Abstract
Hydrogen sulfide acts as an environmental toxin across a range of concentrations and as a cellular signaling molecule at very low concentrations. Despite its toxicity, many animals, including the mudflat polychaete Glycera dibranchiata, are periodically or continuously exposed to sulfide in their environment. We tested the hypothesis that a broad range of ecologically relevant sulfide concentrations induces oxidative stress and oxidative damage to RNA and DNA in G. dibranchiata. Coelomocytes exposed in vitro to sulfide (0–3 mmol L−1 for 1 h) showed dose-dependent increases in oxidative stress (as 2′,7′-dichlorofluorescein fluorescence) and superoxide production (as dihydroethidine fluorescence). Coelomocytes exposed in vitro to sulfide (up to 0.73 mmol L−1 for 2 h) also acquired increased oxidative damage to RNA (detected as 8-oxo-7,8-dihydroguanosine) and DNA (detected as 8-oxo-7,8-dihydro-2′-deoxyguanosine). Worms exposed in vivo to sulfide (0–10 mmol L−1 for 24 h) acquired elevated oxidative damage to RNA and DNA in both coelomocytes and body wall tissue. While the consequences of RNA and DNA oxidative damage are poorly understood, oxidatively damaged deoxyguanosine bases preferentially bind thymine, causing G-T transversions and potentially causing heritable point mutations. This suggests that sulfide can be an environmental mutagen in sulfide-tolerant invertebrates.
Introduction
Investigations into the physiological effects of hydrogen sulfide (representing here the sum of H2S, HS−, and S2−) have broadly fallen into two categories: ecophysiological studies of sulfide toxicity in aquatic animals and predominantly biomedical studies of sulfide’s role as a gasotransmitter in vertebrates and invertebrates. The ecophysiological studies typically utilize invertebrates from the large assortment of sulfidic habitats, including hydrothermal vents and seeps, coastal mudflats, and sewage outfall sites (Fenchel and Riedl 1970; Cavanaugh 1983). In these habitats, sulfide can reach high millimolar concentrations (up to 30 mmol; Childress and Lowell 1982; Grieshaber and Völkel 1998; Urcuyo et al. 2003). Because sulfide diffuses freely across respiratory surfaces (Denis and Reed 1927; Julian and Arp 1992), animals in sulfidic habitats may experience reversible inhibition of cytochrome c oxidase (Lovatt Evans 1967; Nicholls 1975), decreased hemoglobin oxygen affinity (Carrico et al. 1978), sulfhemoglobin formation (Bagarinao 1992; Kraus et al. 1996), mitochondrial depolarization (Julian et al. 2005a), coelomocyte death and decreased cell proliferation (Hance et al. 2008), and inhibition of almost 20 enzymes involved in aerobic metabolism (Bagarinao 1992). Animals in these habitats typically employ physiological mechanisms to reduce sulfide toxicity (Grieshaber and Völkel 1998), including sulfide-oxidizing enzymes and sulfide-binding amino acids (Joyner et al. 2003; Brand et al. 2007). In contrast to ecophysiological studies, a rapidly growing body of literature examines the role of sulfide as a cellular signaling molecule when present in low concentrations. Sulfide is produced endogenously from cysteine in both vertebrates (Abe and Kimura 1996) and invertebrates (Julian et al. 2002, 2005b; Gainey and Greenberg 2005). Extracellular sulfide concentrations from vertebrate tissues have been reported as ranging from 50 to 160 μmol L−1 in the brain and blood (Goodwin et al. 1989; Abe and Kimura 1996; Zhao et al. 2001), although more recent measurements indicate that these values are far too high (Whitfield et al. 2008). Sulfide modulates muscle tone and neuronal activity in vertebrates (Kimura 2002) and invertebrates (Julian et al. 2002, 2005b; Gainey and Greenberg 2005) and can protect against ischemia-reperfusion injury in mammalian cardiomyocytes (Elrod et al. 2007). Therefore, sulfide appears to have both regulatory and toxic actions, depending on the context, exposure paradigm, and dose.
A recently discovered and not fully understood consequence of sulfide exposure in both ecophysiological and biomedical contexts results from the relationship between sulfide and free radicals, which are highly reactive atoms or molecules that contain one or more unpaired electrons. Free radicals, which can include reactive oxygen and sulfur species (ROSS), can cause cellular damage, termed “oxidative damage,” by stripping electrons from cellular macromolecules (Halliwell and Gutteridge 1999). Chen and Morris (1972) proposed that sulfide oxidation would generate ROSS, and this was later measured in aqueous solutions via electron paramagnetic resonance (EPR; Tapley et al. 1999) and in animal tissues in vitro via EPR (Tapley 1993) and the oxidation of fluorogenic substrates (Abele-Oeschger and Oeschger 1995; Eghbal et al. 2004; Julian et al. 2005a). Consistent with this finding, sulfide exposure can stimulate a cellular oxidative-stress response in vivo in some sulfide-tolerant bivalves (e.g., Tapley 1993; but see Abele et al. 1998) and in a non-sulfide-adapted bivalve (Joyner-Matos et al. 2006). However, the best evidence that sulfide exposure causes lasting, biologically significant oxidative stress arguably would be the direct detection of oxidative damage. Recently, Baskar et al. (2007) demonstrated that very low sulfide concentrations (10–75 μmol L−1) cause formation of micronuclei in vertebrate lung fibroblasts, and Attene-Ramos et al. (2007) demonstrated that 1–25 μmol L−1 of sulfide causes DNA strand breaks and purine ring opening in a Chinese hamster ovary cell line. These findings are consistent with oxidative damage to DNA but can be caused by other stressors. Although this suggests that sulfide-exposed aquatic invertebrates should also suffer oxidative damage to DNA, organisms adapted to sulfidic habitats may have detoxification, antioxidant, and ROSS-scavenging mechanisms that prevent significant oxidative damage during sulfide exposure.
In this study, we tested the hypothesis that a broad range of ecologically relevant sulfide concentrations increase oxidative stress, superoxide production, and oxidative damage to RNA and DNA in the mudflat polychaete Glycera dibranchiata Ehlers 1868 (Annelida: Polychaeta: Phyllodocida: Glyceridae). We have previously shown that G. dibranchiata are sulfide tolerant (Hance et al. 2008) but that sulfide exposure causes mitochondrial depolarization in coelomocytes in vitro (Julian et al. 2005a) and decreased survival and proliferation of coelomocytes in vivo at sulfide concentrations of 0.25–1 mmol L−1 (Hance et al. 2008). To directly assess RNA and DNA oxidative damage, we measured the oxidation of guanine bases to 8-oxo-7,8-dihydroguanosine (RNA) and 8-oxo-7,8-dihydro-2′-deoxyguanosine (DNA). In DNA, the oxidatively damaged guanine preferentially binds thymine and can cause G-T transversions (Halliwell and Gutteridge 1999; Evans and Cooke 2004). If sulfide exposure does cause oxidative damage to RNA and DNA in an organism that is otherwise considered sulfide tolerant, this would suggest that these organisms must, in the long term, mitigate the risk of heritable deleterious mutations.
Material and Methods
Worm Collection and Maintenance
Glycera dibranchiata were obtained via overnight delivery from commercial suppliers (Eastern Sea Worm Company, Hancock, ME, or Harbor Bait Company, Wiscasset, ME), who collected the worms by hand from local mudflats during low tides. The worms were maintained unfed in the laboratory in filtered, 15°C seawater for no more than 2 wk before being used for experiments. Seawater was obtained from the University of Florida Whitney Marine Laboratory (Marineland, FL).
Sulfide Exposure of Coelomocytes In Vitro
Coelomic fluid was obtained from each worm through an incision in the proboscis, after which the coelomocytes were pelleted by centrifugation. The coelomocyte fraction was then purified by removing the overlying white layer or by centrifuging the cells through a 25% sucrose-seawater cushion for 10 min at 10°C and 9,000 g. The cells were then washed with buffered seawater (BSW; sterile-filtered seawater with 10 mmol L−1 HEPES and 0.1% glucose, pH 7.6, 1,000 mOsm kg−1) and further diluted in cold BSW, as indicated below.
In vitro sulfide exposures were conducted by exposing a thin layer of coelomocytes to a mixture of H2S gas in air. For experiments requiring small sample volumes (i.e., the fluorescence-based measurements of oxidative stress, as described below), exposures were carried out in five gas-tight chambers, each containing a black-wall, clear-bottom, 96-well microplate (Corning Life Sciences, Corning, NY), with each well containing 50 μL of coelomocytes diluted 1 : 450 in BSW. An appropriate volume of H2S gas (from a tank of compressed 99% pure H2S gas) was then added by syringe to the gas space of each chamber to obtain 0% (control), 0.10%, 0.30%, 1.0%, and 3.0% H2S in air (yielding 0, 0.29, 0.5, 0.73, and 1.2 mmol L−1 dissolved sulfide, respectively). Coelomocytes were exposed to these conditions at 20°C in the dark for 2 h, after which the cells were labeled with dyes, as described below. This experiment was repeated with cells from five worms, with a coelomocyte sample from each worm being exposed to each level of H2S treatment.
For experiments requiring larger sample volumes (i.e., measurement of RNA and DNA oxidation, as described below), the exposures were carried out in sealed 20-mL glass scintillation vials, as described by Julian et al. (2005a). Briefly, 1 mL of coelomocytes diluted 1 : 45 in BSW was added to each vial, and an appropriate volume of H2S gas was added by syringe to the gas space of each vial to obtain 0% (control), 0.1%, and 1.0% H2S in air (yielding 0, 0.29, and 0.73 mmol L−1 dissolved sulfide, respectively). Cells were exposed to these conditions at 15°C in the dark for 1 h. This experiment was repeated with cells from six worms, with a sample from each worm being exposed to each level of H2S treatment.
In Vivo Sulfide Exposure
Glycera dibranchiata were placed individually in 250-mL incubation flasks and exposed to continuous sulfide for 24 h in a flow-through exposure system under normoxic conditions, as described by Hance et al. (2008). Briefly, sulfide stock solutions of NaHS in ultrapure H2O were prepared at concentrations of 0 (control), 0.0012, 0.038, 0.12, 0.38, 1.2, 3.8, and 12 mmol L−1 and were pumped into the flasks by a syringe pump at 7.0 μL min−1. Seawater was pumped into the flasks at 8.4 mL min−1, and the resulting 1,200 : 1 ratio of seawater flow to stock solution flow produced target sulfide concentrations of 0 (control), 0.0010, 0.032, 0.10, 0.32, 1.0, 3.2, and 10 mmol L−1, respectively. Flasks were held in a recirculating water bath that was maintained at 15°C during exposures. The total dissolved sulfide concentration in each flask was determined immediately after adding a worm and at the end of the exposure by using the methylene blue method (Cline 1969). This experiment was repeated five times (40 worms in total).
Measurement of Oxidative Stress
Intracellular oxidative stress and superoxide production were quantified by using the cell-permeant, vital fluorescent dyes 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA; Invitrogen, Carlsbad, CA), which converts to the fluorescent product 2′,7′-dichlorofluorescein (DCF) upon oxidation by oxygen-centered free radicals, and dihydroethidine (DHE; Invitrogen), which fluoresces upon oxidation by superoxide. After 2 h of exposure to sulfide in 96-well plates, as described above, the coelomocytes were stained for 30 min with H2DCF-DA at 1 μg mL−1 (from a 3-mg-mL−1 stock solution in DMSO) or with DHE at 200 μmol L−1 (from a 20-mmol-L−1 stock solution in water). Fluorescence in each well was measured with a microplate reader (Synergy HT; Bio-Tek Instruments, Winooski, VT) in bottom-reading mode with 485/20-nm excitation and 530/25-nm emission for DCF and 485/20-nm excitation and 590/35-nm emission for DHE.
Measurement of Oxidative Damage to Nucleic Acids
RNA guanine base oxidation produces 8-oxo-7,8-dihydroguanosine (8-oxoGuo), and DNA guanine base oxidation produces 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo). The proportions of oxidized RNA and DNA bases were assayed in coelomocytes from the in vitro sulfide exposures and in whole coelomic fluid and body wall tissue collected from the in vivo sulfide exposures. In the in vitro exposures, coelomocytes were exposed to H2S for 1 h and then immediately flash frozen in liquid N2. In the in vivo exposures, worms were exposed to sulfide for 24 h, after which whole coelomic fluid was obtained through an incision and the coelomocytes were washed and resuspended in BSW, as described above, and flash frozen in liquid N2. An incision was then made along the length of the worm, and the proboscis, esophagus, and intestine were removed. The remaining body wall tissue was then rinsed in BSW, blotted dry, and flash frozen in liquid N2. After flash freezing, the samples were homogenized in liquid N2 and stored at −80°C until assayed.
Samples of 100–200 mg of homogenized coelomocytes or 40–60 mg of homogenized body wall were processed with a method designed for vertebrate tissue (Hofer et al. 2006) and optimized for invertebrate tissues (Joyner-Matos et al. 2007). Briefly, RNA and DNA were simultaneously extracted from the homogenates by using guanidine thiocyanate and phenol/chloroform at neutral pH, after which nucleic acids were hydrolyzed with nuclease P1 and alkaline phosphatase. Hydrolytic enzymes were then removed by filtration, and the hydrolysate was analyzed by high-performance liquid chromatography coupled to electrochemical detection (HPLC-ECD; ESA, Chelmsford, MA). An electrochemical filter (Coulochem III; ESA) detected 8-oxoGuo and 8-oxodGuo, while guanosine (RNA) and deoxyguanosine (DNA) were measured with SpectraSYSTEM UV1000 detector (Thermo Electron, San Jose, CA) set at 290 nm. For some samples from the in vivo sulfide exposures, RNA and DNA yield were not sufficient for analysis. This resulted in lower sample sizes for certain sulfide concentrations, as noted in the figure legends.
Statistical Analyses
The design for the in vitro oxidative stress experiment was fully balanced, with DCF and DHE each being assayed in samples from each worm at control conditions and at each H2S concentration. The data were analyzed by a split-plot, one-way ANOVA, with each worm serving as the blocking variable. This was followed by Dunnett’s post hoc comparison against the control. Fluorescence data in the figures are presented as the intensity of fluorescence relative to the control fluorescence for each worm, but all statistics were run on the raw fluorescence data.
The design for the in vitro RNA and DNA oxidation experiment was fully balanced, with RNA and DNA oxidation being assayed in samples from each worm at control conditions and each H2S concentration. Since the oxidation data failed the assumption of homogeneity of variance (Levene’s test P < 0.05), they were first log transformed before being analyzed by a split-plot, one-way ANOVA, with each worm serving as the blocking variable, as above. This was followed by Dunnett’s post hoc comparison against the control. In the in vivo studies, each worm was exposed to one of eight H2S concentrations (i.e., there was no within-subject replication). For the statistical analysis, the relatively small sample size did not provide sufficient power to perform ANOVA, which would have required a Bonferroni correction. Therefore, sulfide concentration was treated as a continuous variable, and the data were analyzed by the nonparametric Spearman rank correlation and simple linear regression.
Results
In Vitro Sulfide Exposure Increases Endogenous ROSS Production
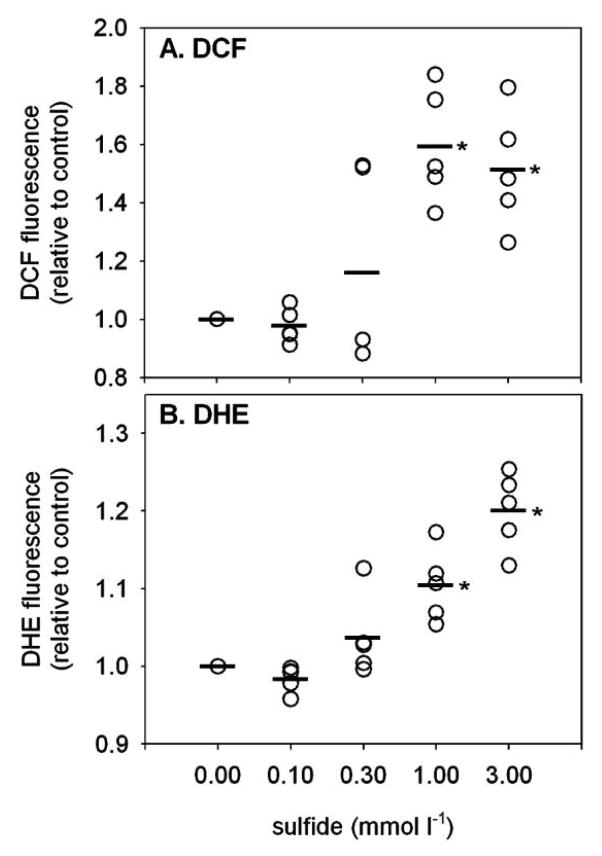
Oxidative stress was increased in coelomocytes exposed to sulfide for 1 h, as indicated by increased DCF fluorescence (Fig. 1A; sulfide: F4 = 48, P < 0.0001; worm: F4 = 3.9, P = 0.022). Coelomocytes exposed to 0.73 and 1.2 mmol L−1 sulfide had significantly higher DCF fluorescence than the coelomocytes exposed to air (P ≤ 0.0001 for each), whereas exposure to 0.29 and 0.5 mmol L−1 total sulfide had no significant effect (P = 0.18 and 0.78, respectively). Similarly, sulfide exposure caused increased superoxide production, as indicated by increased DHE fluorescence (Fig. 1B; sulfide: F4 = 19.4, P < 0.0001; worm: F4 = 2.9, P = 0.056). As with DCF, coelomocytes exposed to 0.73 and 1.2 mmol L−1 sulfide had significantly higher fluorescence than the coelomocytes exposed to air (P ≤ 0.0002 for each), whereas exposure to 0.29 and 0.50 mmol L−1 sulfide had no significant effect (P = 0.39 and 0.99, respectively).
Figure 1.
Fluorescence intensity of 2′,7′-dichlorofluorescein (DCF; A) and dihydroethidine (DHE; B) in Glycera dibranchiata coelomocytes exposed to sulfide in vitro. Fluorescence is presented relative to the mean of the control samples (0 sulfide) for each worm, with circles representing individual data and horizontal lines representing the means for each treatment (coelomocytes from five worms). Asterisks indicate significant difference in the mean compared to the control.
In Vitro Sulfide Exposure Causes Oxidative Damage to Nucleic Acids
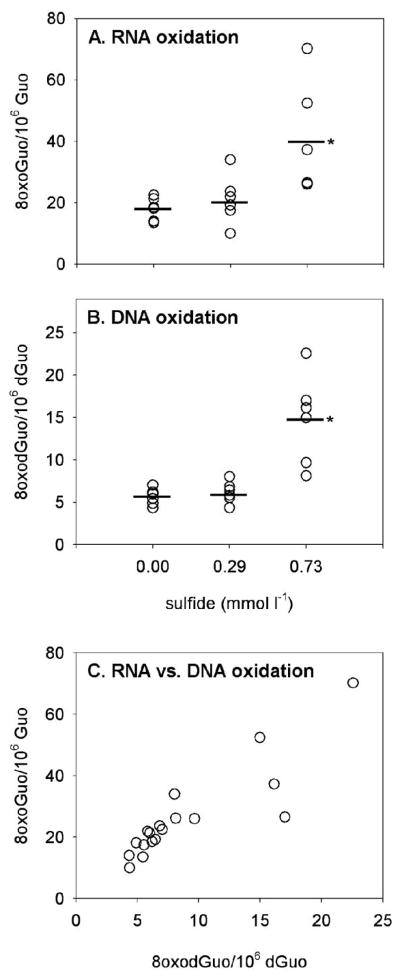
Oxidative damage to RNA and DNA was increased in coelomocytes exposed to sulfide in vitro for 1 h. The concentration of 8-oxoGuo, which indicates oxidative damage to RNA guanine nucleosides, increased significantly with sulfide exposure (Fig. 2A; sulfide: F2 = 16.9, P = 0.0006; worm: F5 = 4.9, P = 0.015), with the most oxidation detected in the high sulfide treatment (P = 0.0006). Similarly, the concentration of 8-oxodGuo, which indicates oxidative damage to DNA, increased significantly with sulfide exposure (Fig. 2B; sulfide: F2 = 31.67, P < 0.0001; worm: F5 = 2.51, P = 0.101), with the most oxidation detected in the high sulfide treatment (P < 0.0001). RNA and DNA oxidation were positively correlated within each sample (r = 0.878, t16 = 7.35, P < 0.0001), and in all cases, oxidative damage to RNA was greater than that to DNA.
Figure 2.
A, B, Oxidative damage to RNA nucleosides (8-oxoGuo, A) and DNA nucleosides (8-oxodGuo, B) in Glycera dibranchiata coelomocytes exposed to sulfide in vitro. Concentrations of oxidized nucleosides are presented per 106 undamaged nucleosides, with circles representing individual data and horizontal lines representing the means for each treatment (coelomocytes from six worms). Asterisks indicate significant differences versus control (0 sulfide). C, Ratio of oxidatively damaged nucleosides from RNA and DNA. 8-oxoGuo = 8-oxo-7,8-dihydroguanosine; 8-oxodGuo = 8-oxo-7,8-dihydro-2′-deoxyguanosine; Guo = guanosine; dGuo = deoxyguanosine.
In Vivo Sulfide Exposure Increases Oxidative Damage to Nucleic Acids
The flow-through apparatus for in vivo sulfide exposure reliably maintained a stable, predictable sulfide concentration (Fig. 3). Although sulfide was undetectable in the outflow water of chambers with the lowest sulfide target concentration of 0.001 mmol L−1, this was almost certainly due to the limited sensitivity of the sulfide assay rather than a failure of the flow-through system. Therefore, data in figures 4 and 5 are presented with the target sulfide concentration rather than the measured sulfide concentration.
Figure 3.
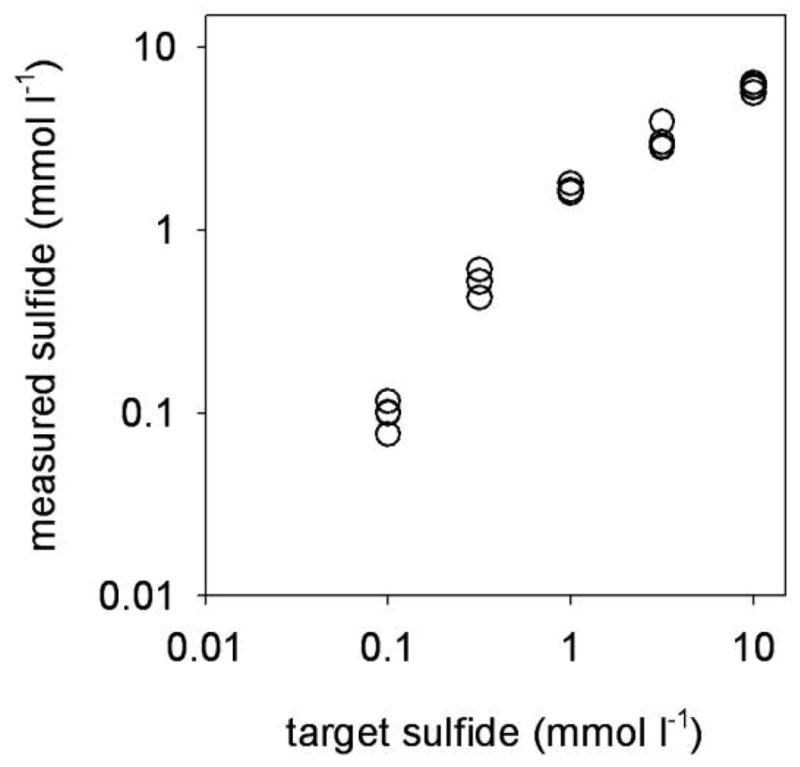
Measured sulfide concentration versus target sulfide concentrations in the flow-through system used for in vivo exposures.
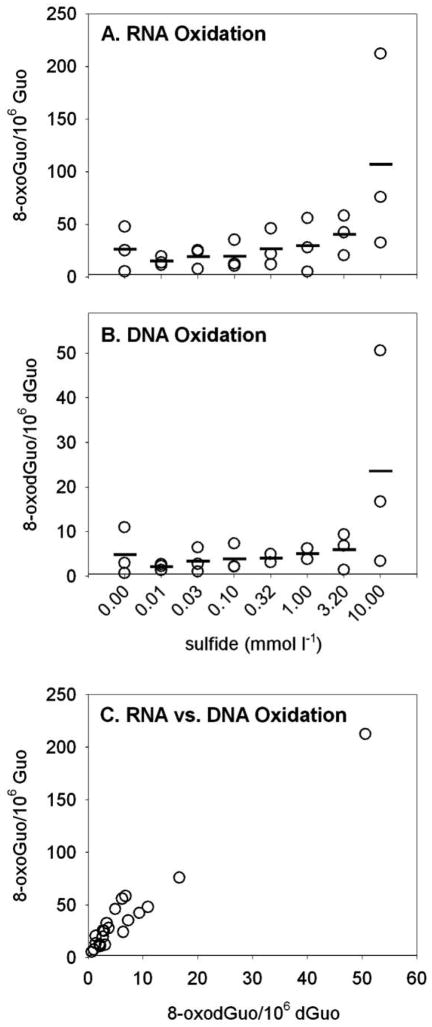
Figure 4.
A, B, Oxidative damage to RNA nucleosides (8-oxoGuo, A) and DNA nucleosides (8-oxodGuo, B) in coelomocytes from Glycera dibranchiata exposed to sulfide in vivo. Concentrations of oxidized nucleosides are presented per 106 undamaged nucleosides, with circles representing individual data and horizontal lines representing the means for each treatment (two or three worms per treatment for DNA, three worms per treatment for RNA). C, Ratio of oxidatively damaged nucleosides from RNA and DNA. Note that although the data in A and B were analyzed by linear regression, the abscissas in the plots are log scaled. 8-oxoGuo = 8-oxo-7,8-dihydroguanosine; 8-oxodGuo = 8-oxo-7,8-dihydro-2′-deoxyguanosine; Guo = guanosine; dGuo = deoxyguanosine.
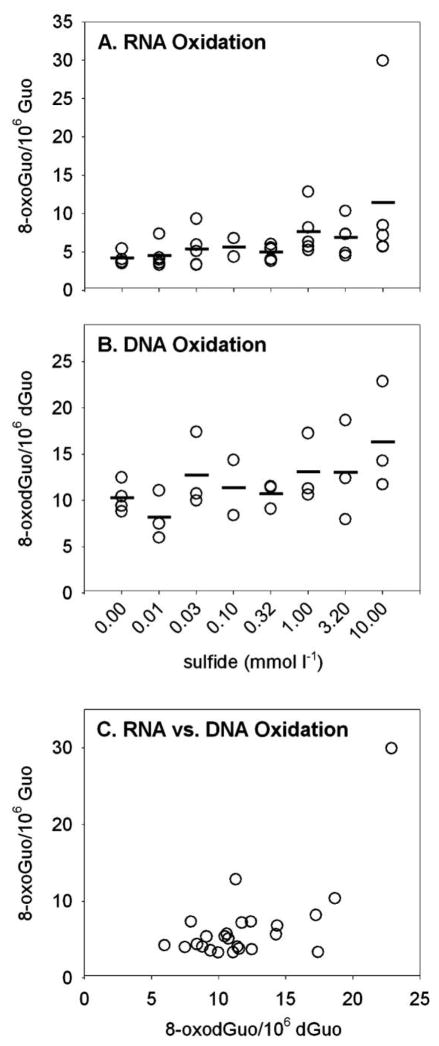
Figure 5.
A, B, Oxidative damage to RNA nucleosides (8-oxoGuo, A) and DNA nucleosides (8-oxodGuo, B) in body wall of Glycera dibranchiata coelomocytes exposed to sulfide in vivo. Concentrations of oxidized nucleosides are presented per 106 undamaged nucleosides, with circles representing individual data and horizontal lines representing the means for each treatment (three worms per treatment for RNA, two or three worms per treatment for DNA). Note that although the data in A and B were analyzed by linear regression, the abscissas in the plots are log scaled. C, Ratio of oxidatively damaged nucleosides from RNA and DNA. 8-oxoGuo = 8-oxo-7,8-dihydroguanosine; 8-oxodGuo = 8-oxo-7,8-dihydro-2′-deoxyguanosine; Guo = guanosine; dGuo = deoxyguanosine.
Of the five worms exposed to the highest sulfide concentration (10 mmol L−1), one worm exhibited a particularly strong oxidative damage response. The data from this worm were assumed to be valid because (1) the extent of RNA oxidation was correlated with that of DNA oxidation, and (2) this worm showed the largest change in body wall color, from pink to gray-green, consistent with high sulfide toxicity (Hance et al. 2008). Therefore, data from this “high-responding” worm were not excluded. Nonetheless, to be maximally conservative, all analyses that included data from this worm were performed with nonparametric statistics.
Worms exposed to sulfide in vivo for 24 h had significant increases in RNA and DNA oxidation in their coelomocytes, as indicated by a positive correlation of target sulfide concentration with both 8-oxoGuo (Fig. 4A; rs = 0.564, P < 0.05) and 8-oxodGuo (Fig. 4B; rs = 0.579, P < 0.05). When these trends were analyzed by simple linear regression without the data point from the high-responding worm, the positive relationships between sulfide and RNA and DNA oxidation remained significant (RNA: r = 0.661, F1, 20 = 11.22, P = 0.0032; DNA: r = 0.632, F1, 17 = 7.51, P = 0.014). RNA oxidation was significantly correlated with DNA oxidation (Fig. 4C; rs = 0.89, P < 0.05).
Sulfide exposure in vivo also caused elevated oxidative damage in the body wall of the worms, as indicated by a positive correlation of target sulfide concentration with both 8-oxoGuo (Fig. 5A; rs = 0.696, P < 0.05) and 8-oxodGuo (Fig. 5B; rs = 0.625, P < 0.05). When these trends were analyzed by simple linear regression without the data point from the high-responding worm, the positive relationships between sulfide and RNA and DNA oxidation remained significant (RNA: r = 0.569, F1, 21 = 10.08, P = 0.0046; DNA: r = 0.421, F1, 21 = 4.54, P = 0.045). In contrast to the pattern found in coelomocytes exposed to sulfide in vitro and in vivo, RNA and DNA oxidation were not correlated in body wall tissue (Fig. 5C; rs = 0.351, P > 0.05). There was a significant positive correlation between tissues in the extent of oxidative damage to RNA (8-oxoGuo in coelomocytes vs. 8-oxoGuo in body wall: rs = 0.86, P < 0.05; data not shown) and DNA (8-oxodGuo in coelomocytes vs. 8-oxodGuo in body wall: rs = 0.67, P < 0.05; data not shown).
Discussion
Extensive investigations of sulfide have established that it can act as a toxin and as a signaling molecule, depending on its concentration, the organism or tissue being investigated, and even the context in which it is being studied. Sulfide exerts multiple toxic effects across a wide range of concentrations relevant to sulfidic environments (for a review, see Somero et al. 1989; Bagarinao 1992; Grieshaber and Völkel 1998) and thus influences aquatic-community structure and dynamics (Gamenick et al. 1996; Levesque et al. 2006; Levin et al. 2006). However, at low concentrations sulfide may serve a physiological role as a gasotransmitter (Maclean and Kraus 2004; Kimura et al. 2005) and even a cytoprotective agent (for a review, see Szabó 2007). To investigate whether sulfide has the potential for lasting, sublethal toxicity in a sulfide-tolerant organism, we tested the hypothesis that a broad range of sulfide concentrations induces evidence of dose-dependent, oxygen-centered free radical production and causes oxidative damage to RNA and DNA in the sulfide-tolerant polychaete Glycera dibranchiata. We first determined that in vitro sulfide exposure increased oxidative stress and oxidative damage to nucleic acids in G. dibranchiata coelomocytes. We then determined that in vivo sulfide exposure also increased oxidative damage to nucleic acids in coelomocytes and body wall tissue.
Sulfide Induces Cellular Oxidative Stress
Several studies have documented a link between sulfide and ROSS production. Sulfide oxidizes spontaneously in the presence of divalent metals, generating oxygen-centered and sulfur-centered radicals in aqueous solutions (Chen and Morris 1972; Tapley et al. 1999) in the tissues of sulfide-tolerant marine invertebrates (Tapley 1993; Abele-Oeschger and Oeschger 1995; Julian et al. 2005a) and in rat hepatocytes (Eghbal et al. 2004). We confirmed this by finding that sulfide exposure causes dose-dependent increases in oxidative stress (DCF fluorescence) and superoxide production (DHE fluorescence) in coelomocytes in vitro. These results suggest, but do not prove, that sulfide-tolerant invertebrates can experience cellular oxidative stress when exposed to concentrations of sulfide that can occur in mudflats during low tide (Fenchel and Riedl 1970).
Sulfide Induces Oxidative Damage
To determine whether sulfide causes cellular oxidative stress, most investigators measure the cellular oxidative stress response, a suite of protective mechanisms that minimize or repair oxidative damage. In particular, the upregulated expression of stress proteins, including antioxidants (superoxide dismutase, catalase, and glutathione peroxidase), have received much attention. However, studies of antioxidant activities or expression patterns in sulfide-exposed invertebrates (Morrill et al. 1988; Tapley 1993; Abele-Oeschger 1996; Abele et al. 1998; Joyner-Matos et al. 2006) and mammals (Kimura et al. 2006; Truong et al. 2006) have provided inconsistent results. The lack of consensus is perhaps not surprising, given limitations inherent in inferring stress based solely on the interpretation of stress protein expression or activity data (Bierkens 2000).
A more direct approach to determining whether sulfide causes cellular oxidative stress is to measure markers of oxidative damage. To our knowledge, the only documentation of sulfide-induced oxidative damage to DNA has been from mammalian cells in vitro. In naked nuclei and whole Chinese hamster ovary cells, low levels of sodium sulfide (<25 μmol L−1) caused genomic DNA damage in the form of strand breaks (Attene-Ramos et al. 2006, 2007) and imidazole ring opening (Attene-Ramos et al. 2007). Similarly, Baskar et al. (2007) showed that lung fibroblasts exposed to up to 75 μmol L−1 sulfide acquired DNA damage, as indicated by the formation of micronuclei. Although these results are consistent with oxidative damage, DNA strand breaks also can result from ionizing radiation (Khanna and Jackson 2001), and micronucleus formation can have multiple causes, including strand breaks (Mateuca et al. 2006).
To document oxidative damage from in vitro and in vivo sulfide exposure, we selected a marker that results only from free radical damage. Although all components of DNA are vulnerable to ROSS attack (Halliwell and Gutteridge 1999), the oxidation of guanine is the best studied, in part because 8-oxodGuo is an easily identifiable intermediate and its formation has, at least thus far, been attributed only to oxidation by reactive oxygen species (Shigenaga et al. 1989; Cooke and Evans 2007). Deoxyguanosine oxidation results from a variety of oxidative stressors (Kim et al. 2001; Risom et al. 2003) and increases significantly with aging (for a review, see Sanz et al. 2006). Production of 8-oxodGuo tends to be higher in mitochondrial DNA than in nuclear DNA, since mitochondria are the site of much of a cell’s free radical production (Sanz et al. 2006). Oxidation of deoxyguanosine into 8-oxodGuo is the predominant form of oxidative damage that is linked to mutation. The oxidized base, if not removed by repair enzymes, readily binds adenine, causing GC-TA transversions and/or GC-AT transitions (Tkeshelashvili et al. 1991; Cheng et al. 1992; Maki and Sekiguchi 1992).
We found that steady state levels of oxidative damage to RNA and DNA increased in coelomocytes exposed to sulfide in vitro and in coelomocytes and body wall exposed to sulfide in vivo. Coelomocytes exposed to sulfide in vitro had slightly (but not significantly) higher oxidation levels than those exposed to equivalent sulfide concentrations in vivo. This may have resulted from differences in the isolated cell fractions, which for the in vitro exposures were purified heme-rich erythrocytes, while in vivo exposures contained a mixed cell population of erythrocytes and white cells (collectively called coelomocytes). Although in vitro exposure to 0.73 mmol L−1 sulfide increased oxidative damage to RNA and DNA in coelomocytes, we did not detect a significant increase in oxidative damage at 0.29 mmol L−1 sulfide, which is an order of magnitude greater than the concentration of sulfide that induces DNA damage in mammalian cells (Attene-Ramos et al. 2006, 2007; Baskar et al. 2007). Furthermore, coelomocytes exposed to sulfide in vitro showed a significant increase in cellular oxidative stress and superoxide production at 0.29 mmol L−1 sulfide. These data suggest that although G. dibranchiata coelomocytes (including erythrocytes) experienced oxidative stress at the lower sulfide concentration, the cells were capable of minimizing or repairing oxidative damage to RNA and DNA. Whether this represents an adaptation to sulfide exposure requires further study.
The sulfide concentrations that caused oxidative damage to coelomocytes in vivo were an order of magnitude higher than the concentrations that caused oxidative damage in vitro, suggesting that aspects of whole-animal physiology reduce sulfide toxicity. Typical sulfide concentrations experienced by G. dibranchiata are unknown, but in this study and a previous one (Hance et al. 2008), the worms tolerated exposure to millimolar levels of sulfide in vivo for at least 24 h. A wide range of protective strategies have been documented in sulfide-tolerant invertebrates, some of which can maintain the internal sulfide concentration below the ambient sulfide concentration (Oeschger and Vetter 1992; Grieshaber and Völkel 1998). Whether these occur in G. dibranchiata is unknown. Nonetheless, the results of this study suggest that markers of oxidative stress and damage may have utility for examining the relationship between sulfide exposure and cellular injury in natural populations.
In the only measurement of 8-oxoGuo in a wild population of animals to date, there was a positive correlation between environmental dissolved O2 concentration and RNA oxidation in a hypoxia-tolerant bivalve (Joyner-Matos et al. 2007). RNA oxidation has otherwise not been extensively studied outside of its key role in Alzheimer’s disease (e.g., Nunomura et al. 1999) and Parkinson’s disease (e.g., Zhang et al. 1999) and its tendency to increase with age in mammals (Hofer et al. 2005, 2006). We found that steady state levels of RNA oxidation in G. dibranchiata increased significantly with in vitro and in vivo sulfide exposure. In all cases, RNA oxidation was greater than DNA oxidation, which is consistent with the findings of previous studies (Hofer et al. 2005, 2006; Joyner-Matos et al. 2007). Interestingly, we also found that RNA oxidation was correlated with DNA oxidation in coelomocytes but not in the body wall tissue. Although a correlation between RNA oxidation and DNA oxidation is not unexpected, it had not been reported in any system. The dynamics of RNA oxidative damage and repair processes are much less studied than those of DNA and likely differ among the three RNA pools (Evans and Cooke 2004). Nonetheless, our results suggest that both processes respond similarly to sulfide exposure.
Understanding the toxic effects of sulfide on natural populations of aquatic animals has been an active area of research for more than 3 decades. Much of this work has focused on biochemical processes of sulfide toxicity and detoxification strategies. Our results, in combination with those showing that micromolar levels of sulfide cause strand breaks and purine ring opening in mammalian cells (Attene-Ramos et al. 2006, 2007; Baskar et al. 2007), strongly suggest that sulfide exposure could be mutagenic, even for sulfide-tolerant organisms. Genetic variation generally is higher in stressful environments (Hoffmann and Parsons 1991), and extensive studies on microorganisms have shown that stress-induced increases in mutation rate can be adaptive (for a review, see Galhardo et al. 2007). Whether animals in sulfidic environments experience elevated mutation rates is not known, but sulfide-induced oxidative damage to DNA is one mechanism by which mutation rates could be elevated in sulfidic environments. This may be especially important in G. dibranchiata, since reproductively mature worms store gametes in the coelomic cavity (Simpson 1962), where the cells would clearly be at risk of sulfide exposure, and therefore may be susceptible to mutagenic effects of sulfide-induced DNA damage.
Acknowledgments
We thank Jocelyn Ortega and Jessica Ortega for performing the H2DCF-DA and DHE assays, Bill Rossignol for help with the in vitro sulfide exposures, and Dr. Tim Hofer for help with the RNA and DNA oxidation assays, which we performed in the Genomics, Metabolism, and Biomarkers Core of the University of Florida’s Claude D. Pepper Older Americans Independence Center (supported by NIH grant 1 P30 AG028740 to C.L.). This research was supported by NSF grant IBN-0422139 to D.J.
Literature Cited
- Abe K, Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abele D, Groβpietsch H, Pörtner HO. Temporal fluctuations and spatial gradients of environmental PO2, temperature, H2O2 and H2S in its intertidal habitat trigger enzymatic antioxidant protection in the capitellid worm Heteromastus filiformis. Mar Ecol Prog Ser. 1998;163:179–191. [Google Scholar]
- Abele-Oeschger D. A comparative study of superoxide dismutase activity in marine benthic invertebrates with respect to environmental sulphide exposure. J Exp Mar Biol Ecol. 1996;197:39–49. [Google Scholar]
- Abele-Oeschger D, Oeschger R. Hypoxia-induced autoxidation of haemoglobin in the benthic invertebrates Arenicola marina (Polychaeta) and Astarte borealis (Bivalvia) and the possible effects of sulphide. J Exp Mar Biol Ecol. 1995;187:63–80. [Google Scholar]
- Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- Bagarinao T. Sulfide as an environmental factor and toxicant: tolerance and adaptations in aquatic organisms. Aquat Toxicol. 1992;24:21–62. [Google Scholar]
- Baskar R, Li L, Moore PK. Hydrogen sulfide induces DNA damage and changes in apoptotic gene expression in human lung fibroblast cells. FASEB J. 2007;21:247–255. doi: 10.1096/fj.06-6255com. [DOI] [PubMed] [Google Scholar]
- Bierkens JGEA. Applications and pitfalls of stress-proteins in biomonitoring. Toxicology. 2000;153:61–72. doi: 10.1016/s0300-483x(00)00304-8. [DOI] [PubMed] [Google Scholar]
- Brand GL, Horak RV, Le Bris N, Goffredi SK, Carney SL, Govenar B, Yancey PH. Hypotaurine and thiotaurine as indicators of sulfide exposure in bivalves and vestimentiferans from hydrothermal vents and cold seeps. Mar Ecol. 2007;28:208–218. [Google Scholar]
- Carrico RJ, Blumberg WE, Peisach J. The reversible binding of oxygen to sulfhemoglobin. J Biol Chem. 1978;253:7212–7215. [PubMed] [Google Scholar]
- Cavanaugh CM. Symbiotic chemoautotrophic bacteria in marine invertebrates from sulphide-rich habitats. Nature. 1983;302:58–61. [Google Scholar]
- Chen KY, Morris JC. Oxidation of sulfide by O2: catalysis and inhibition. J Sanit Eng Div Proc Am Soc Civ Eng. 1972;98:215–227. [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Childress JJ, Lowell W. The abundance of a sulfide-oxidizing symbiosis (the clam Solemya reidi) in relation to interstitial water chemistry. EOS: Trans Am Geol Union. 1982;63:957. [Google Scholar]
- Cline JD. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- Cooke MS, Evans MD. 8-oxo-deoxyguanosine: reduce, reuse, recycle? Proc Natl Acad Sci USA. 2007;104:13535–13536. doi: 10.1073/pnas.0706878104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis W, Reed L. The action of blood on sulfides. J Biol Chem. 1927;72:385–394. [Google Scholar]
- Eghbal MA, Pennefather PS, O’Brien PJ. H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochondrial depolarisation. Toxicology. 2004;203:69–76. doi: 10.1016/j.tox.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Elrod JW, Calvert JW, Chow CW, Morrison J, Doeller JE, Kraus DW, Kimura H, Lefer DJ. Cardiomyocyte overexpression of the hydrogen sulfide producing enzyme cystathioine gamma-lyase attenuates myocardial ischemia-reperfusion injury. Abstract 843. Circulation. 2007;116(suppl II):164. [Google Scholar]
- Evans MD, Cooke MS. Factors contributing to the outcome of oxidative damage to nucleic acids. Bioessays. 2004;26:533–542. doi: 10.1002/bies.20027. [DOI] [PubMed] [Google Scholar]
- Fenchel TM, Riedl RJ. The sulfide system: a new biotic community underneath the oxidized layer of marine sand bottoms. Mar Biol. 1970;7:255–268. [Google Scholar]
- Gainey LF, Jr, Greenberg MJ. Hydrogen sulfide is synthesized in the gills of the clam Mercenaria mercenaria and acts seasonally to modulate branchial muscle contraction. Biol Bull. 2005;209:11–20. doi: 10.2307/3593138. [DOI] [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamenick I, Jahn A, Vopel K, Giere O. Hypoxia and sulphide as structuring factors in a macrozoobenthic community on the Baltic Sea shore: colonisation studies and tolerance experiments. Mar Ecol Prog Ser. 1996;144:73–85. [Google Scholar]
- Goodwin LR, Francom D, Dieken FP, Taylor JD, Warenycia MW, Reiffenstein RJ, Dowling G. Determination of sulfide in brain tissue by gas dialysis/ion chromatography: postmortem studies and two case reports. J Anal Toxicol. 1989;13:105–109. doi: 10.1093/jat/13.2.105. [DOI] [PubMed] [Google Scholar]
- Grieshaber MK, Völkel S. Animal adaptations for tolerance and exploitation of poisonous sulfide. Annu Rev Physiol. 1998;60:33–53. doi: 10.1146/annurev.physiol.60.1.33. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford University Press; New York: 1999. [Google Scholar]
- Hance JM, Andrzejewski JE, Predmore BL, Dunlap KJ, Misiak KL, Julian D. Cytoxicity from sulfide exposure in a sulfide-tolerant marine invertebrate. J Exp Mar Biol Ecol. 2008;359:102–109. [Google Scholar]
- Hofer T, Badouard C, Bajak E, Ravanat J-L, Mattsson Å, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- Hofer T, Seo AY, Prudencio M, Leeuwenburgh C. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biol Chem. 2006;387:103–111. doi: 10.1515/BC.2006.014. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Parsons PA. Evolutionary Genetics and Environmental Stress. Oxford University Press; New York: 1991. [Google Scholar]
- Joyner JL, Peyer SM, Lee RW. Possible roles of sulfur-containing amino acids in a chemoautotrophic bacterium-mollusc symbiosis. Biol Bull. 2003;205:331–338. doi: 10.2307/1543296. [DOI] [PubMed] [Google Scholar]
- Joyner-Matos J, Chapman LJ, Downs CA, Hofer T, Leeuwenburgh C, Julian D. Stress response of a freshwater clam along an abiotic gradient: too much oxygen may limit distribution. Funct Ecol. 2007;21:344–355. [Google Scholar]
- Joyner-Matos J, Downs CA, Julian D. Increased expression of stress proteins in the surf clam Donax variabilis following hydrogen sulfide exposure. Comp Biochem Physiol A. 2006;145:245–257. doi: 10.1016/j.cbpa.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Julian D, April KL, Patel S, Stein JR, Wohlgemuth SE. Mitochondrial depolarization following hydrogen sulfide exposure in erythrocytes from a sulfide-tolerant marine invertebrate. J Exp Biol. 2005a;208:4109–4122. doi: 10.1242/jeb.01867. [DOI] [PubMed] [Google Scholar]
- Julian D, Arp AJ. Sulfide permeability in the marine invertebrate Urechis caupo. J Comp Physiol. 1992;162B:59–67. [Google Scholar]
- Julian D, Statile J, Roepke TA, Arp AJ. Sodium nitroprusside potentiates hydrogen-sulfide-induced contractions in body wall muscle from a marine worm. Biol Bull. 2005b;209:6–10. doi: 10.2307/3593137. [DOI] [PubMed] [Google Scholar]
- Julian D, Statile JL, Wohlgemuth SE, Arp AJ. Enzymatic hydrogen sulfide production in marine invertebrate tissues. Comp Biochem Physiol A. 2002;133:105–115. doi: 10.1016/s1095-6433(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Kim H-N, Morimoto Y, Tsuda T, Ootsuyama Y, Hirohashi M, Hirano T, Tanaka I, Lim Y, Yun I-G, Kasai H. Changes in DNA 8-hydroxyguanine levels, 8-hydroxyguanine repair activity, and hOGG1 and hMTH1 mRNA expression in human lung alveolar epithelial cells induced by crocidolite asbestos. Carcinogenesis. 2001;22:265–269. doi: 10.1093/carcin/22.2.265. [DOI] [PubMed] [Google Scholar]
- Kimura H. Hydrogen sulfide as a neuromodulator. Mol Neurbiol. 2002;26:13–19. doi: 10.1385/MN:26:1:013. [DOI] [PubMed] [Google Scholar]
- Kimura H, Nagai Y, Umemura K, Kimura Y. Physiological roles of hydrogen sulfide: synaptic modulation, neuroprotection, and smooth muscle relaxation. Antioxid Redox Signal. 2005;7:795–803. doi: 10.1089/ars.2005.7.795. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Dargusch R, Schubert D, Kimura H. Hydrogen sulfide protects HT22 neuronal cells from oxidative stress. Antioxid Redox Signal. 2006;8:661–670. doi: 10.1089/ars.2006.8.661. [DOI] [PubMed] [Google Scholar]
- Kraus DW, Doeller JE, Powell CS. Sulfide may directly modify cytoplasmic hemoglobin deoxygenation in Solemya reidi gills. J Exp Biol. 1996;199:1343–1352. doi: 10.1242/jeb.199.6.1343. [DOI] [PubMed] [Google Scholar]
- Levesque C, Juniper SK, Limén H. Spatial organization of food webs along habitat gradients at deep-sea hydrothermal vents on Axial Volcano, northeast Pacific. Deep-Sea Res Part I Oceanogr Res Pap. 2006;53:726–739. [Google Scholar]
- Levin LA, Ziebis W, Mendoza GF, Growney-Cannon V, Walther S. Recruitment response of methane-seep macrofauna to sulfide-rich sediments: an in situ experiment. J Exp Mar Biol Ecol. 2006;330:132–150. [Google Scholar]
- Lovatt Evans C. The toxicity of hydrogen sulphide and other sulphides. Q J Exp Physiol. 1967;52:231–248. doi: 10.1113/expphysiol.1967.sp001909. [DOI] [PubMed] [Google Scholar]
- Maclean KN, Kraus JP. Hydrogen sulfide production and metabolism in mammalian tissues. In: Wang R, editor. Signal Transduction and the Gasotransmitters: NO, CO, and H2S in Biology and Medicine. Humana; Totowa, NJ: 2004. pp. 275–292. [Google Scholar]
- Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Mateuca R, Lombaert N, Aka PV, Decordier I, Kirsch-Volders M. Chromosomal changes: induction, detection methods and applicability in human biomonitoring. Biochimie. 2006;88:1515–1531. doi: 10.1016/j.biochi.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Morrill AC, Powell EN, Bidigare RR, Shick JM. Adaptations to life in the sulfide system: a comparison of oxygen detoxifying enzymes in thiobiotic and oxybiotic meiofauna (and freshwater planarians) J Comp Physiol B. 1988;158:335–344. [Google Scholar]
- Nicholls P. The effect of sulphide on cytochrome aa3: isoteric and allosteric shifts of the reduced alpha-peak. Biochim Biophys Acta. 1975;396:24–35. doi: 10.1016/0005-2728(75)90186-3. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Perry G, Pappolla MA, Wade R, Hirai K, Chiba S, Smith MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer’s disease. J Neurosci. 1999;19:1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeschger R, Vetter RD. Sulfide detoxification and tolerance in Halicryptus spinulosus (Priapulida): a multiple strategy. Mar Ecol Prog Ser. 1992;86:167–179. [Google Scholar]
- Risom L, Dybdahl M, Bornholdt J, Vogel U, Wallin H, Møller P, Loft S. Oxidative DNA damage and defence gene expression in the mouse lung after short-term exposure to diesel exhaust particles by inhalation. Carcinogenesis. 2003;24:1847–1852. doi: 10.1093/carcin/bgg144. [DOI] [PubMed] [Google Scholar]
- Sanz A, Pamplona R, Barja G. Is the mitochondrial free radical theory of aging intact? Antioxid Redox Signal. 2006;8:582–599. doi: 10.1089/ars.2006.8.582. [DOI] [PubMed] [Google Scholar]
- Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2′-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci USA. 1989;86:9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M. Reproduction of the polychaete Glycera dibranchiata at Solomons, Maryland. Biol Bull. 1962;123:396–411. [Google Scholar]
- Somero GN, Childress JJ, Anderson AE. Transport, metabolism, and detoxification of hydrogen sulfide in animals from sulfide-rich marine environments. Crit Rev Aquat Sci. 1989;1:591–614. [Google Scholar]
- Szabó C. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 2007;6:917–935. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- Tapley DW. PhD diss. University of Maine; Orono: 1993. Sulfide-dependent oxidative stress in marine invertebrates, especially thiotrophic symbioses. [Google Scholar]
- Tapley DW, Beuttner GR, Shick JM. Free radicals and chemiluminescence as products of the spontaneous oxidation of sulfide in seawater, and their biological implications. Biol Bull. 1999;196:52–56. doi: 10.2307/1543166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkeshelashvili LK, McBride T, Spence K, Loeb LA. Mutation spectrum of copper-induced DNA damage. J Biol Chem. 1991;266:6401–6406. [PubMed] [Google Scholar]
- Truong DH, Eghbal MA, Hindmarsh W, Roth SH, O’Brien PJ. Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab Rev. 2006;38:733–744. doi: 10.1080/03602530600959607. [DOI] [PubMed] [Google Scholar]
- Urcuyo IA, Massoth GJ, Julian D, Fisher CR. Habitat, growth and physiological ecology of a basaltic community of Ridgeia piscesae from the Juan de Fuca Ridge. Deep-Sea Res Part I Oceanogr Res Pap. 2003;50:763–780. [Google Scholar]
- Whitfield NL, Kreimier EL, Verdial FC, Skovgaard N, Olson KR. Reappraisal of H2S/sulfide concentration in vertebrate blood and its potential significance in ischemic preconditioning and vascular signaling. Am J Physiol. 2008;294:R1930–R1937. doi: 10.1152/ajpregu.00025.2008. [DOI] [PubMed] [Google Scholar]
- Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Zhang J, Lu Y, Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. EMBO (Eur Mol Biol Organ) J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]