Abstract
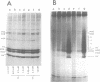
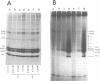
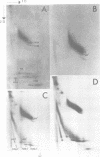
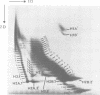
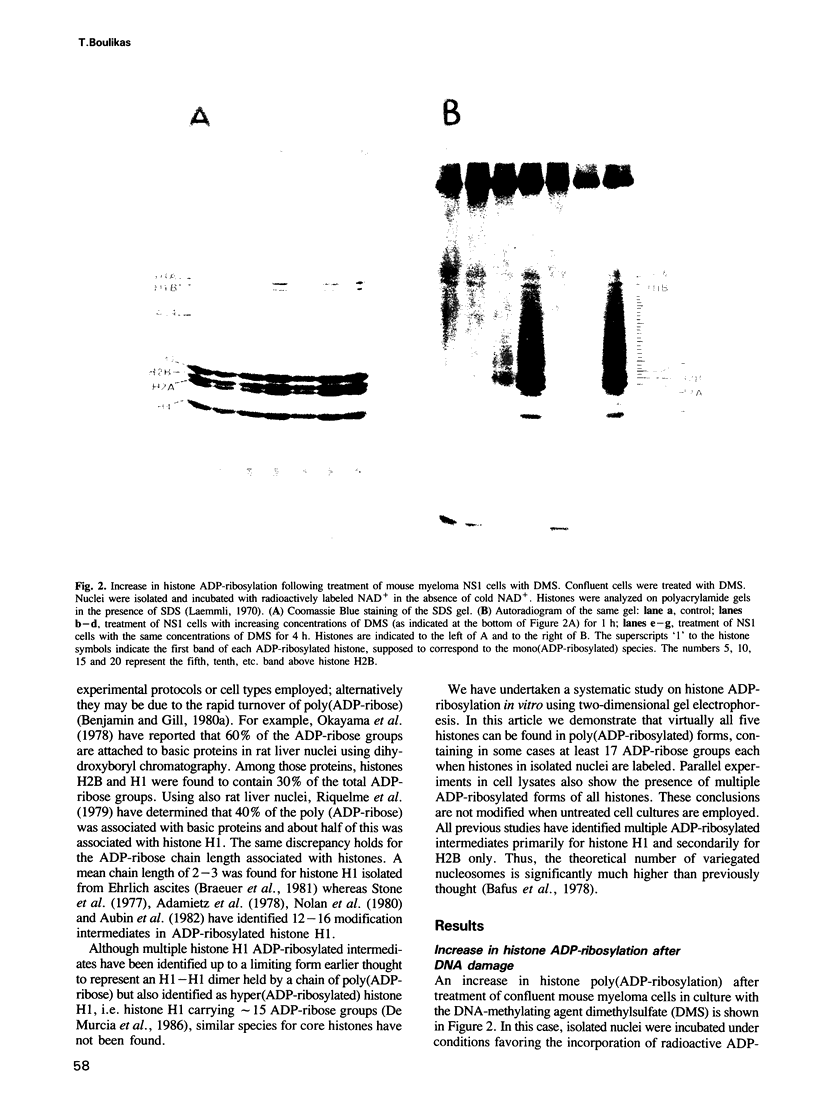
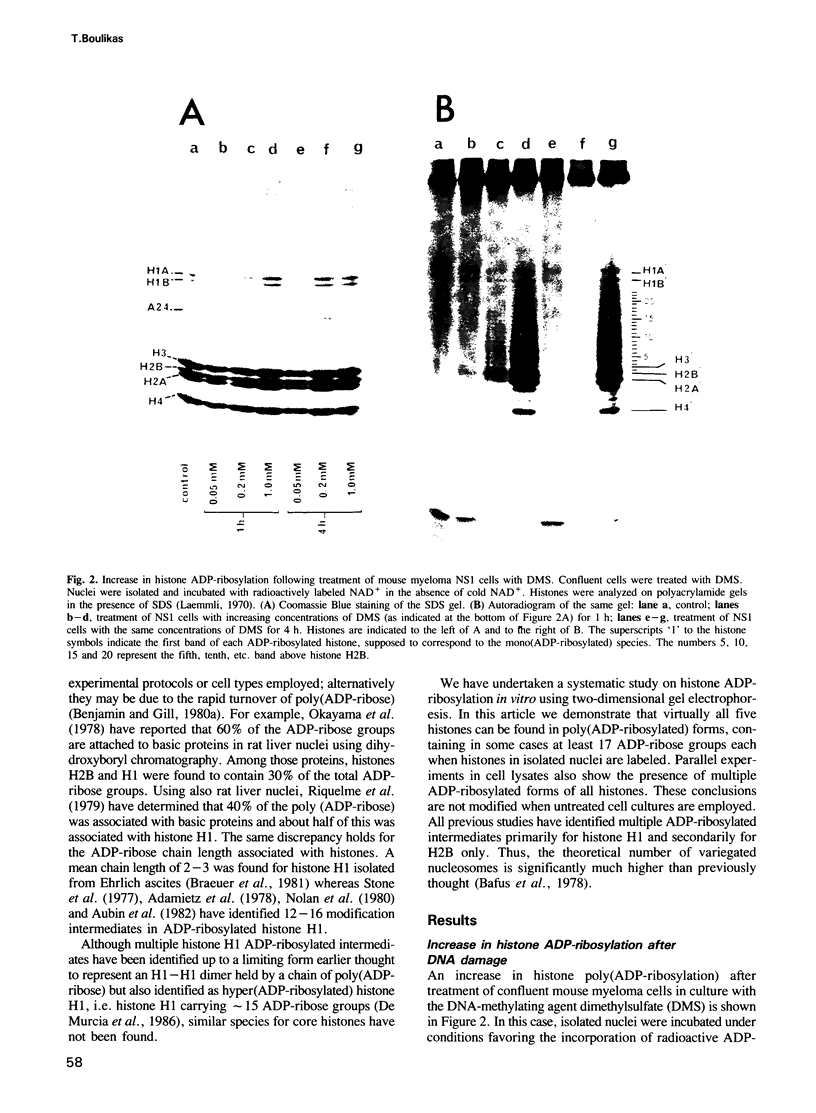
The level of histone adenosine diphospho (ADP) ribosylation was studied in isolated nuclei from mouse myeloma cells in culture. The cells were treated with dimethylsulfate (DMS), a DNA-methylating agent, and histones were analyzed using two-dimensional gel electrophoresis. Seventeen or more bands probably representing mono-to heptadeca (ADP-ribosylated) histones could be visualized for each major variant histone. DMS treatment, by increasing the number of chromatin sites undergoing repair, greatly enhanced histone ADP-ribosylation. When histones were labeled in a cell lysate rather than in isolated nuclei, mono- and oligo(ADP-ribosylated) histone forms prevailed. The presence of approximately 87 ADP-ribosylated variant histone forms in cell lysates and of approximately 170 in isolated nuclei is shown for the first time in this work. Previous studies show multiple ADP-ribosylated forms for only histone H1. The theoretical number of variegated nucleosomes is thus much higher than previously thought, provided that histone-histone contacts are not disrupted at up to a certain level of histone ADP-ribosylation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamietz P., Bredehorst R., Hilz H. ADP-ribosylated histone H1 from HeLa cultures. Fundamental differences to (ADP-ribose)n-histone H1 conjugates formed into vitro. Eur J Biochem. 1978 Nov 15;91(2):317–326. doi: 10.1111/j.1432-1033.1978.tb12682.x. [DOI] [PubMed] [Google Scholar]
- Adamietz P., Rudolph A. ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J Biol Chem. 1984 Jun 10;259(11):6841–6846. [PubMed] [Google Scholar]
- Aubin R. J., Dam V. T., Miclette J., Brousseau Y., Huletsky A., Poirier G. G. Hyper(ADP-ribosyl)ation of histone H1. Can J Biochem. 1982 Dec;60(12):1085–1094. doi: 10.1139/o82-139. [DOI] [PubMed] [Google Scholar]
- Aubin R. J., Fréchette A., de Murcia G., Mandel P., Lord A., Grondin G., Poirier G. G. Correlation between endogenous nucleosomal hyper(ADP-ribosyl)ation of histone H1 and the induction of chromatin relaxation. EMBO J. 1983;2(10):1685–1693. doi: 10.1002/j.1460-2075.1983.tb01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafus N. L., Albright S. C., Todd R. D., Garrard W. T. A method for mapping the distributions of modified and variant histones among mono- and polynucleosomes. J Biol Chem. 1978 Apr 25;253(8):2568–2574. [PubMed] [Google Scholar]
- Benjamin R. C., Gill D. M. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem. 1980 Nov 10;255(21):10493–10501. [PubMed] [Google Scholar]
- Benjamin R. C., Gill D. M. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem. 1980 Nov 10;255(21):10502–10508. [PubMed] [Google Scholar]
- Berger N. A., Sikorski G. W., Petzold S. J., Kurohara K. K. Association of poly(adenosine diphosphoribose) synthesis with DNA damage and repair in normal human lymphocytes. J Clin Invest. 1979 Jun;63(6):1164–1171. doi: 10.1172/JCI109410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N. A., Weber G., Kaichi A. S. Characterization and comparison of poly(adenosine dephosphoribose) synthesis and DNA synthesis in nucleotide-permeable cells. Biochim Biophys Acta. 1978 Jun 22;519(1):87–104. doi: 10.1016/0005-2787(78)90064-3. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., West M. H., Stedman J. D. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980 Aug;109(1):17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Electrophoretic separation of histones and high-mobility-group proteins on acid-urea-Triton gels. Anal Biochem. 1985 Sep;149(2):379–386. doi: 10.1016/0003-2697(85)90586-x. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Interaction of histone H1 with oligonucleosomes in vitro provides a convenient method for isolating mononucleosomes. Can J Biochem Cell Biol. 1985 Sep;63(9):1022–1032. doi: 10.1139/o85-127. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeuer H. C., Adamietz P., Nellessen U., Hilz H. ADP-ribosylated histone H1. Isolation from Ehrlich-ascites-tumor-cell nuclei and partial characterization. Eur J Biochem. 1981;114(1):63–68. [PubMed] [Google Scholar]
- Burzio L. O., Riquelme P. T., Koide S. S. ADP ribosylation of rat liver nucleosomal core histones. J Biol Chem. 1979 Apr 25;254(8):3029–3037. [PubMed] [Google Scholar]
- Butt T. R., Smulson M. Relationship between nicotinamide adenine dinucleotide concentration and in vitro synthesis of poly(adenosine diphosphate ribose) on purified nucleosomes. Biochemistry. 1980 Nov 11;19(23):5235–5242. doi: 10.1021/bi00564a013. [DOI] [PubMed] [Google Scholar]
- Carlsson L., Lazarides E. ADP-ribosylation of the Mr 83,000 stress-inducible and glucose-regulated protein in avian and mammalian cells: modulation by heat shock and glucose starvation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4664–4668. doi: 10.1073/pnas.80.15.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Cohen J. J., Catino D. M., Petzold S. J., Berger N. A. Activation of poly(adenosine diphosphate ribose) polymerase by SV 40 minichromosomes: effects of deoxyribonucleic acid damage and histone H1. Biochemistry. 1982 Sep 28;21(20):4931–4940. doi: 10.1021/bi00263a016. [DOI] [PubMed] [Google Scholar]
- DeLange R. J., Fambrough D. M., Smith E. L., Bonner J. Calf and pea histone IV. 3. Complete amino acid sequence of pea seedling histone IV; comparison with the homologous calf thymus histone. J Biol Chem. 1969 Oct 25;244(20):5669–5679. [PubMed] [Google Scholar]
- Durkacz B. W., Omidiji O., Gray D. A., Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980 Feb 7;283(5747):593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Ferro A. M., Higgins N. P., Olivera B. M. Poly(ADP-ribosylation) of a DNA topoisomerase. J Biol Chem. 1983 May 25;258(10):6000–6003. [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Gaudreau A., Ménard L., de Murcia G., Poirier G. G. Poly(ADP-ribose) accessibility to poly(ADP-ribose) glycohydrolase activity on poly(ADP-ribosyl)ated nucleosomal proteins. Biochem Cell Biol. 1986 Feb;64(2):146–153. doi: 10.1139/o86-023. [DOI] [PubMed] [Google Scholar]
- Goding C. R., Shaw C. H., Blair G. E., Russell W. C. ADP-ribosylation in in vitro systems synthesizing adenovirus DNA. J Gen Virol. 1983 Feb;64(Pt 2):477–483. doi: 10.1099/0022-1317-64-2-477. [DOI] [PubMed] [Google Scholar]
- Goldman N., Brown M., Khoury G. Modification of SV40 T antigen by poly ADP-ribosylation. Cell. 1981 May;24(2):567–572. doi: 10.1016/0092-8674(81)90347-0. [DOI] [PubMed] [Google Scholar]
- Halldorsson H., Gray D. A., Shall S. Poly (ADP-ribose) polymerase activity in nucleotide permeable cells. FEBS Lett. 1978 Jan 15;85(2):349–352. doi: 10.1016/0014-5793(78)80489-x. [DOI] [PubMed] [Google Scholar]
- Huletsky A., Niedergang C., Fréchette A., Aubin R., Gaudreau A., Poirier G. G. Sequential ADP-ribosylation pattern of nucleosomal histones. ADP-ribosylation of nucleosomal histones. Eur J Biochem. 1985 Jan 15;146(2):277–285. doi: 10.1111/j.1432-1033.1985.tb08650.x. [DOI] [PubMed] [Google Scholar]
- Jacobson E. L., Antol K. M., Juarez-Salinas H., Jacobson M. K. Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts. J Biol Chem. 1983 Jan 10;258(1):103–107. [PubMed] [Google Scholar]
- Jacobson M. K., Levi V., Juarez-Salinas H., Barton R. A., Jacobson E. L. Effect of carcinogenic N-alkyl-N-nitroso compounds on nicotinamide adenine dinucleotide metabolism. Cancer Res. 1980 Jun;40(6):1797–1802. [PubMed] [Google Scholar]
- James M. R., Lehmann A. R. Role of poly(adenosine diphosphate ribose) in deoxyribonucleic acid repair in human fibroblasts. Biochemistry. 1982 Aug 17;21(17):4007–4013. doi: 10.1021/bi00260a016. [DOI] [PubMed] [Google Scholar]
- Jongstra-Bilen J., Ittel M. E., Niedergang C., Vosberg H. P., Mandel P. DNA topoisomerase I from calf thymus is inhibited in vitro by poly(ADP-ribosylation). Eur J Biochem. 1983 Nov 2;136(2):391–396. doi: 10.1111/j.1432-1033.1983.tb07754.x. [DOI] [PubMed] [Google Scholar]
- Juarez-Salinas H., Sims J. L., Jacobson M. K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979 Dec 13;282(5740):740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- Kawaichi M., Ueda K., Hayaishi O. Initiation of poly(ADP-ribosyl) histone synthesis by poly(ADP-ribose) synthetase. J Biol Chem. 1980 Feb 10;255(3):816–819. [PubMed] [Google Scholar]
- Kawaichi M., Ueda K., Hayaishi O. Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J Biol Chem. 1981 Sep 25;256(18):9483–9489. [PubMed] [Google Scholar]
- Kawashima K., Izawa M. Poly(ADP-ribose) synthesis in nucleoli and ADP-ribosylation of nucleolar proteins in mouse ascites tumor cells in vitro. J Biochem. 1981 Jun;89(6):1889–1901. doi: 10.1093/oxfordjournals.jbchem.a133391. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B. Glycosylation, ADP-ribosylation, and methylation of Tetrahymena histones. Biochemistry. 1983 Jan 18;22(2):484–489. doi: 10.1021/bi00271a035. [DOI] [PubMed] [Google Scholar]
- Louie A. J., Dixon G. H. Synthesis, acetylation, and phosphorylation of histone IV and its binding to DNA during spermatogenesis in trout. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1975–1979. doi: 10.1073/pnas.69.7.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M., Ishihara M., Takishima S., Takasuka N., Maeda M., Yamaizumi Z., Sugimura T., Yokoyama S., Miyazawa T. The branching and linear portions of poly(adenosine diphosphate ribose) have the same alpha(1 leads to 2) ribose-ribose linkage. J Biol Chem. 1981 Mar 25;256(6):2916–2921. [PubMed] [Google Scholar]
- Miwa M., Tanaka M., Matsushima T., Sugimura T. Purification and properties of glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose). J Biol Chem. 1974 Jun 10;249(11):3475–3482. [PubMed] [Google Scholar]
- Morgan W. F., Cleaver J. E. Effect of 3-aminobenzamide on the rate of ligation during repair of alkylated DNA in human fibroblasts. Cancer Res. 1983 Jul;43(7):3104–3107. [PubMed] [Google Scholar]
- Müller W. E., Zahn R. K. Poly ADP-ribosylation of DNA-dependent RNA polymerase I from quail oviduct. Dependence on progesterone stimulation. Mol Cell Biochem. 1976 Sep 30;12(3):147–159. doi: 10.1007/BF01741713. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y., Ueda K., Nakazawa K., Hayaishi O. Studies on the polymer of adenosine diphosphate ribose. I. Enzymic formation from nicotinamide adenine dinuclotide in mammalian nuclei. J Biol Chem. 1967 Jul 10;242(13):3164–3171. [PubMed] [Google Scholar]
- Nolan N. L., Butt T. R., Wong M., Lambrianidou A., Smulson M. E. Characterization of poly(ADP-ribose)--histone H1 complex formation in purified polynucleosomes and chromatin. Eur J Biochem. 1980 Dec;113(1):15–25. doi: 10.1111/j.1432-1033.1980.tb06133.x. [DOI] [PubMed] [Google Scholar]
- Ogata N., Ueda K., Hayaishi O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J Biol Chem. 1980 Aug 25;255(16):7610–7615. [PubMed] [Google Scholar]
- Ogata N., Ueda K., Kagamiyama H., Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980 Aug 25;255(16):7616–7620. [PubMed] [Google Scholar]
- Oka J., Ueda K., Hayaishi O., Komura H., Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J Biol Chem. 1984 Jan 25;259(2):986–995. [PubMed] [Google Scholar]
- Okayama H., Ueda K., Hayaishi O. Purification of ADP-ribosylated nuclear proteins by covalent chromatography on dihydroxyboryl polyacrylamide beads and their characterization. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1111–1115. doi: 10.1073/pnas.75.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier G. G., Niedergang C., Champagne M., Mazen A., Mandel P. Adenosine diphosphate ribosylation of chicken-erythrocyte histones H1, H5 and high-mobility-group proteins by purified calf-thymus poly(adenosinediphosphate-ribose) polymerase. Eur J Biochem. 1982 Oct;127(3):437–442. doi: 10.1111/j.1432-1033.1982.tb06891.x. [DOI] [PubMed] [Google Scholar]
- Poirier G. G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M., Hillyard D., Olivera B. M. Magnitude and significance of NAD turnover in human cell line D98/AH2. Nature. 1976 Feb 26;259(5545):695–696. doi: 10.1038/259695a0. [DOI] [PubMed] [Google Scholar]
- Reeves R., Chang D., Chung S. C. Carbohydrate modifications of the high mobility group proteins. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6704–6708. doi: 10.1073/pnas.78.11.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme P. T., Burzio L. O., Koide S. S. ADP ribosylation of rat liver lysine-rich histone in vitro. J Biol Chem. 1979 Apr 25;254(8):3018–3028. [PubMed] [Google Scholar]
- Sims J. L., Sikorski G. W., Catino D. M., Berger S. J., Berger N. A. Poly(adenosinediphosphoribose) polymerase inhibitors stimulate unscheduled deoxyribonucleic acid synthesis in normal human lymphocytes. Biochemistry. 1982 Apr 13;21(8):1813–1821. doi: 10.1021/bi00537a017. [DOI] [PubMed] [Google Scholar]
- Smulson M. E., Schein P., Mullins D. W., Jr, Sudhakar S. A putative role for nicotinamide adenine dinucleotide-promoted nuclear protein modification in the antitumor activity of N-methyl-N-nitrosourea. Cancer Res. 1977 Sep;37(9):3006–3012. [PubMed] [Google Scholar]
- Stone P. R., Lorimer W. S., 3rd, Kidwell W. R. Properties of the complex between histone H1 and poly(ADP-ribose synthesised in HeLa cell nuclei. Eur J Biochem. 1977 Nov 15;81(1):9–18. doi: 10.1111/j.1432-1033.1977.tb11921.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanev R., Sendov B. Possible molecular mechanism for cell differentiation in multicellular organisms. J Theor Biol. 1971 Feb;30(2):337–393. doi: 10.1016/0022-5193(71)90059-2. [DOI] [PubMed] [Google Scholar]
- Ueda K., Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- Ueda K., Kawaichi M., Okayama H., Hayaishi O. Poly(ADP-ribosy)ation of nuclear proteins. Enzymatic elongation of chemically synthesized ADP-ribose-histone adducts. J Biol Chem. 1979 Feb 10;254(3):679–687. [PubMed] [Google Scholar]
- Ueda K., Oka J., Naruniya S., Miyakawa N., Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem Biophys Res Commun. 1972 Jan 31;46(2):516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- Ueda K., Omachi A., Kawaichi M., Hayaishi O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc Natl Acad Sci U S A. 1975 Jan;72(1):205–209. doi: 10.1073/pnas.72.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Holt C., Strickland W. N., Brandt W. F., Strickland M. S. More histone structures. FEBS Lett. 1979 Apr 15;100(2):201–218. doi: 10.1016/0014-5793(79)80337-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Flint S. J., Leffak I. M., Groudine M., Grainger R. M. The generation and propagation of variegated chromosome structures. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):401–407. doi: 10.1101/sqb.1978.042.01.042. [DOI] [PubMed] [Google Scholar]
- de Murcia G., Huletsky A., Lamarre D., Gaudreau A., Pouyet J., Daune M., Poirier G. G. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J Biol Chem. 1986 May 25;261(15):7011–7017. [PubMed] [Google Scholar]