ellis TM, Plant SL, Gregory AR, Wilcox GE. Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet Microbiol. 1999;68(1–2):119–25. doi: 10.1016/S0378-1135(99)00067-X. [DOI] [PubMed] [Google Scholar]
- 2.Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J Gen Virol. 2001;82(Pt 10):2449–62. doi: 10.1099/0022-1317-82-10-2449. [DOI] [PubMed] [Google Scholar]
- 3.Meng XJ. Hepatitis E, virus: animal reservoirs and zoonotic risk. Vet Microbiol. 2010;140(3–4):256–65. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke JK, Allan GM, Bryson DG, Williams W, Todd D, Mackie DP, et al. Big liver and spleen disease of broiler breeders. Avian Pathol. 1990;19(1):41–50. doi: 10.1080/03079459008418654. [DOI] [PubMed] [Google Scholar]
- 5.Handlinger JH, Williams W. An egg drop associated with splenomegaly in broiler breeders. Avian Dis. 1988;32(4):773–8. doi: 10.2307/1590997. [DOI] [PubMed] [Google Scholar]
- 6.Morrow CJ, Samu G, Matrai E, Klausz A, Wood AM, Richter S, et al. Avian hepatitis E virus infection and possible associated clinical disease in broiler breeder flocks in Hungary. Avian Pathol. 2008;37(5):527–35. doi: 10.1080/03079450802356946. [DOI] [PubMed] [Google Scholar]
- 7.Troxler S, Pac K, Prokofieva I, Liebhart D, Chodakowska B, Furmanek D, et al. Subclinical circulation of avian hepatitis E virus within a multiple-age rearing and broiler breeder farm indicates persistence and vertical transmission of the virus. Avian Pathol. 2014;43(4):310–8. doi: 10.1080/03079457.2014.924616. [DOI] [PubMed] [Google Scholar]
- 8.Sun ZF, Larsen CT, Dunlop A, Huang FF, Pierson FW, Toth TE, et al. Genetic identification of avian hepatitis E virus (HEV) from healthy chicken flocks and characterization of the capsid gene of 14 avian HEV isolates from chickens with hepatitis-splenomegaly syndrome in different geographical regions of the United States. J Gen Virol. 2004;85(Pt 3):693–700. doi: 10.1099/vir.0.19582-0. [DOI] [PubMed] [Google Scholar]
- 9.Peralta B, Biarnes M, Ordonez G, Porta R, Martin M, Mateu E, et al. Evidence of widespread infection of avian hepatitis E virus (avian HEV) in chickens from Spain. Vet Microbiol. 2009;137(1–2):31–6. doi: 10.1016/j.vetmic.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Hsu IW, Tsai HJ. Avian hepatitis E virus in chickens, Taiwan, 2013. Emerg Infect Dis. 2014;20(1):149–51. doi: 10.3201/eid2001.131224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng Y, Zhao C, Song A, Wang J, Zhang X, Harrison TJ, et al. The serological prevalence and genetic diversity of hepatitis E virus in farmed rabbits in China. Infect Genet Evol. 2011;11(2):476–82. doi: 10.1016/j.meegid.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Huang FF, Sun ZF, Emerson SU, Purcell RH, Shivaprasad HL, Pierson FW, et al. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J Gen Virol. 2004;85(Pt 6):1609–18. doi: 10.1099/vir.0.79841-0. [DOI] [PubMed] [Google Scholar]
- 13.Billam P, Sun ZF, Meng XJ. Analysis of the complete genomic sequence of an apparently avirulent strain of avian hepatitis E virus (avian HEV) identified major genetic differences compared with the prototype pathogenic strain of avian HEV. J Gen Virol. 2007;88(Pt 5):1538–44. doi: 10.1099/vir.0.82754-0. [DOI] [PubMed] [Google Scholar]
- 14.Bilic I, Jaskulska B, Basic A, Morrow CJ, Hess M. Sequence analysis and comparison of avian hepatitis E viruses from Australia and Europe indicate the existence of different genotypes. J Gen Virol. 2009;90(Pt 4):863–73. doi: 10.1099/vir.0.007179-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Q, Zhou EM, Dong SW, Qiu HK, Zhang L, Hu SB, et al. Analysis of avian hepatitis E virus from chickens, China. Emerg Infect Dis. 2010;16(9):1469–72. doi: 10.3201/eid1609.100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marek A, Bilic I, Prokofieva I, Hess M. Phylogenetic analysis of avian hepatitis E virus samples from European and Australian chicken flocks supports the existence of a different genus within the Hepeviridae comprising at least three different genotypes. Vet Microbiol. 2010;145(1–2):54–61. doi: 10.1016/j.vetmic.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Huang FF, Haqshenas G, Shivaprasad HL, Guenette DK, Woolcock PR, Larsen CT, et al. Heterogeneity and seroprevalence of a newly identified avian hepatitis e virus from chickens in the United States. J Clin Microbiol. 2002;40(11):4197–202. doi: 10.1128/JCM.40.11.4197-4202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon HM, Sung HW, Meng XJ. Serological prevalence, genetic identification, and characterization of the first strains of avian hepatitis E virus from chickens in Korea. Virus Genes. 2012;45(2):237–45. doi: 10.1007/s11262-012-0761-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Q, Sun Y, Zhao J, Hu S, Zhao F, Chen F, et al. Development and application of an indirect ELISA for detection of antibodies against avian hepatitis E virus. J Virol Methods. 2013;187(1):32–6. doi: 10.1016/j.jviromet.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Troxler S, Marek A, Prokofieva I, Bilic I, Hess M. TaqMan real-time reverse transcription-PCR assay for universal detection and quantification of avian hepatitis E virus from clinical samples in the presence of a heterologous internal control RNA. J Clin Microbiol. 2011;49(4):1339–46. doi: 10.1128/JCM.01626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke GM, Cheng HL, Ke LY, Ji WT, Chulu JL, Liao MH, et al. Development of a quantitative Light Cycler real-time RT-PCR for detection of avian reovirus. J Virol Methods. 2006;133(1):6–13. doi: 10.1016/j.jviromet.2005.09.011. [DOI] [PubMed] [Google Scholar]


