SUMMARY
Despite great advances in understanding the mechanisms underlying blood production, lineage specification at the level of multipotent progenitors (MPPs) remains poorly understood. Here, we show that MPP2 and MPP3 are distinct myeloid-biased MPP subsets that work together with lymphoid-primed MPP4 cells to control blood production. We find that all MPPs are produced in parallel by hematopoietic stem cells (HSCs), but with different kinetics and at variable levels depending on hematopoietic demands. We also show that the normally rare myeloid-biased MPPs are transiently overproduced by HSCs in regenerating conditions, hence supporting myeloid amplification to rebuild the hematopoietic system. This shift is accompanied by a reduction in self-renewal activity in regenerating HSCs and reprogramming of MPP4 fate towards the myeloid lineage. Our results support a dynamic model of blood development in which HSCs convey lineage specification through independent production of distinct lineage-biased MPP subsets that, in turn, support lineage expansion and differentiation.
INTRODUCTION
Blood production is a highly regulated process that tailors the output of the myeloid and lymphoid lineages based on hematopoietic demands and the needs of the organism (Ema et al., 2014). Blood development starts with rare self-renewing HSCs that produce a series of increasingly more abundant and lineage-committed progenitor cells, ultimately giving rise to all types of mature blood cells. While the overall structure of the blood system and its hierarchical nature is well established, many questions still remain regarding how HSCs specify lineage fate in non self-renewing MPPs prior to the generation of lineage-committed progenitors and the separation of the myeloid and lymphoid lineages.
HSCs are defined functionally by their ability to serially engraft transplanted recipients and regenerate the entire blood system. This unique property is used as a direct measurement of HSC self-renewal activity, and to identify HSCs based on phenotypic markers. In the mouse, HSCs are found in the Lin−/Sca-1+/c-Kit+ (LSK) fraction of the bone marrow (BM), and are usually defined as CD150+/CD48− LSK cells (Kiel et al., 2005) although other surface markers can be used to enrich for more quiescent and/or functionally distinct subsets including Flk2, CD34, EPCR, rhodamine, the other SLAM markers CD229 and CD244, and CD41 (Wilson et al., 2008; Kent et al., 2009; Oguro et al., 2013; Yamamoto et al., 2013; Miyawaki et al., 2015). Transplantation experiments have shown that markers enriching for the most quiescent and metabolically inert HSC subsets will directly favor engraftment and self-renewal activity (Pietras et al., 2011; Kohli and Passegué, 2014). Single cell transplantation experiments have further demonstrated that even HSCs with identical surface phenotypes are heterogeneous in their engraftment behaviors, with different stabilities over time and variable degrees of myeloid vs. lymphoid lineage output (Dykstra et al., 2007; Yamamoto et al., 2013). Several models are currently proposed to explain this heterogeneity, including the sequential loss of lineage potential in differentiating HSCs (Adolfsson et al., 2005), the existence of long-lived myeloid bypass pathways (Yamamoto et al., 2013), and the presence of functionally distinct clones of lineage-biased HSCs with different biological activities and hard-wired lineage potentials (Muller-Sieburg et al., 2004; Dykstra et al., 2007). However, it remains unclear whether the behavior of transplanted HSCs accurately reflects steady state hematopoiesis and HSC function in native conditions. In fact, it is possible that the distinct behaviors exhibited by single transplanted HSCs represent reversible activities that span a continuum of surface markers and activation states. The identification of MPP1 as a metabolically active subset of HSCs directly supports this idea (Cabezas-Wallscheid et al., 2014). Moreover, two exciting lineage tracking studies directly marking HSCs in their native environment using either sleeping beauty transposons (Sun et al., 2014) or Tie2-Cre endogenous labeling (Busch et al., 2015) have recently shown a limited contribution of HSCs to steady state hematopoiesis and, conversely, a major role for MPPs and lineage-committed progenitors to ongoing blood production.
MPPs are currently a poorly defined hematopoietic compartment, and the term itself is used rather indiscriminately to refer to cells within the LSK fraction that have limited to no engraftment ability in transplantation experiments. The best-characterized and most abundant MPP subset is defined as Flk2+ LSK cells and is now considered as a fully multipotent but lineage-biased population, with low megakaryocyte/erythroid (MegE) and high lymphoid potentials (Adolfsson et al., 2005; Forsberg et al., 2006; Boyer et al., 2011; Buza-Vidas et al., 2011). In fact, the top 25% of the most highly expressing Flk2+ LSK cells have been called lymphoid-primed MPPs or LMPPs (Adolfsson et al., 2005). Recently, two other MPP subsets have been described in the Flk2− LSK fraction and termed MPP2 and MPP3, with Flk2+ LSK cells re-named MPP4 (Wilson et al., 2008). While preliminary investigations suggest that MPP2 and MPP3 have myeloid-biased outputs (Cabezas-Wallscheid et al., 2014), little is known about their biological function. Here, we directly compared the function of MPP2, MPP3 and MPP4 in blood production at steady state and in regenerating conditions following HSC transplantation. We propose a model wherein HSCs produce in parallel distinct subsets of lineage-biased MPPs, which together coordinate the output of the myeloid and lymphoid lineages in response to hematopoietic demands.
RESULTS
Lineage-biased MPP subsets
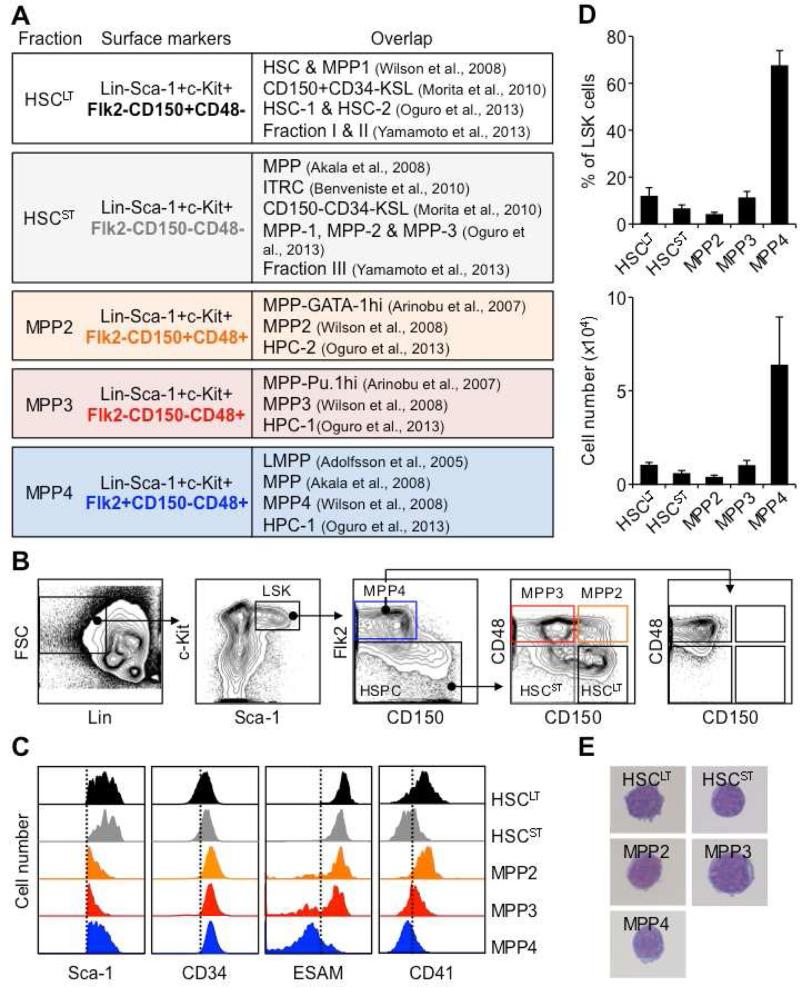
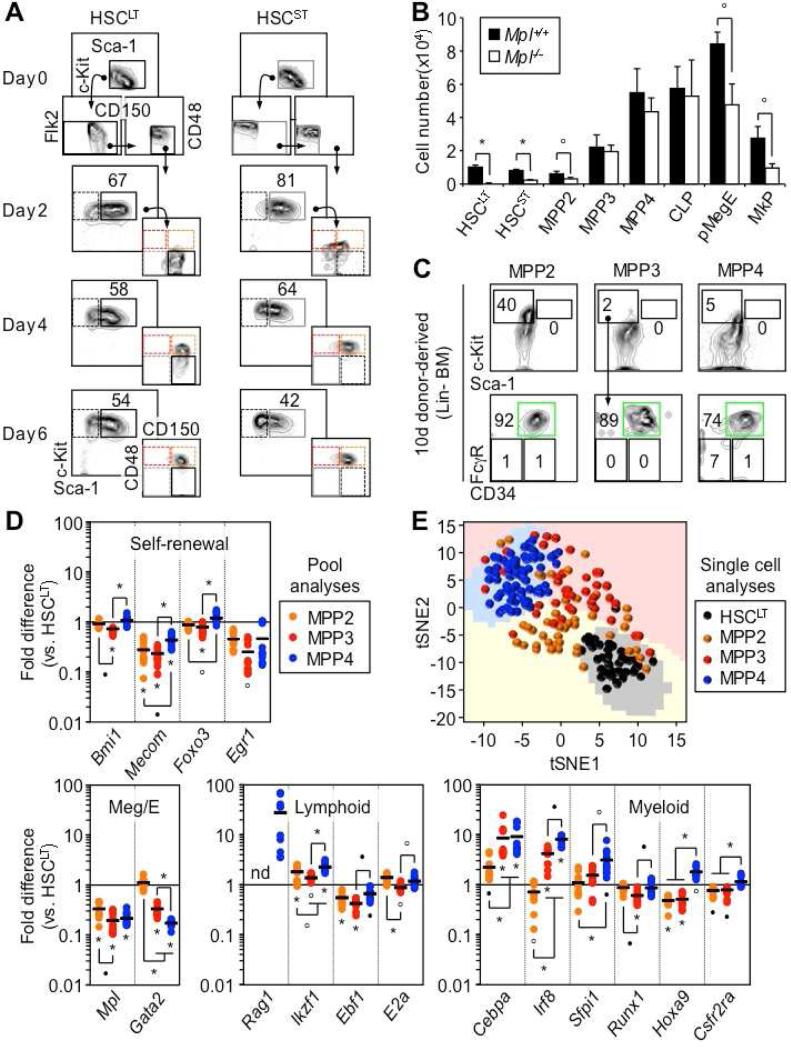
Many names and phenotypic definitions are currently used to describe the spectrum of MPP subsets present in the mouse LSK BM compartment (Figure 1A). One of the most broadly applicable schemes separates the most quiescent HSCs (CD34−/Flk2−/CD150+/CD48− LSK) from the more metabolically active MPP1 (CD34+/Flk2−/CD150+/CD48− LSK), and divides MPPs into three further distinct subsets: MPP2 (Flk2−/CD150+/CD48+ LSK), MPP3 (Flk2−/CD150−/CD48+ LSK) and MPP4 (Flk2+/CD150−/CD48+/− LSK) (Figure 1B) (Wilson et al., 2008; Cabezas-Wallscheid et al., 2014). Importantly, these populations overlap with other MPP definitions based on reporter gene combinations and different surface markers (Arinobu et al., 2007; Akala et al., 2008; Oguro et al., 2013). Since our focus was on MPP biology, we did not use CD34 separation and referred to the combination of HSC and MPP1 as HSCLT. We also included an additional sub-population of LSK cells defined as CD34+/Flk2−/CD150−/CD48− LSK (Figure 1B) that we named short-term HSCs (HSCST), and referred to the combination of HSCLT and HSCST as HSCs.
Figure 1. Re-investigating MPP subsets.
(A) Table showing overlap of MPP subsets with previously published definitions.
(B) Representative gating strategy used to identify and isolate HSCLT, HSCST, MPP2, MPP3 and MPP4 based on expression of Flk2, CD48 and CD150 in BM LSK.
(C) Representative histograms of Sca-1, CD34, ESAM and CD41 expression in the indicated LSK subsets.
(D) Average percentage in BM LSK and absolute numbers of each population (8 mice/group).
(E) Wright-Giemsa staining of the indicated LSK subsets.
Results are expressed as mean ± SD.
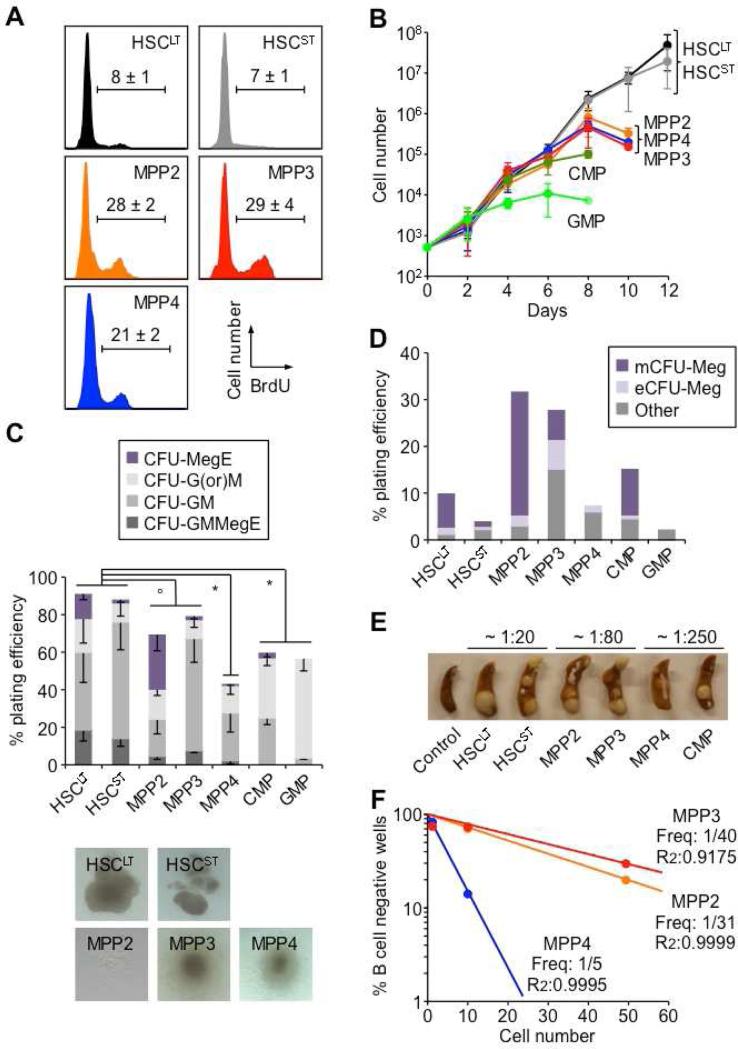
Both MPP2 and MPP3 showed low surface expression of Sca-1 compared to HSCs and MPP4 and, like MPP4, expressed high levels of CD34 (Figure 1C). However, unlike MPP4, they also expressed the HSC markers ESAM (Ooi et al., 2009) and, to various degrees, CD41 (Figure 1C). In steady state conditions, both MPP2 and MPP3 were as rare as HSCs (Figure 1D), and were morphologically undistinguishable from other LSK populations (Figure 1E). As expected, short-term BrdU incorporation experiments showed higher proliferation rates in MPP2 and MPP3 than in HSCs, which were in the range of MPP4 (Figure 2A) (Wilson et al., 2008). Liquid culture experiments confirmed similar expansion rates from all three MPP subsets, which distinguished them from the long-lived HSCs and growth-restricted common myeloid progenitors (CMPs) and granulocyte/macrophage progenitors (GMPs) (Figure 2B). Flow cytometry analyses for Mac-1 and FcγR myeloid differentiation markers also demonstrated persistence of granulocyte/macrophage (GM) potential in cultured MPP2 and MPP3 compared to the transient myeloid output observed with MPP4 (Figure S1A). In clonogenic methylcellulose assays, both MPP2 and MPP3 gave rise to all myeloid lineages but with plating efficiency and pattern of differentiation distinct from HSCs, MPP4 and myeloid progenitors (Figure 2C). Strikingly, MPP3 showed a dominant GM output while MPP2 displayed extensive Meg potential, which was confirmed in collagen-based MegaCult assays (Figure 2D). Spleen colony-forming-unit (CFU-S) assays also indicated similarly strong erythroid potential from both MPP2 and MPP3, which was intermediate to that of HSCs and MPP4/CMPs (Figure 2E). When cultured on OP9/IL-7 to assess B cell potential, both MPP2 and MPP3 generated CD19+ B cells, but with kinetics that again were intermediary between HSCLT and MPP4 (Figure S1B). Furthermore, single cell analyses revealed ~ 8-fold lower B cell clonogenic potential in MPP2 and MPP3 compared to MPP4, and a unique ability to produce B/myeloid mix colonies (Figures 2F and S1C). Similarly, when cultured on OP9-DL1 stromal cells for T cell potential, both MPP2 and MPP3 produced CD25+/CD44+ and CD25+/CD44− immature thymocytes with kinetics intermediary between HSCLT and MPP4 (Figure S1D). Taken together, these in vitro analyses indicate that MPP2 and MPP3 retain full multipotentiality but display specific myeloid biases (i.e., Meg for MPP2 and GM for MPP3) analogous to the lymphoid bias exhibited by MPP4. In addition, based on surface marker expression, MPP2 could be positioned upstream of Pre-MegE and MPP3 upstream of Pre-GM (Figure S1E) (Pronk et al., 2007), thereby establishing a connection between these MPP subsets and myeloid progenitors committed to their dominant lineage fate, similar to the link between MPP4 and Flk2-expressing common lymphoid progenitors.
Figure 2. Co-existence of functionally distinct MPP subsets.
(A) Proliferation rates. Cells were pulsed for 1h with BrdU before analysis (n ≥ 3).
(B) Expansion in liquid culture. Cells were counted every other day (n = 2).
(C) Methylcellulose clonogenic assays and pictures of representative colonies. Single cells were used to measure plating efficiency and colony forming unit (CFU) activity (n ≥ 3).
(D) Meg differentiation potential in collagen-based MegaCult assays (n = 2). mCFU-Meg: small mature colony of ≤ 6 Meg; eCFU-Meg: large early colony of ≥ 6 Meg.
(E) CFU-S assays. Representative photograph of spleen colonies obtained after transplantation of the indicated populations in lethally irradiated mice (n = 2). CFU-S frequency is given at day12.
(F) Clonogenic B-cell differentiation potential on OP9/IL-7 stromal cells. Single, 10 and 50 cells were grown for 16 days and analyzed by flow cytometry for production of CD19+ B cells (10-34 wells/cell dose).
Results are expressed as mean ± SEM; op < 0.05, • p < 0.01, * p < 0.001.
See also Figure S1.
Functional validation
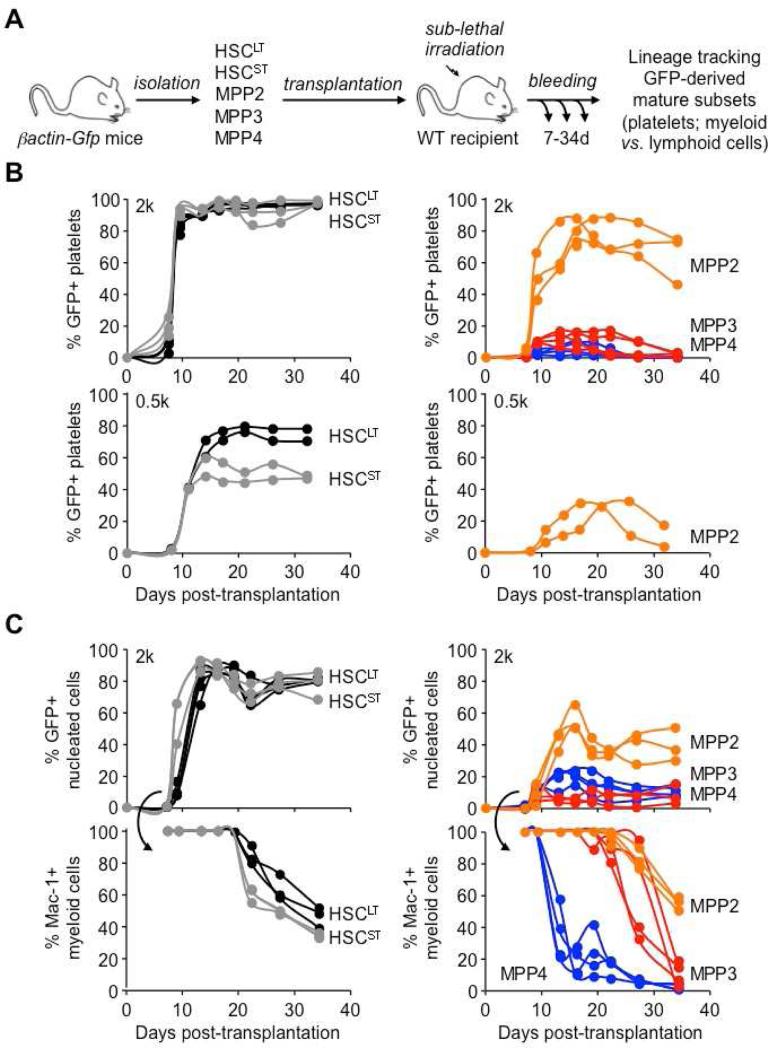
To further demonstrate the intrinsic myeloid bias of these two MPP subsets, we transplanted HSCLT, HSCST, MPP2, MPP3 and MPP4 isolated from β-actin-Gfp mice into sublethally irradiated recipients and followed their multilineage output in peripheral blood (PB) as we have previously done (Forsberg et al., 2006) (Figure 3A). These lineage-tracking experiments confirmed that MPP2 and MPP3 were both devoid of self-renewal potential (Santaguida et al., 2009; Cabezas-Wallscheid et al., 2014) since, like MPP4, they only exhibited short-term myeloid reconstitution ability (≤1 months) in contrast to the sustained potential displayed by HSCs (Figure 3B). Interestingly, differences in HSCLT and HSCST reconstitution activity were only revealed by transplanting a low number of cells (Figure 3B) and tracking the persistence of myeloid output for up to 4 months (Figure S2A). In fact, HSCST behaved similarly to the previously described intermediate-term HSCs (ITRC) (Benveniste et al., 2010). Importantly, these lineage-tracking experiments directly showed the multipotent nature of MPP2 and MPP3, and their ability to generate low levels of B and T cells in vivo (Figure S2B). They also highlighted the considerable but transient ability of MPP2 to produce platelets (Figures 3B and S2C), and the extensive GM potential of both MPP2 and MPP3, which were able to maintain production of mature myeloid cells for two weeks longer than MPP4 (Figure 3C). Taken together, these in vivo experiments confirm that MPP2 and MPP3 are two myeloid-biased MPP subsets that are functionally distinct from the lymphoid-primed MPP4.
Figure 3. Specific lineage biases in MPP subsets.
(A) Experimental scheme for the in vivo lineage tracking experiments. GFP+ populations were transplanted into sub-lethally irradiated recipients and followed over time for their reconstitution activity and lineage potential in PB.
(B) Platelet chimerism following transplantation of 2,000 (upper graphs) or 500 (lower graphs) cells of the indicated donor GFP+ population. Each line represents individual mice (2-4/group).
(C) Nucleated cell chimerism (upper graphs) and percent of donor-derived Mac1+ myeloid cells (lower graphs) following transplantation of 2,000 cells of the indicated donor GFP+ population. Each line represents individual mice (2-4/group).
See also Figure S2.
Molecular validation
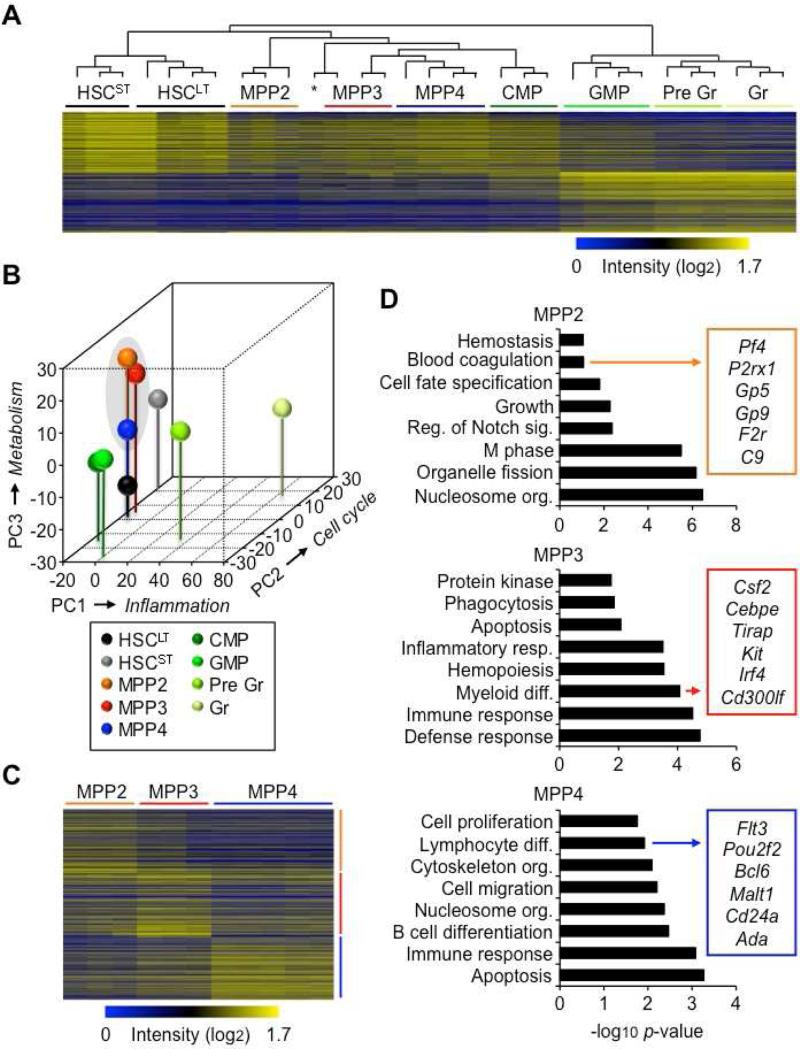
To understand how MPP2 and MPP3 fit into the hematopoietic continuum, we conducted genome-wide microarray analyses. Hierarchical clustering based on gene signatures derived from the 1000 most highly differentially expressed genes in HSCLT and GM lineage-committed cells (Figures S3A, S3B, and Table S1), divided the hematopoietic hierarchy into three main groups: (1) highly immature HSCLT and HSCST; (2) multipotent progenitors including MPP2, MPP3, MPP4 and early myeloid-committed CMPs; and (3) GM lineage-committed cells including GMPs, Gr precursors (Pre Gr) and Gr (Figure 4A). Interestingly, principal component (PC) and gene ontology (GO) analyses identified inflammation genes as main drivers of PC1, which set apart the entire GM-committed myeloid differentiation axis, while cell cycle genes drove PC2, which segregated HSCST from HSCLT, and metabolism genes PC3, which separated MPP2 and MPP3 from MPP4 (Figure 4B, S3C and Table S2). Moreover, GO analyses performed on gene signatures representing the 1000 most highly differentially expressed genes in each MPP relative to the other two underscored their unique molecular features (Figures 4C, 4D and Table S3). As expected, the MPP4 signature was enriched in lymphoid differentiation genes, while MPP3 signature was enriched for myeloid differentiation genes and MPP2 signature for genes related to hemostasis. Consistently, MPP2 showed features of an active MegE transcription program, with high expression levels of Gfi1b, Gata1 and Fog1 (Figure S3D). These distinct gene expression signatures allowed recognition of MPP2, MPP3 and MPP4 in other published gene expression datasets (Table S4). Taken together, these molecular studies corroborate our functional analyses by positioning the rare myeloid-biased MPP2 and MPP3 together with the more abundant lymphoid-primed MPP4 in a multipotent compartment located downstream of HSCs and upstream of the already lineage-committed myeloid and lymphoid progenitors.
Figure 4. Molecular biases in MPP subsets.
(A) Hierarchical clustering analysis based on the 1000 most highly expressed genes in HSCLT and GM lineage-committed cells (GMP, Gr precursors: pre Gr; Gr). * Indicates one MPP4 sample clustering independently.
(B) Principal component (PC) analysis of the microarray results shown in (A). Axis labels indicate the primary gene signature driving each PC separation.
(C) Individual gene signatures representing the 1000 most highly expressed genes in MPP2, MPP3 and MPP4 relative to the other two populations.
(D) Gene ontology (GO) analyses of the gene signatures shown in (C).
Hierarchical organization
To confirm the hierarchy between MPP2, MPP3 and MPP4, we first performed short-term in vitro differentiation experiments, and followed changes in early stem and progenitor surface marker expression by flow cytometry (Figure 5A and S4A). While robust c-Kit and Sca-1 expression allowed tracking of LSK differentiation, the lack of Flk2 induction in vitro, or its quick down-regulation following exposure to Flt-3 ligand, did not permit the separation of newly generated MPP3 and MPP4. In both HSCLT and HSCST cultures, we observed a persistent LSK compartment, which first produced MPP2 and then MPP3/4 (Figure 5A). In contrast, in MPP cultures, the LSK compartment quickly differentiated into c-Kit+/Sca-1− myeloid progenitors and c-Kit−/Sca-1− mature myeloid cells, with the few remaining LSK cells preserving their initial identity despite some fluctuation in marker expression (Figure S4A). While these in vitro results establish that MPP2 is the first MPP subset produced by differentiating HSCs, they do not distinguish whether MPP2 then make MPP3/4, or whether the other MPP subsets are directly generated from HSCs but with slower kinetics. To further clarify this relationship, we took advantage of mice lacking the thrombopoietin receptor (Mpl) gene (Gurney et al., 1994) to determine how defects in platelet production could affect the genesis of the different MPP subsets (Figure 5B). We confirmed the major decrease in HSCLT and HSCST in Mpl−/− mice (Qian et al., 2007), and found a consistent ~ 2-fold reduction in the numbers of MPP2 and all lineage-committed progenitors with Meg activity including Pre-MegE, MkP and CMP (Figure 5B and S4B). In sharp contrast, the numbers of MPP3, MPP4 and other progenitors devoid of Meg activity remained unchanged. These results highlight the key contribution of the Meg-biased MPP2 to megakaryopoiesis, and suggest that MPP3 and MPP4 are produced independently from MPP2 by HSCLT. Finally, we transplanted MPP2, MPP3 and MPP4 isolated from wild type mice into sub-lethally irradiated recipients, to evaluate their ability to regenerate other MPP subsets shortly after transplantation (Figure 5C). However, despite injecting high cell numbers, we were unable to detect production of donor-derived LSK cells in BM from any MPP subsets, while in the same conditions HSCLT regenerated a robust LSK population within 14 days post-transplantation (Figure 5C and S4C). In contrast, after 10 days, we observed strong production of donor-derived GMPs from all MPP subsets, which persisted 14 days after transplantation for MPP2 and MPP3, but not for MPP4, which, by then, was mostly producing lymphoid-derived progeny (Figure S4D). These results confirm the enhanced production of GMPs by myeloid-biased MPP2 and MPP3, and demonstrate that no MPPs are able to generate other MPPs in vivo. Taken together, these experiments establish a hierarchy where HSCs independently generate all three types of lineage-biased MPPs, but with faster production of Meg-biased MPP2 over GM-biased MPP3 and lymphoid-primed MPP4.
Figure 5. Hierarchical organization and molecular priming.
(A) Differentiation in vitro. Representative FACS plots showing HSCLT and HSCST differentiation kinetics in myeloid conditions (n = 2).
(B) Size of the indicated BM populations in Mpl−/− and littermate control mice (3 mice/group).
(C) Differentiation in vivo. Representative FACS plots of LSK and myeloid progenitor output 10 days following transplantation of 5,000 cells of the indicated donor population (3 mice/group).
(D) Fluidigm gene expression analyses at steady state. Results are expressed as mean (bar) and individual fold differences compared to HSCLT (8-12 pools of 100 cells /population; nd: not detectable).
(E) t-distributed stochastic neighbor embedding (tSNE) analysis of Fluidigm gene expression data acquired from single cells (30-58 cells/population).
Results are expressed as mean ± SD; op < 0.05, •p < 0.01, *p < 0.001.
See also Figures S4 and S5.
Molecular priming at steady state
To gain insights into the molecular pathways controlling the production of these different MPP subsets, we used a custom-made Fluidigm dynamic PCR array platform to analyze the expression of 57 genes in pools of 100 HSCLT, MPP2, MPP3 and MPP4 (Figure 5D). Importantly, we detected the expected changes in expression of the surface markers Slamf1 (CD150), Cd48 and Flt3 (Flk2) that define the identity of these populations (Figure S5A). Overall, we found a significant reduction in expression of self-renewal genes in MPPs compared to HSCLT, which was more pronounced in MPP3 than in MPP4 (Figure 5D), and was confirmed at the protein level using cells isolated from Bmi1-eGFP reporter mice (Figure S5B) (Hosen et al., 2007). We also documented the Meg poising of MPP2, with maintenance of Gata2 expression to levels similar to HSCLT, and the lymphoid priming of MPP4, with induction of Rag1 and higher expression of Ikzf1 (Ikaros) compared to HSCLT and other MPP subsets (Figure 5D). In addition, both MPP3 and MPP4 showed clear evidence of GM poising compared to HSCLT and MPP2, with induction of Cebpa, Irf8 and Sfpi1 (Pu.1) gene expression and protein levels as measured in cells isolated from Pu.1-eYFP reporter mice (Figure 5D and S5B). Consistent with their active cell cycle status, all MPPs displayed higher expression levels of specific G1-to-S and G2-to-M phase cyclin/CDK complex genes compared to HSCLT, and a major down-regulation of the quiescence-enforcer Cdkn1c (p57) (Figure S5C). All MPPs also showed elevated expression levels of some DNA repair pathway components, especially Rad51 and Rpa1 (Figure S5C). Moreover, we performed single cell analyses with the same Fluidigm platform, and investigated the 49 most robustly expressed genes using PC analyses and t-distributed stochastic neighbor embedding (tSNE) to pinpoint the key molecular drivers segregating these different populations (Figure 5E and S5D). As expected, Cd48, Slamf1 and Flt3 were all important for PC1 separation. In addition, Cebpa, Irf8, Ccne1/2 (cyclin E1 and E2) and Rad51 emerged as important drivers for both PC1 and PC2 separation, which together with Slamf1 distinguished MPP2 and MPP3 from each other, and separated them from HSCLT and MPP4. Collectively, these analyses demonstrate that, at steady state, all three MPP subsets are part of an actively proliferating continuum of differentiation with molecular priming towards their respective lineage biases and, in the case of the most abundant MPP4, persistence of a significant GM poising alongside its lymphoid priming.
Contribution to blood regeneration
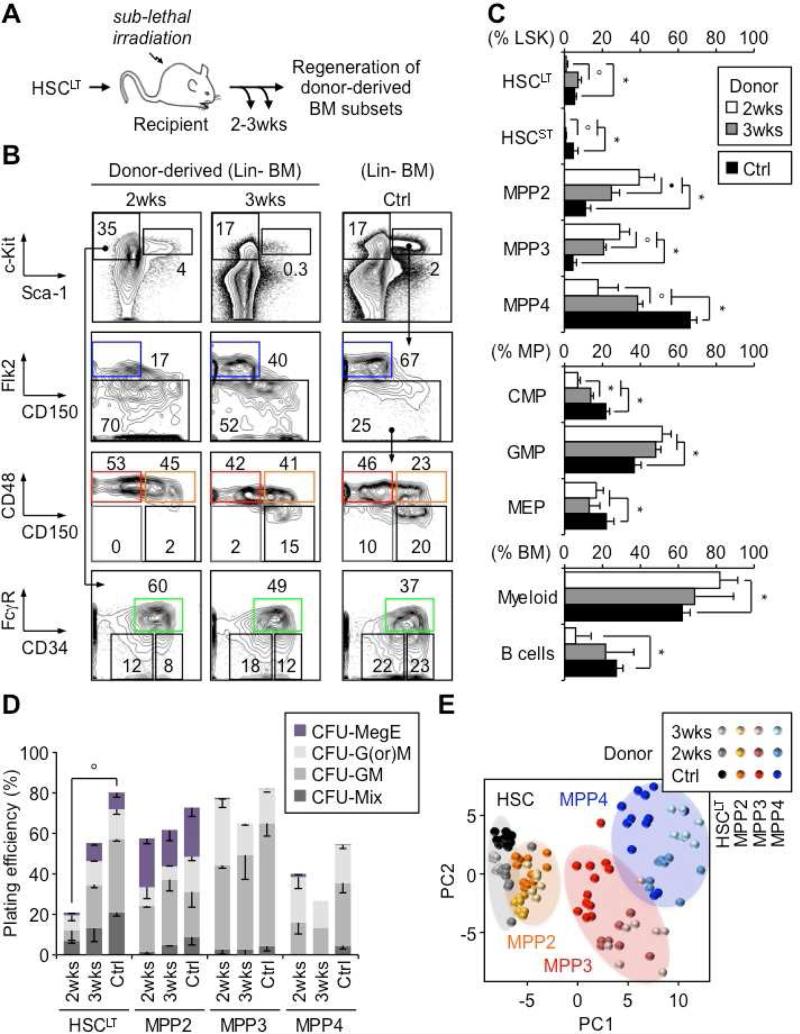
To understand how each MPP subset emerges from HSCs and contributes to blood regeneration, we injected CD45.2 donor HSCLT into sub-lethally irradiated CD45.1 congenic recipients and followed the production of donor-derived BM populations at 2 and 3 weeks post-transplantation (Figure 6A). Strikingly, at 2 weeks, both MPP2 and MPP3 were massively expanded and accounted for most of the reforming LSK compartment, with HSCLT and HSCST being exceedingly rare and MPP4 just starting to be generated (Figure 6B and 6C). By 3 weeks, this process began to revert, with MPP2 and MPP3 contracting toward their steady state levels as HSCLT and MPP4 expanded. In addition, at 2 weeks, all donor-derived BM cells were essentially myeloid, while by 3 weeks, lymphopoiesis had recovered, with a BM lineage composition becoming similar to steady state levels (Figure 6C and S5E). Consistently, we observed a transient expansion of GMPs at 2 weeks, with kinetics matching the initial burst of MPP2/MPP3 expansion and myeloid cell production (Figure 6C). These results indicate that regenerating HSCs first produce myeloid-biased MPPs to quickly establish myeloid output, followed by lymphoid-primed MPP4 to rebuild the lymphoid lineages. Functional assessment in methylcellulose showed similar plating efficiency and overall lineage distribution between newly produced MPP subsets at 2 and 3 weeks, compared to steady state (Figure 6D). In contrast, regenerating HSCLT at 2 weeks displayed profoundly impaired differentiation potential in vitro, which was in large part recovered by 3 weeks (Figure 6D).
Figure 6. Contribution to blood regeneration.
(A) Experimental scheme for in vivo blood regeneration experiments. Donor HSCLT were transplanted into sub-lethally irradiated congenic recipients (2,000 HSCLT/mouse) and regeneration of donor BM subsets was followed at 2 and 3 weeks post-transplantation and compared to steady state control (Ctrl) mice.
(B) Representative FACS plots of regenerating BM subsets.
(C) Frequency of the indicated BM subsets (6-10 mice/group).
(D) Methylcellulose clonogenic assays for the indicated populations (n = 1-4)
(E) PC analysis of Fluidigm gene expression data from the indicated populations (8-12 pools of 100 cells/condition).
Results are expressed as mean ± SD or SEM (D); op < 0.05, •p < 0.01, *p < 0.001.
See also Figure S5, S6.
We also performed Fluidigm gene expression analyses on pools of 100 donor-derived cells re-isolated at 2 and 3 weeks post-transplantation to gain a molecular understanding of this regeneration process (Figure 6E and S6). PC analyses indicated that regenerating HSCLT and newly generated MPP2 preserved their overall molecular identity, while MPP3 and MPP4 showed greater dispersion, mainly reflecting more pronounced changes in expression levels of certain lineage commitment genes (Figure 6E). In particular, Meg/E lineage determinants were significantly downregulated in MPP3 and MPP4, while being preserved in both HSCLT and MPP2 (Figure S6). In addition, MPP4 showed changes in both lymphoid lineage determinants, with transient up-regulation of Rag1 and Ikzf1 at 2 weeks, and GM lineage determinants. In fact, we discerned two reciprocal patterns of changes in myeloid commitment genes in regenerating HSCLT and newly produced MPP4. At 2 weeks, both populations showed increased expression of Pu.1 and Runx1 and decreased expression of Cebpa and Irf8, which completely reversed at 3 weeks (Figure S6). In contrast, newly produced MPP2 and MPP3 displayed limited changes in GM lineage determinants aside from increased expression of Irf8 at 3 weeks and fluctuations in Hoxa9 expression. These results show that MPP4 undergo a significant molecular reprogramming during regeneration, while newly produced MPP2 and MPP3 are able to maintain a strong Meg and GM potential, respectively, hence highlighting the stability of their intrinsic lineage priming. Altogether, they identify MPP2/MPP3 as a compartment of myeloid amplification emerging rapidly from regenerating HSCLT, and suggest that the molecular reprogramming of MPP4 also likely contributes to the initial burst of myeloid cell production
Functional reprogramming in regenerating HSCs
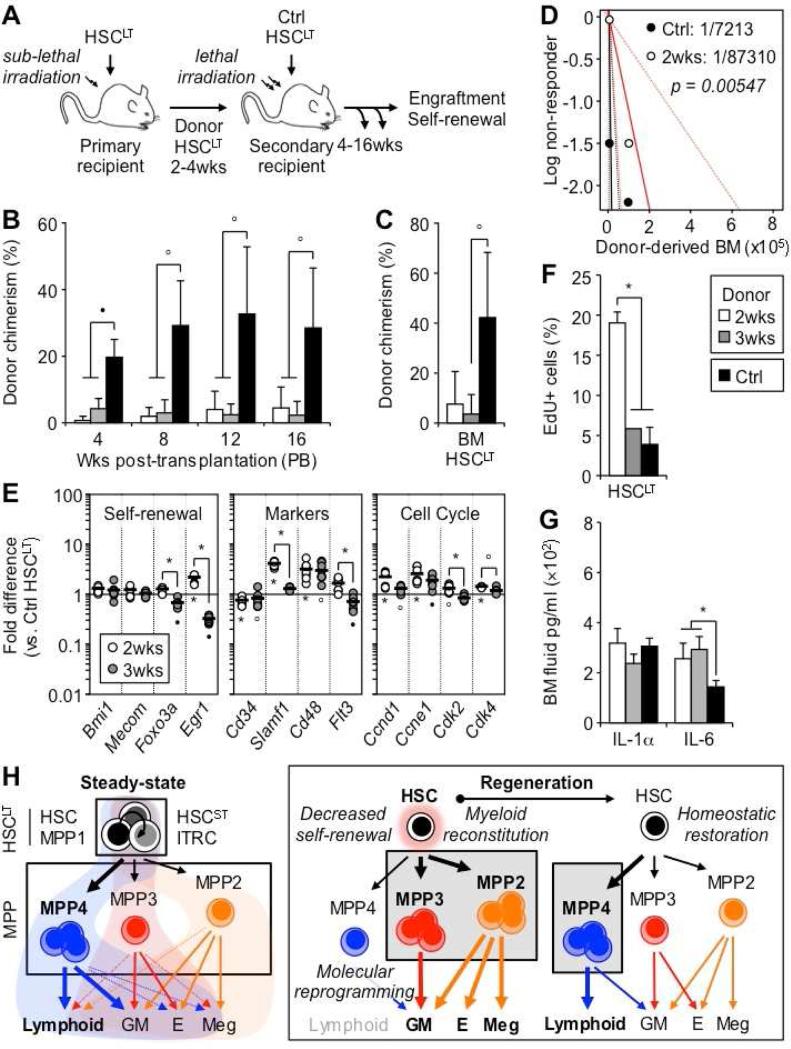
To further investigate the changes in self-renewal activity occurring in regenerating HSCLT, we re-transplanted donor-derived HSCLT isolated at 2 and 3 weeks post-transplantation into lethally-irradiated secondary recipients, and compared their engraftment to freshly isolated control HSCLT (Figure 7A). Strikingly, both 2 and 3 week regenerating HSCLT showed a nearly complete lack of reconstitution activity in PB output and BM engraftment compared to control HSCLT (Figures 7B, 7C, and S7A). Limit dilution analyses (LDA) performed with unfractionated BM cells directly confirmed the impaired engraftment ability of 2 week regenerating HSCLT independently of surface markers (Figures 7D and S7B). Both 2 and 3 week regenerating HSCLT also clearly displayed an activated status, characterized by decreased expression of the quiescence-enforcing transcription factors Foxo3a and Egr1, increased expression of Slamf1 and the MPP markers Cd48 and Flt3, and induction of the cell cycle machinery with increased expression of G2-to-M phase cyclin/CDK complex genes (Figure 7E). Short-term in vivo EdU incorporation experiments directly confirmed the highly proliferative status of 2 week regenerating HSCLT, which had essentially returned to steady state levels by 3 weeks (Figure 7F). In fact, by 4 weeks post-transplantation, donor-derived HSCLT had also restored normal engraftment ability in vivo and plating activity in methylcellulose (Figure S7C and S7D). Finally, cytokine array analyses showed minimal perturbation in cytokine levels (including M-CSF and IL-1) except for increased IL-6 production in the BM cavity of transplanted mice (Figure 7G, S7E and data not shown). Taken together, these results demonstrate that regenerating HSCLT temporarily lose their self-renewal activity as they overproduce myeloid-biased MPPs, and only recover their normal engraftment when blood homeostasis and adequate production of lymphoid-primed MPP4 are restored.
Figure 7. Myeloid-biased MPPs are a transient compartment of myeloid amplification.
(A) Experimental scheme for re-transplantation of regenerating HSCLT. Donor CD45.2 HSCLT isolated from primary recipients at 2 and 3 weeks post-transplantation or from Ctrl mice were injected into lethally irradiated secondary CD45.1 recipients (100 HSCLT/mouse) together with 3×105 Sca-1-depleted CD45.1 BM cells (4-13 mice/group).
(B) Engraftment over time in PB.
(C) Engraftment in BM HSCLT at 16 weeks post-transplantation.
(D) Limit dilution analyses (LDA) of Ctrl (black) and 2 weeks post-transplantation (red) BM cells. Dotted lines represent confidence intervals and values the estimated HSC frequency.
(E) Fluidigm gene expression analyses of key self-renewal determinant, surface marker and cell cycle genes in regenerating HSCLT at 2 and 3 weeks post-transplantation. Results are expressed as mean (bar) and individual fold compared to steady state Ctrl HSCLT (8-12 pools of 100 cells/condition).
(F) Proliferation rates in mice pulsed for 1h with EdU (n = 1-3).
(G) IL-1 and IL-6 levels in BM fluids (5 mice/group).
(H) Revised model of blood production at steady state and in regenerating conditions.
Results are expressed as mean ± SD or SEM (F); op < 0.05, •p < 0.01, *p < 0.001.
See also Figure S7.
DISCUSSION
Here, we show that the MPP compartment is composed of at least two distinct subsets of myeloid-biased MPPs (i.e., MPP2 and MPP3), which work together with lymphoid-primed MPP4 to regulate blood production. We find that all three subsets of lineage-biased MPPs are independently produced by HSCs, but with different kinetics and at varying levels depending on hematopoietic demands. We demonstrate that myeloid-biased MPPs are important for maintaining blood homeostasis at steady state, and for rebuilding the myeloid lineages in regenerative conditions. Taken together, our results support a model of blood development that highlights the key role of the MPP compartment, and place its various lineage-biased subsets at the center stage of lineage specification. They also support the idea that the HSC compartment is functionally plastic, with HSCs committed to differentiation to support blood regeneration temporarily losing their engraftment ability. This updated scheme has important implications for understanding HSC self-renewal and blood production in various physiological contexts.
Competing models of blood development
HSCs are extremely efficient at sustaining long-term multilineage hematopoietic reconstitution in vivo, and are the only blood cells amenable to single cell transplantation assays. In contrast, the functional characterization of multipotent or lineage-committed progenitors remains challenging due to their low yield and transient output in transplantation assays, which preclude most clonal analyses except for the recent attempt at lentiviral barcoding of MPP4 (Naik et al., 2013). As a consequence, current models of blood development place significantly more emphasis on HSC function as measured by transplantation rather than on the activity of non-self-renewing MPPs to explain lineage specification. In the “clonal composition” model, the distinct patterns of blood reconstitution in single cell transplantation assays are explained by selection of particular HSC clones (Copley et al., 2012), and the myeloid bias associated with old age by a shift in clonal dominance with over-representation of myeloid-biased HSCs (Beerman et al., 2010). Consistent with this idea, myeloid-biased HSCs have been prospectively identified based on Hoechst dye exclusion (Challen et al., 2010) or high CD150 expression (Morita et al., 2010). Both the “graded differentiation” and “myeloid bypass” models directly argue against the existence of true MPPs and propose instead either a gradual loss of lineage potential with differentiation, starting with loss of Meg potential at the HSC stage (Adolfsson et al., 2005) and dendritic cell (DC) potential at the MPP4 stage (Naik et al., 2013), or the existence of long-lived myeloid-restricted repopulating progenitors that produce myeloid cells independently from other lineages (Yamamoto et al., 2013). However, all these models are interpretations of reconstitution patterns in transplanted mice that are unlikely to reflect HSC function and lineage specification in native conditions as directly suggested by two recent lineage tracking studies, which both showed limited contribution of HSCs to ongoing hematopoiesis (Sun et al., 2014; Busch et al., 2015). It is also clear that engraftment and quiescence are intrinsically connected, and that more metabolically active MPP1 are less engrafting than quiescent HSCs (Cabezas-Wallscheid et al., 2014). However, it remains unclear whether MPP1 are truly separable from HSCs or whether they represents a reversible stage of HSC activation. This unfortunately cannot be directly tested using transplantation approaches.
To reconcile these observations, we propose a “dynamic model” of blood development in which the HSC compartment is, in fact, composed of distinct subsets of quiescent, activated and lineage-primed HSCs that represent a continuum of likely reversible states and encompass the currently described HSCLT (HSC/MPP1) and HSCST/ITRC populations. In turn, this plastic HSC compartment conveys lineage specification through the independent production of distinct subsets of lineage-biased MPPs, in proportions that are based on hematopoietic demands (Figure 7H). This differs substantially from the prevailing views that consider HSCs solely as an engrafting population rather than a continuum of functional activity, and MPPs as a single linear progression of differentiation rather than parallel pathways that independently control the output of the myeloid and lymphoid lineages. The capacity of HSCs to independently produce specific subsets of lineage-biased MPPs is likely to contribute to the clonal heterogeneity observed upon single cell transplantations. Hence, myeloid-biased HSCs and long-lived myeloid-restricted repopulating progenitors could reflect the reconstitution pattern of HSCs that are primed to produce myeloid-biased MPP2 and MPP3, with balanced HSCs reflecting the behavior of uncommitted HSCs and lymphoid-biased HSCs of HSCs that are poised to produce lymphoid-primed MPP4.
Parallel MPP compartments
Our results considerably expand on the functional characterization of the MPP compartment. We show that while fully multipotent in permissive culture conditions, both myeloid-biased MPPs display specific lineage preferences (i.e., Meg for MPP2 and GM for MPP3). We confirm these biases at the molecular level, with lineage priming reflective of the dominant blood output for each MPP subset, but otherwise only modest differences in their global transcriptional networks as expected for multipotent progenitors. In the case of the lymphoid-primed MPP4, it is also likely that their strong GM poising directly contributes to their ability to produce myeloid cells at steady state and to be quickly reprogrammed towards exclusive myeloid output in regenerative conditions. We validate these lineage biases in vivo by showing that MPP2 and MPP3 are more efficient at producing GMPs than MPP4, and by establishing the importance of MPP2 for megakaryopoiesis using Mpl−/− mice. Moreover, we show that HSCs independently produce all MPP subsets, but at different levels and with different kinetics as exemplified by the delayed emergence of the lymphoid-primed MPP4 in regenerative conditions and the faster production of the Meg-biased MPP2 in vitro. This is interesting in the context of the strong Meg potential described for HSCs further purified with Von Willebrand factor (Sanjuan-Pla et al., 2013), which could directly reflect in vivo enrichment of HSCs that are primed to produce Meg-biased MPP2. Our results also indicate that Flk2− myeloid-biased MPPs are produced independently of Flk2+ MPP4, which contradicts recent fate-mapping results suggesting that all blood cells are derived from Flk2-expressing progenitors (Boyer et al., 2011). However, since MPP2 and MPP3 both express Flk2 mRNA at low levels at steady state and at increasing levels during regeneration, it is possible that the transgenic Flk2-Cre used in these experiments is activated in these MPP subsets despite their lack of Flk2 surface protein expression. Another possibility is that the early window during which MPP2 and MPP3 rebuild the myeloid lineage, before re-establishment of homeostasis and the dominant contribution of Flk2+ MPP4, has been missed in this study. It will therefore be important to use additional in vivo tracking approaches, such as those recently published in native conditions (Sun et al., 2014; Busch et al., 2015), to precisely map the differentiation paths of these MPP subsets.
Molecular reprogramming in regenerative conditions
Consistent with their lineage biases and specific roles in myeloid cell production, we show that the lineage programming of MPP2 and MPP3 remains largely unchanged during regeneration. In contrast, HSCs and MPP4 show transient reprogramming of their lineage fate, with altered expression of similar myeloid lineage genes during blood regeneration. This is characterized by downregulation of Cebpa/Irf8 and concomitant upregulation of Runx1/Pu.1, suggesting that the rebalancing of these two sets of transcription factors plays an important role in priming myeloid lineage output in regenerative conditions. Indeed, C/EBPα is dispensable for regenerative ‘emergency’ granulopoiesis (Hirai et al., 2006), and its downregulation may represent a mechanism to activate HSC proliferation (Ye et al., 2013). Recent evidence also suggests that elevated levels of Pu.1, a direct target of Runx1, are crucial for promoting rapid myeloid differentiation in HSCs (Mossadegh-Keller et al., 2013). These reciprocal changes could therefore enforce HSC proliferation and transiently favor HSC differentiation along a myeloid-biased MPP pathway, with decreased expression of Irf8 eventually limiting monocyte/DC specification in the context of elevated Pu.1 activity (Tamura and Ozato, 2002). The transient induction of this program in MPP4 could directly enforce myeloid output in a population that, despite its GM priming, is also beginning to upregulate key lymphoid differentiation genes. The increased IL-6 levels detected in the BM cavity of transplanted mice could be part of this mechanism, as IL-6 is known to reprogram MPP4 away from lymphoid differentiation and to amplify myeloid differentiation (Reynaud et al., 2011).
Implications for blood production
Our results support the idea that myeloid-biased MPPs serve as a transient compartment of myeloid expansion that can be rapidly activated by HSCs to ensure appropriate production of myeloid cells (Figure 7H). They also demonstrate that regenerating HSCs temporarily lose self-renewal activity as they overproduce myeloid-biased MPPs and rebuild the blood system, a phenotype perhaps linked to their increased cell cycle activity, activated metabolic status and/or myeloid priming (Kohli and Passegué, 2014). This finding raises further caution about defining HSC identity solely based on transplantation experiments, as regenerating HSCs do not perform well in this assay despite being fully capable of maintaining blood production over time. It also raises the interesting possibility that increased CD150 expression observed in both poorly engrafting myeloid-biased HSC clones (Morita et al., 2010; Beerman et al., 2010) and regenerating HSCs is linked to the production of myeloid-biased MPPs, and is essentially a reflection of their activated/primed status. It will now be interesting to explore whether differential production of lineage-biased MPPs by HSCs could account for other long-lasting changes in blood production such as the predominant lymphoid/GM output of fetal hematopoiesis (Kawamoto, 2006), the GM skewing, immunosenescence and anemia of the old blood system (Geiger and Rudolph, 2009), or the aberrant overproduction of different myeloid lineages in hematological malignancies (Tefferi and Gilliland, 2007). It will also be exciting to determine whether similar populations of myeloid-biased MPPs exist in humans. In addition, one of the main complications of anti-cancer therapies or BM transplantation protocols is their deleterious effect on the blood system, leading to prolonged neutropenia and increased risk of infections. It will therefore be compelling to test whether HSC differentiation pathways can be manipulated to favor production of specific lineage-biased MPPs, and thereby optimize blood recovery following hematopoietic injury or rebalance lineage output in an aging or deregulated blood system.
EXPERIMENTAL PROCEDURES
Mice and flow cytometry
Six- to 8-week-old CD45.2 C57Bl/6 wild type or β-actin-Gfp (Forsberg et al., 2006) mice were used as donor for cell isolation, and 8-12 week old CD45.1 C57Bl/6-Boy/J wild type mice as recipients for cell transplantation. Transplanted mice were kept on antibiotic-containing water for 4 weeks. All mice were maintained at UCSF in accordance with IACUC approved protocols. Staining and enrichment procedures for flow cytometry were performed as previously described (Santaguida et al., 2009; Reynaud et al., 2011). Cells were sorted on a FACS ARIAII and analyzed on an LSRII (Becton Dickinson) upon PI exclusion of dead cells. Each population was double sorted to ensure maximum purity.
In vitro analyses
Proliferation, BrdU incorporation, differentiation and OP9/OP9-DL1 co-culture experiments, Microarray and Fluidigm analyses were performed as described in the Extended Experimental Procedures. Three to five independent biological replicates were used for each population. Methylcellulose and MegaCult assays were performed according to the manufacturer's protocols (Stem Cell Technologies).
In vivo analyses
Transplantations, lineage tracking and spleen colony-forming unit (CFU-S) assay were performed as previously described (Forsberg et al., 2006; Reynaud et al., 2011). Congenic recipient mice (CD45.1) were either lethally (1100 rad, split dose 3h apart) or sub-lethally (950 rad, split dose 3h apart) irradiated. Purified donor cells were injected into the retro-orbital plexus and hematopoietic reconstitution was monitored over time in the peripheral blood based on CD45.2 or GFP expression. EdU incorporation experiments and cytokine analyses were performed according to the manufacturer's protocols (Life Technologies).
Statistics
All data are expressed as mean ± standard deviation (SD) or standard error of the mean (SEM) as indicated. P values were generated using unpaired Student's t-test or a Mann-Whitney u-test (Fluidigm), and considered significant when ≤ 0.05. N indicates the numbers of independent experiments performed.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. C. Nerlov (Oxford University) for Pu.1-eYFP mice; Dr. F. de Savage (Genentech) for Mpl−/− mice; M. Sánchez-Castillo for help with t-SNE and discriminant classification analyses; M. Kissner and M. Lee for management of our Flow Cytometry core facility; B. King for initial analyses of Mpl−/− mice; S. Wang for help with MegaCult experiments; Gladstone Genomics Core for microarray processing; and all members of the Passegué laboratory for critical insights and suggestions. This work was supported by NIH awards F32HL106989 and K01DK098315 to E.M.P, grants from Leukaemia and Lymphoma Research, Cancer Research UK and core support by the Wellcome Trust to B.G.; and NIH grant R01HL092471, Rita Allen Scholar Award and Leukemia Lymphoma Society Scholar Award to E.P. The authors have no financial interests to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, E.P.; Methodology, E.M.P., D.R. and E.P.; Investigation, E.M.P., D.R. and Y-A.K.; Formal Analyses - Microarrays, D.C. and J.M.S.; Formal Analyses - Fluidigm, F.J.C-N. and B.G.; Resources, A.D.L.; Writing - Original Draft, E.M.P., D.R. and E.P.; Writing - Review & Editing, E.M.P., Y-A.K. and E.P.; Funding Acquisition, E.M.P., B.G., E.P.
ACCESSION NUMBERS
The accession number for microarray data reported in this paper is GEO GSE68529. Reanalyzed public datasets are described in Table S4.
SUPPLEMENTAL INFORMATION
Supplemental Information for this article includes seven figures, four tables, and Supplemental Experimental Procedures and can be found with this article online.
REFERENCES
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, Rossi DJ. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci USA. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, Iscove NN. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell. 2010;6:48–58. doi: 10.1016/j.stem.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9:64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald HR. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Buza-Vidas N, Woll P, Hultquist A, Duarte S, Lutteropp M, Bouriez-Jones T, Ferry H, Luc S, Jacobsen SE. FLT3 expression initiates in fully multipotent mouse hematopoietic progenitor cells. Blood. 2011;118:1544–1548. doi: 10.1182/blood-2010-10-316232. [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, et al. Identification of Regulatory Networks in HSCs and Their Immediate Progeny via Integrated Proteome, Transcriptome, and DNA Methylome Analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;6:690–697. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Ema H, Morita Y, Suda T. Heterogeneity and hierarchy of hematopoietic stem cells. Exp Hematol. 2014;42:74–82. doi: 10.1016/j.exphem.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegué E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- Geiger H, Rudolph KL. Aging in the lympho-hematopoietic stem cell compartment. Trends Immunol. 2009;30:360–365. doi: 10.1016/j.it.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Gurney AL, Carver-Moore K, de Sauvage FJ, Moore MW. Thrombocytopenia in c-mpl-deficient mice. Science. 1994;265:1445–1447. doi: 10.1126/science.8073287. [DOI] [PubMed] [Google Scholar]
- Hirai H, Zhang P, Dayaram T, Heterington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- Hosen N, Yamane T, Muijtjens M, Pham K, Clarke MF, Weissman IL. Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells. 2007;25:1635–1644. doi: 10.1634/stemcells.2006-0229. [DOI] [PubMed] [Google Scholar]
- Kawamoto H. A close developmental relationship between the lymphoid and myeloid lineages. Trends Immunol. 2006;27:169–175. doi: 10.1016/j.it.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Kent DG, Copley MR, Benz C, Wohrer S, Dykstra BJ, Ma E, Cheyne J, Zhao Y, Bowie MB, Zhao Y, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113:6342–6350. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kohli L, Passegué E. Surviving change: the metabolic journey of hematopoietic stem cells. Trends Cell Biol. 2014;24:479–487. doi: 10.1016/j.tcb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Perié L, Swart E, Gerlach C, van Rooij N, de Boer RJ, Schumacher TN. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Arinobu Y, Iwasaki H, Kohno K, Tsuzuki H, Iino T, Shima T, Kikushige Y, Takenaka K, Miyamoto T, et al. CD41 marks the initial myelo-erythroid lineage specification in adult mouse hematopoiesis: redefinition of murine common myeloid progenitor. Stem Cells. 2015;33:976–987. doi: 10.1002/stem.1906. [DOI] [PubMed] [Google Scholar]
- Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, Moore J, Sieweke MH. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497:239–243. doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Sieburg CE, Cho RH, Karlsson L, Huang JF, Sieburg HB. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood. 2004;103:4111–4118. doi: 10.1182/blood-2003-10-3448. [DOI] [PubMed] [Google Scholar]
- Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi AG, Karsunky H, Majeti R, Butz S, Vestweber D, Ishida T, Quertermous T, Weissman IL, Forsberg EC. The adhesion molecule esam1 is a novel hematopoietic stem cell marker. Stem Cells. 2009;27:653–661. doi: 10.1634/stemcells.2008-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, Passegué E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Qian H, Buza-Vidas N, Hyland CD, Jensen CT, Antonchuk J, Mansson R, Thoren LA, Ekblom M, Alexander WS, Jacobsen SE. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, Sala-Torra O, Radich JP, Passegue E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan-Pla A, Macaulay IC, Jensen CT, Woll PS, Luis TC, Mead A, Moore S, Carella C, Matsuoka S, Bouriez JT, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- Santaguida M, Schepers K, King B, Sabnis AJ, Forsberg EC, Attema JL, Braun BS, Passegue E. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15:341–352. doi: 10.1016/j.ccr.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Gilliland DG. Oncogenes in myeloproliferative disorders. Cell Cycle. 2007;6:550–566. doi: 10.4161/cc.6.5.3919. [DOI] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant C, Eshkind L, Bockamp E, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, Levantini E, Alberich-Jorda M, Zhang J, Kawasaki A, et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol. 2013;15:385–394. doi: 10.1038/ncb2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.