Abstract
Methyl green has long been known as a histological stain with a specific affinity for DNA, although its fluorescent properties have remained unexplored until recently. In this article, we illustrate the method for preparing a methyl green aqueous stock solution, that when diluted can be used as a very convenient fluorescent nuclear label for fixed cells and tissues. Easy procedures to label whole zebrafish and chick embryos are detailed, and examples of images obtained shown. Methyl green is maximally excited by red light, at 633 nm, and emits with a relatively sharp spectrum that peaks at 677 nm. It is very inexpensive, non-toxic, highly stable in solution and very resistant to photobleaching when bound to DNA. Its red emission allows for unaltered high resolution scanning confocal imaging of nuclei in thick specimens. Finally, this methyl green staining protocol is compatible with other cell staining procedures, such as antibody labeling, or actin filaments labeling with fluorophore-conjugated phalloidin.
Keywords: Developmental Biology, Issue 99, DNA, methyl green, laser scanning confocal microscopy, fluorescence microscopy, nucleus, whole-mount, zebrafish, embryo
Introduction
Fluorescent staining of DNA is broadly and routinely used both in biological research and in clinical diagnosis, either for the detailed study of nuclear and chromosomal structure, for counterstaining tissues and cells, or for studying cell cycle parameters1. Although several DNA stains are available, the most widely used have been those excited by UV light and emitting in blue, which fit in the usual three-filter systems used in most conventional epifluorescence microscopes when combined with green and orange-red-emitting fluorophores, such as fluorescent proteins or small molecules attached to antibodies. Among these blue-emitting DNA stains, the most common are DAPI and Hoechst minor groove-binding agents2,3. Nonetheless, the incorporation of a wide variety of laser emission lines and spectral detection in laser scanning microscopes, has allowed researchers to choose for the use of far-red-emitting nuclear stains such as propidium iodide (PI) or TO-PRO 34. An advantage of these stains is that they overcome the problem of autofluorescence found in many cells and tissues, particularly in embryos5, but many of them have different problems such as having wide emission profiles or being very sensitive to photobleaching6.
The Swiss researcher Friedrich Miescher discovered DNA in 1869, extracting it from the nuclei of white blood cells found in pus. He called it nuclein, and showed a few years later that it formed precipitates with the relatively new stain methyl green. Methyl green is composed by three aniline rings, with different degrees of methylation, and has two positive charges that allow it to strongly bind to the major groove of DNA7,8. DNA staining by methyl green was very early shown to be specific, leading to the development by Unna and Pappenheim of a combined stain with pyronin, which specifically labels RNA. Methyl green-pyronin has since then been extensively used in routine histological technique9.
Until recently, very little was known about the fluorescent properties of methyl green. Our previous work, however, has uncovered some of methyl green spectral advantages6 such as its excitation at 633 nm, a convenient Stokes shift, its emission on the near-infrared portion of the electromagnetic spectrum, higher photostability than commercially available DNA stains with similar spectral properties and at a very low cost. This is why we decided to test it for being used as a very high affinity and specific fluorescent label for DNA on embryonic tissues and in whole embryos. Furthermore, until now methyl green nuclear staining was performed at low pH, making it difficult to combine with antibody staining. The goal of the method detailed here is to have a protocol for fluorescent nuclear DNA staining which overcomes the aforementioned inconvenience by performing the staining procedure at physiological pH, thus making it a suitable counterstain for immunolabeling on sections and whole tissues.
Protocol
1. Purification of the Dye
Prepare a 4% aqueous solution of methyl green by dissolving 0.4 g of stain powder in 10 ml distilled water. Mix thoroughly until completely dissolving it.
Under a fume-hood, mix it with at least 2 parts of chloroform. It is important to check beforehand whether the tubes are chloroform resistant.
Mix thoroughly and centrifuge for 1 min at 2,000 x g to accelerate phase separation.
After centrifugation, an upper aqueous phase with methyl green and a lower organic phase with chloroform and the usual contaminant crystal violet are obtained.
Recover the upper phase and repeat this step until lower phase appears completely devoid of crystal violet.
At the end, the upper phase is recovered and diluted in water to a final concentration of 2% methyl green. This stock solution is stable on the workbench for months and does not need to be protected from light.
2. Fluorescent Nuclear Staining of Whole Zebrafish Embryos using Methyl Green
Fix the embryos overnight in 4% formaldehyde (from paraformaldehyde) in phosphate buffered saline (PBS). NOTE: This can take from a few hours to overnight depending on the thickness of the embryos. In the example, we fixed 48 hr post-fertilization (hpf) zebrafish embryos overnight.
Wash the embryos three times in PBS-T (1% Triton X-100 in PBS) for 5 min. The elevated concentration of detergent permeabilizes the cuticle.
Prepare a 1:5,000-1:10,000 solution of methyl green (from 2% stock solution) in PBS-T. The presence of detergent during the incubation is useful for thick specimens, such as zebrafish embryos and larvae.
Incubate the embryos in the solution for at least 6 hr at 4 °C, with gentle rocking. Again, the thickness of the embryo is the calibrating parameter for the length of the incubation.
Wash the embryos 3 times with PBS for 30 min to remove the detergent excess. Methyl green fluorescence is only evident when bound to DNA, hence background from the unbound molecule is negligible.
Mount on an excavated slide or a homemade chamber with mounting solution (75% glycerol, 0.1 M Tris·HCl pH 8). NOTE: If the nuclear staining is to be performed after immunolabeling, methyl green can be incorporated to the mounting solution at the working concentration, with no need to wash it away.
Store refrigerated or start imaging immediately. Perform imaging with a laser scanning confocal (or other fluorescence) microscope with excitation at 633 nm and emission filters set to register at 650-750 nm taking into account that methyl green emission reaches its maximum at 677 nm. NOTE: We provide the measured data of the methyl green emission profile, to be loaded into the acquisition software, as supplementary material.
3. Fluorescent Nuclear Staining of Whole Chick Embryos using Methyl Green
Incubate fertilized hen eggs to the stage of interest. Fix the embryos overnight in 4% formaldehyde (from paraformaldehyde) in PBS at 4 °C. Wash embryos 3 times in PBS-T for 2 hr.
Prepare a 1:5,000-1:10,000 solution of methyl green (from 2% stock solution) in PBS-T. Incubate the embryos in the staining solution overnight at 4 °C. Wash the embryos 3 times in PBS, 1 hr each. Mount in chamber with 75% glycerol, 0.1 M Tris, pH 8.
4. Imaging Fluorescently Labeled Nuclei in Whole-mounted Embryos
Prepare an imaging chamber by sticking several layers of electrical tape over a microscope slide and carefully cut a hole into it with a scalpel to place the embryos within. This will prevent embryos from crushing during imaging.
Carefully transfer the embryos to the imaging chamber and fill the chamber to the top with 75% glycerol, 0.1 M Tris-HCl, pH=8 to avoid bubble formation.
Cover with a #0 coverslip avoiding or eliminating medium overflowing. Seal it with nail polish.
Acquire images of the specimen on a fluorescence microscope equipped with a 633 excitation filter or laser line. NOTE: Emission filters should cover the 677 emission maximum of methyl green.
Representative Results
Nuclear staining of thick specimens. The protocol herein described allows for the achievement of homogeneous staining of deep structures in whole embryos. Developing lens nuclei form 48 hpf zebrafish embryos are homogeneously labeled (Figure 1).
Methyl green DNA staining allows discrimination of cell cycle dependent morphological differences such as the identification of mitotic figures or apoptotic nuclei (Figure 2A). Subnuclear structures at high resolution are also evident as shown for epidermal nuclei of zebrafish embryos (Figure 2B).
Methyl green is resistant to photobleaching under high intensity irradiation for prolonged periods of time. When irradiated with 633 nm laser light at the same intensity, methyl green nuclear staining does not bleach out as it is the case of another DNA-specific stain sharing similar spectral characteristics, TO-PRO-34. A confocal plane of the retina of a whole-mounted zebrafish embryo was selected and zoomed in with a 63x 1.4NA objective to achieve high irradiation. When zoomed out, the specimens stained with TO-PRO-3 showed bleaching within 60 seconds of continuous irradiation (Figure 3, upper panels), while the specimens stained with methyl green showed no visible evidence of photobleaching even at 120 seconds in the same conditions (Figure 3, lower panels). Images acquired in LASAF software were further processed (brightness/contrast; 3D reconstruction; maximum intensity projection) using Fiji open source software (http://fiji.sc/).
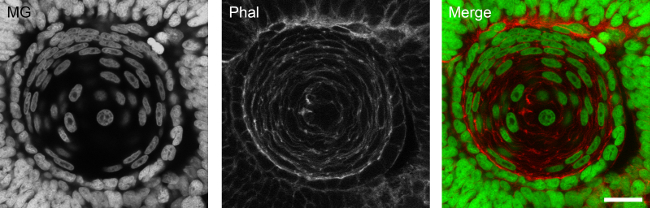 Figure 1: Homogeneous nuclear staining of deep structures with methyl green. Developing lens of a 48 hpf zebrafish embryo stained with methyl green (MG) in a z-projection of 20 confocal planes. Embryos were counterstained for F-actin with TRITC-conjugated phalloidin (Phal). Scale bar: 15 µm.
Figure 1: Homogeneous nuclear staining of deep structures with methyl green. Developing lens of a 48 hpf zebrafish embryo stained with methyl green (MG) in a z-projection of 20 confocal planes. Embryos were counterstained for F-actin with TRITC-conjugated phalloidin (Phal). Scale bar: 15 µm.
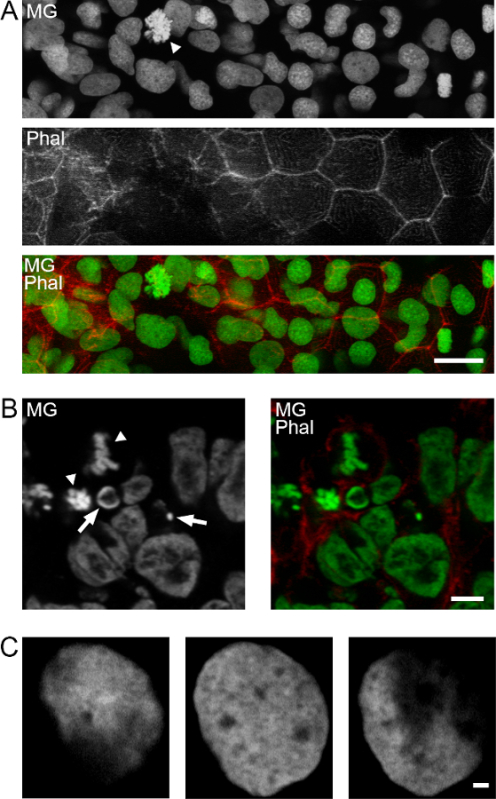 Figure 2: Nuclear morphology and subnuclear resolution with methyl green staining. A) Different nuclear morphologies in the epidermis of a 48 hpf zebrafish embryo. MG, methyl green. Specimens were counterstained for F-actin with TRITC-phalloidin. Arrowhead: mitotic cell. Maximum-intensity projection of 10 confocal planes. B) Chick embryo neural plate labeled in the same way, displaying mitotic cells (arrowheads) and pyknotic nuclei (arrows). Single confocal plane. C) Three single confocal planes of a zebrafish embryo epidermal nucleus at high magnification, showing a high degree of subnuclear resolution. Scale bars: A, 15 µm; B, 5 µm; C, 1 µm.
Figure 2: Nuclear morphology and subnuclear resolution with methyl green staining. A) Different nuclear morphologies in the epidermis of a 48 hpf zebrafish embryo. MG, methyl green. Specimens were counterstained for F-actin with TRITC-phalloidin. Arrowhead: mitotic cell. Maximum-intensity projection of 10 confocal planes. B) Chick embryo neural plate labeled in the same way, displaying mitotic cells (arrowheads) and pyknotic nuclei (arrows). Single confocal plane. C) Three single confocal planes of a zebrafish embryo epidermal nucleus at high magnification, showing a high degree of subnuclear resolution. Scale bars: A, 15 µm; B, 5 µm; C, 1 µm.
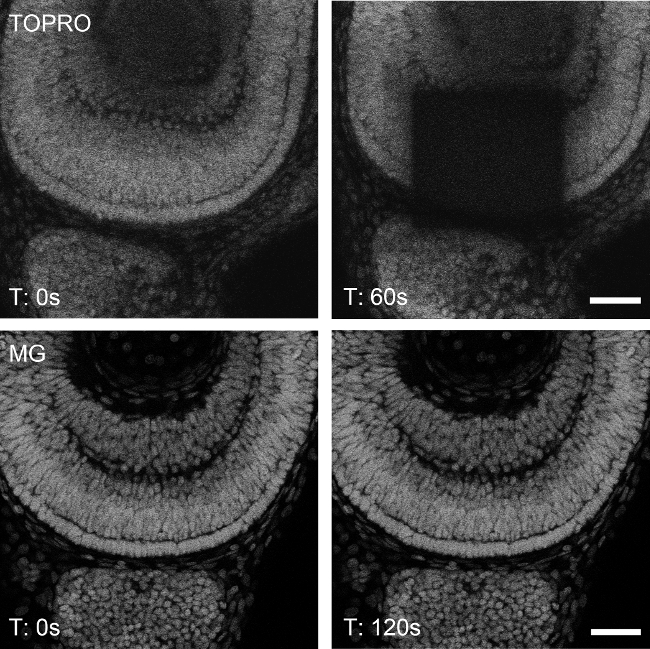 Figure 3: Photostability of methyl green under continuous excitation. TO-PRO-3-stained nuclei of a 48 hpf zebrafish retina was bleached upon 60 sec of continuous excitation at 633 nm (upper panels). Under the same conditions, a methyl green-stained embryo (MG) did not show perceptible bleaching, even when duplicating the exposure time (lower panels). Bleaching and acquisition was made by scanning in single planes, at 8,000 Hz, 30% laser power and line (x2) and frame (x4) averaging. Scale bar: 30 µm.
Figure 3: Photostability of methyl green under continuous excitation. TO-PRO-3-stained nuclei of a 48 hpf zebrafish retina was bleached upon 60 sec of continuous excitation at 633 nm (upper panels). Under the same conditions, a methyl green-stained embryo (MG) did not show perceptible bleaching, even when duplicating the exposure time (lower panels). Bleaching and acquisition was made by scanning in single planes, at 8,000 Hz, 30% laser power and line (x2) and frame (x4) averaging. Scale bar: 30 µm.
| Wavelength (nm) | Percent emission |
| 640 | 21.37 |
| 643 | 27.5 |
| 646 | 36.69 |
| 649 | 44.47 |
| 652 | 53.92 |
| 655 | 61.82 |
| 658 | 67.86 |
| 661 | 83.2 |
| 664 | 85.22 |
| 667 | 89.45 |
| 670 | 87.33 |
| 673 | 92.3 |
| 676 | 100 |
| 679 | 97.41 |
| 682 | 90.34 |
| 685 | 83.32 |
| 688 | 78.37 |
| 691 | 76.09 |
| 694 | 73.95 |
| 697 | 67.36 |
| 700 | 64.4 |
| 703 | 61.26 |
| 706 | 56.39 |
| 709 | 55.06 |
| 712 | 49.56 |
| 715 | 46.12 |
| 718 | 41.4 |
| 721 | 38.8 |
| 724 | 35.97 |
| 727 | 33.27 |
| 730 | 30.95 |
| 733 | 28.78 |
| 736 | 26.28 |
| 739 | 24.96 |
| 742 | 23.3 |
| 745 | 21.12 |
| 748 | 19.86 |
| 751 | 17.47 |
| 754 | 16.14 |
| 757 | 14.21 |
| 760 | 12.45 |
| 763 | 11.49 |
| 766 | 9.97 |
| 769 | 7.55 |
| 772 | 7.75 |
| 775 | 6.46 |
| 778 | 5.76 |
| 781 | 4.38 |
| 784 | 2.6 |
| 787 | 2.75 |
| 790 | 2.27 |
| 793 | 1.52 |
| 796 | 0 |
Supplementary material 1: Emission spectrum of methyl green nuclear staining under 633 nm excitation.
Discussion
The progressive development of microscopy techniques in the last decades, with a particular focus in variations of fluorescent microscopy, have led to the possibility of exploring the dynamics and structure of intact tissue samples, including whole embryos10,11. One major limitation has been, however, the availability of good quality fluorophores that can be used to directly and quickly stain subcellular structures.
The most frequently used fluorescent DNA stains, such as DAPI or Hoechst, are excited with UV light and emit in the blue area of the visible spectrum2,3, making them inadequate for imaging of deep structures in whole tissues. Red light penetrates much better into biological samples, making red-emitting fluorophores more suitable for these studies12. Autofluorescence is a common problem in embryonic tissues that can also be overcome by using red-emitting fluorophores5.
We have shown that the very common histological stain methyl green is a highly efficient red-emitting fluorophore when bound to DNA, and it can be applied to the detection of DNA in microscopy, flow cytometry, fluorometry or gel electrophoresis6. The protocol for fluorescent nuclear labeling of whole embryos with methyl green described here is relatively fast and straightforward. It is an inexpensive technique and a stock solution suitable for hundreds of experiments can be prepared with minimal effort in the lab.
The critical steps of this method are stain purification, specimen permeabilization and incubation times for whole embryos. Crystal violet contamination can easily affect methyl green fluorescent properties thus making it essential to have a highly pure methyl green solution. Methyl green is safer than ethidium bromide as it is a non-intercalant dye which cannot enter cells unless when membrane integrity is compromised, and this is the basis for its application as a vital dye. However, when nuclear DNA is to be stained in fixed embryos, it is required that cuticles and cells go through a permeabilization step such as the ones described here, i.e. incubation with a detergent. Thicker embryos require longer incubation times, as methyl green will enter the embryos by diffusion and minimal incubation times described herein can be used as a reference for other types of specimens. Although we have not tested methyl green in thicker diaphanized tissues as whole mounted organs, due to its spectral properties we do not forsee major limitations to its use provided that chemical clarification (i.e. CLARITY, methysalicilate, Sca/e, etc) does not react with methyl green. An interesting point would be to test its suitability for multi-photon excitation, as this illumination allows for deeper specimen penetration. A simple and useful modification of the protocol can be used for cultured cells by incubating them for 5 minutes with methyl green.
Finally, methyl green emission spectrum is relatively narrow when compared with other red-emitting DNA stains (such as propidium iodide), it presents a relatively large Stokes shift (44 nm) and it is extremely resistant to photobleaching. All these characteristics make it ideal for high resolution laser confocal imaging of whole embryos, as it can be combined with several other fluorophores at the same time and it allows for long exposures.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors would like to acknowledge Patxi Jaso for the production of the video, PEDECIBA and Agencia Nacional de Investigación e Innovación (ANII) for partial funding.
References
- Klonisch T, Wark L, Hombach-Klonisch S, Mai S. Nuclear imaging in three dimensions: a unique tool in cancer research. Annals of anatomy. 2010;192(5):292–301. doi: 10.1016/j.aanat.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Kapuscinski J. DAPI: a DNA-specific fluorescent probe. Biotechnic. 1995;70(5):220–233. doi: 10.3109/10520299509108199. [DOI] [PubMed] [Google Scholar]
- Latt SA, Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. The journal of histochemistry and cytochemistry. 1976;24(1):24–33. doi: 10.1177/24.1.943439. [DOI] [PubMed] [Google Scholar]
- Van Hooijdonk CA, Glade CP, Van Erp PE. TO-PRO-3 iodide: a novel HeNe laser-excitable DNA stain as an alternative for propidium iodide in multiparameter flow cytometry. Cytometry. 1994;17(2):185–189. doi: 10.1002/cyto.990170212. [DOI] [PubMed] [Google Scholar]
- Beumer TL, Veenstra GJ, Hage WJ, Destrée OH. Whole-mount immunohistochemistry on Xenopus embryos using far-red fluorescent dyes. Trends in genetics. 1995;11(1):9. doi: 10.1016/s0168-9525(00)88978-7. [DOI] [PubMed] [Google Scholar]
- Prieto D, Aparicio G, Morande PE, Zolessi FR. A fast, low cost, and highly efficient fluorescent DNA labeling method using methyl green. Histochemistry and cell biology. 2014;142(3):335–345. doi: 10.1007/s00418-014-1215-0. [DOI] [PubMed] [Google Scholar]
- Kim SK, Nordén B. Methyl green. A DNA major-groove binding drug. FEBS letters. 1993;315(1):61–64. doi: 10.1016/0014-5793(93)81133-k. [DOI] [PubMed] [Google Scholar]
- Nordén B, Tjerneld F. Binding of methyl green to deoxyribonucleic acid analyzed by linear dichroism. Chemical Physics Letters. 1977;50(3):508–512. [Google Scholar]
- Lyon H, Jakobsen P, Andersen aP. Standardized methyl green-pyronin Y procedures using pure dyes. The Histochemical journal. 1986;18(2-3):90–94. doi: 10.1007/BF01675361. [DOI] [PubMed] [Google Scholar]
- Amat F, Keller PJ. Towards comprehensive cell lineage reconstructions in complex organisms using light-sheet microscopy. Development, growth & differentiation. 2013;55(4):563–578. doi: 10.1111/dgd.12063. [DOI] [PubMed] [Google Scholar]
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nature methods. 2013;10(6):508–5013. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Lenz P. Fluorescence measurement in thick tissue layers by linear or nonlinear long-wavelength excitation. Applied. 1999;38(16):3662–3669. doi: 10.1364/ao.38.003662. [DOI] [PubMed] [Google Scholar]


