ABSTRACT
Type I signal peptidase (SPase) is essential for viability in wild-type bacteria because the terminal step of the bacterial general secretory pathway requires its proteolytic activity to release proteins from their membrane-bound N-terminal leader sequences after translocation across the cytoplasmic membrane. Here, we identify the Staphylococcus aureus operon ayrRABC (SA0337 to SA0340) and show that once released from repression by AyrR, the protein products AyrABC together confer resistance to the SPase inhibitor arylomycin M131 by providing an alternate and novel method of releasing translocated proteins. Thus, the derepression of ayrRABC allows cells to bypass the essentiality of SPase. We demonstrate that AyrABC functionally complements SPase by mediating the processing of the normally secreted proteins, albeit in some cases with reduced efficiency and either without cleavage or via cleavage at a site N-terminal to the canonical SPase cleavage site. Thus, ayrRABC encodes a secretion stress-inducible alternate terminal step of the general secretory pathway.
Importance Addressing proteins for proper localization within or outside a cell in both eukaryotes and prokaryotes is often accomplished with intrinsic signals which mediate membrane translocation and which ultimately must be removed. The canonical enzyme responsible for the removal of translocation signals is bacterial type I signal peptidase (SPase), which functions at the terminal step of the general secretory pathway and is thus essential in wild-type bacteria. Here, we identify a four-gene operon in S. aureus that encodes an alternate terminal step of the general secretory pathway and thus makes SPase nonessential. The results have important implications for protein secretion in bacteria and potentially for protein trafficking in prokaryotes and eukaryotes in general.
Importance
Addressing proteins for proper localization within or outside a cell in both eukaryotes and prokaryotes is often accomplished with intrinsic signals which mediate membrane translocation and which ultimately must be removed. The canonical enzyme responsible for the removal of translocation signals is bacterial type I signal peptidase (SPase), which functions at the terminal step of the general secretory pathway and is thus essential in wild-type bacteria. Here, we identify a four-gene operon in S. aureus that encodes an alternate terminal step of the general secretory pathway and thus makes SPase nonessential. The results have important implications for protein secretion in bacteria and potentially for protein trafficking in prokaryotes and eukaryotes in general.
Observation
The proper localization of many proteins requires their translocation across one or more membranes. The general secretory (Sec) pathway, conserved throughout bacteria, is the canonical translocation pathway and is responsible for translocating the vast majority of secreted proteins across the cytoplasmic membrane. Like other general translocation pathways, Sec requires the synthesis of its cargo as preproteins with N-terminal signal peptides, which target them to the Sec machinery and from which the mature protein must be released after translocation (1, 2). The enzyme responsible for the liberation of most mature proteins translocated by Sec is type I signal peptidase (SPase) (3–5). Accordingly, SPase has been demonstrated to be essential in both Gram-positive and Gram-negative bacteria. Staphylococcus aureus is a striking example of the importance of SPase, as it mediates the secretion of a diverse range of virulence factors, including proteins required for adhesion and colonization, evasion of the host immune response, scavenging of nutrients and minerals from the environment, and dissemination (6).
The arylomycin family of natural products are potent inhibitors of SPase (7–10). As part of an effort to develop the arylomycins as therapeutics, we and others have been exploring the optimization of their spectrum of activity (11–13). An arylomycin with particularly promising activity against S. aureus is arylomycin M131 (Fig. 1A), which was disclosed by Merck in 2012 (13). We have also developed the arylomycins as chemical biology probes to assess secretion in different bacteria, including Staphylococcus epidermidis (14) and S. aureus (6). As part of these efforts, we recently demonstrated that S. aureus responds to arylomycin-mediated SPase inhibition by increasing expression of the four adjacent genes, SA0337 to SA0340, and that arylomycin resistance is conferred by loss-of-function mutations in SA0337 (15).
FIG 1 .
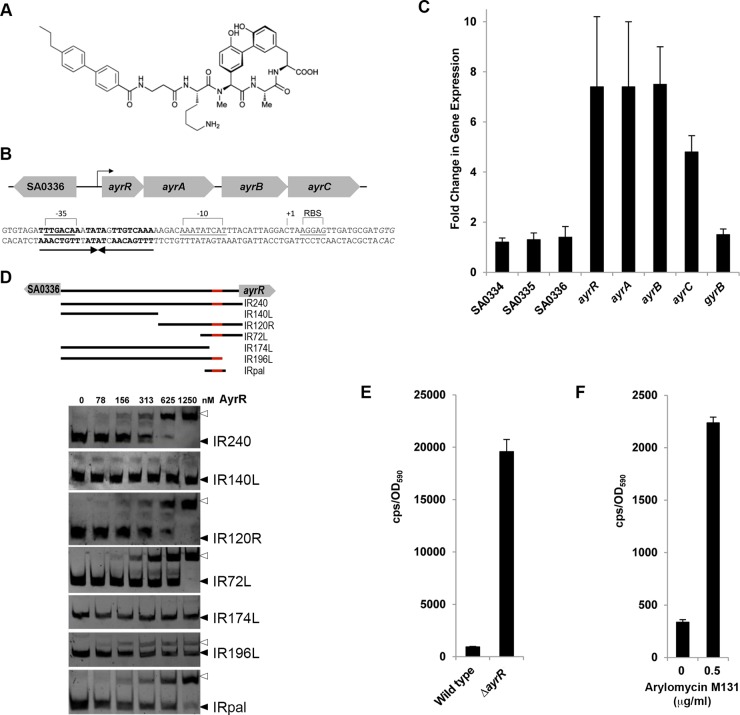
AyrR is a transcriptional repressor of itself and three downstream open reading frames. (A) Chemical structure of the SPase inhibitor arylomycin M131. (B) Genomic organization of the ayrRABC operon (ayrR, SA0337; ayrA, SA0338; ayrB, SA0339; ayrC, SA0340) and the region immediately upstream that includes SA0336. Below is the sequence of the putative promoter region upstream of ayrR ending with its GTG translational start site. A 22-nt palindromic repeat (arrows) is located 43 to 65 bp upstream of the start site. The positions of the putative −35 and −10 regions and ribosome binding site (RBS) are indicated in the coding strand. The transcriptional start site is marked as +1, corresponding to 18 nt upstream from the translational start site. (C) RT-PCR analysis of the ayrRABC operon and divergent genes (SA0334 to SA0336). Fold change in gene expression in ARC0001 with respect to N315. gyrB was used as an external control, and gene expression was normalized using gmk. See Table S1 in the supplemental material for primer sequences. (D) Gel shifts performed with 30 ng of each DNA probe and an increasing concentration of AyrR as indicated at the top. The region of DNA corresponding to each gel shift probe is shown above the gel data, with the AyrR binding site shown in red. Filled triangles denote free DNA probe, and open triangles denote the indicated DNA-AyrR complexes. All DNA probes were amplified from wild-type N315 genomic DNA with the exception of IRpal, which was created by annealing synthetic oligonucleotides. See Table S1 for primer sequences. (E) Luminescence from the AyrR promoter (expressed from pARC1) in wild-type N315 and N315∆SA0336-ayrRABC (labeled ∆ayrR), expressed as counts per second per unit of optical density at 590 nm (cps/OD590). (F) Luminescence from N315 harboring pARC1 in the presence and absence of arylomycin M131 (0.5× MIC).
Here, we demonstrate that the genes SA0337 to SA0340 constitute an operon and that SA0337 is a transcriptional repressor that controls its expression. Remarkably, derepression of these genes bypasses the need for SPase, rendering SPase nonessential and suggesting that the function of the operon is to mediate an alternate process by which translocated proteins are released from the cytoplasmic membrane. We have thus named the repressor gene ayrR and the downstream genes ayrABC for their role in arylomycin resistance. Sequence analysis identifies AyrA as a 6-transmembrane-domain protein of unknown function (DUF3169/PF11368) and AyrB and AyrC as two domains of an ABC transporter, and we demonstrate that all three are required to bypass SPase. Finally, we demonstrate that AyrABC is able to mediate secretion of the proteins normally processed by SPase, although in some cases with reduced efficiency, but it does so either with the signal peptide still attached or after cleavage at a site within the signal peptide.
ayrRABC is an operon, and AyrR is a repressor that regulates its expression.
The stop and start codons of ayrR and ayrA overlap, as do those of ayrB and ayrC, while the stop and start codons of ayrA and ayrB are separated by only 24 bp (Fig. 1B), suggesting that they are all part of a cotranscriptionally regulated operon. Using reverse transcription-PCR (RT-PCR), we examined transcription of ayrRABC in the arylomycin-resistant strain ARC0001, which harbors a nonsense mutation in ayrR (15), and its parental wild-type strain N315. As expected, no differences in transcript levels were observed for control genes (SA0334 to SA0336 and gyrB), but transcript levels of ayrR, ayrA, ayrB, and ayrC in ARC0001 were each increased ~8-fold compared to those in the wild type (Fig. 1C). Moreover, analysis of our previously published RNA-Seq data (15) reveals reads that when overlapped align continuously across the entire ayrRABC locus, including the intergenic sequences (see Fig. S1 in the supplemental material). Collectively, these results suggest that ayrRABC is an operon and demonstrate that the mutation in ayrR that renders S. aureus resistant to SPase inhibition results in operon derepression.
ayrR encodes a helix-turn-helix motif protein annotated as an XRE family transcriptional regulator with homology to the phage λ Cro repressor. Sequence analysis revealed an almost perfect 22-nucleotide (nt) palindrome, TTTGACAAATATAGTTGTCAAA, upstream of ayrR which overlaps the −35 promoter element (Fig. 1B). Such palindromes commonly compose the binding site of transcriptional regulators (16), and Cro regulates its own transcription by binding such a palindrome (17, 18). To determine if AyrR binds its upstream palindrome, a gel shift assay was employed using various DNA fragments (Fig. 1D). The data clearly demonstrate that AyrR selectively binds the palindrome.
To test the functional significance of this binding, we constructed the transcriptional reporter plasmid pARC1, which harbors the intergenic region between SA0336 and ayrR upstream of the genes encoding luciferase (luxCDABE) (19). Wild-type N315 and a strain lacking the entire region from SA0336 to ayrC (N315∆SA0336-ayrRABC) were then transformed with either the empty luxCDABE vector or pARC1, and luminescence was assayed in actively growing cultures (Fig. 1E). Only in the case of N315∆SA0336-ayrRABC transformed with pARC1 did we observe high-intensity luminescence. Luminescence was significantly lower for N315 harboring pARC1, consistent with repression by the genomically encoded AyrR protein. As an additional control, the ability of SPase inhibition to release repression was tested by conducting the same experiment in the presence of a subinhibitory concentration of arylomycin M131 (0.5× MIC). Two hours after arylomycin addition, the luminescence signal observed with wild-type N315 cells harboring pARC1 was 7-fold higher than that observed in the absence of arylomycin (Fig. 1F). These results confirm that AyrR acts as a repressor of ayrRABC and that SPase inhibition induces derepression.
Each gene of the ayrRABC operon contributes to tolerating SPase inhibition.
ayrA encodes a putative membrane protein, predicted to possess a DUF3169 domain of unknown function. Proteins in the DUF3169 family are found in both Staphylococcus and Streptococcus species, with one homolog present per genome in sequenced strains, and while they share only ~30% sequence identity, six predicted transmembrane domains and a D-E(a/g)E motif located in the loop connecting the fourth and fifth predicted transmembrane segments are highly conserved. Interestingly, each ayrA homolog identified appears to be immediately downstream of a gene that is homologous to ayrR. The downstream genes ayrB and ayrC are predicted to encode the two domains of a type 2 family ABC transporter, with ayrC encoding the transporter domain and ayrB encoding the ATP-binding cassette domain (conserved domain cd03230).
The contribution of each gene to arylomycin resistance was assessed via gene deletion. Although ayrA could be deleted in an otherwise wild-type strain (N315), we were unable to delete ayrA in ARC0001. This suggests that the derepression of ayrBC is toxic in the absence of AyrA. Thus, to test whether ayrA is required to tolerate SPase inhibition, we attempted to evolve arylomycin resistance in N315∆ayrA. As a control, we constructed and analyzed a strain lacking SA0336, which is predicted to encode a hypothetical protein and which is not part of the ayrRABC operon. High-level arylomycin M131 resistance (>16 µg/ml) was evolved in N315∆SA0336 with frequencies indistinguishable from the wild-type parental strain (2.0 × 10−8) (15) (see Table S2 in the supplemental material). In addition, sequencing revealed that 8 out of 8 of the resistant strains examined contained mutations in ayrR (see Table S3 in the supplemental material), similar to the behavior observed with wild-type cells (15). In contrast, N315∆ayrA developed high-level resistance with a significantly reduced frequency (2.9 × 10−10) (see Table S2), and for the single resistant clone isolated, sequencing revealed a wild-type ayrR.
Deletion of ayrBC in ARC0001 (ARC0001∆ayrBC) resulted in full resensitization to arylomycin M131, demonstrating that the ABC transporter is required to tolerate SPase inhibition (see Table S2 in the supplemental material). Moreover, deletion of ayrBC in N315 (N315∆ayrBC) results in the same reduced frequency of resistance to arylomycin M131 that was observed with N315∆ayrA (see Table S2), and again, sequencing revealed wild-type ayrR in the single resistant mutant isolated. Collectively, these results demonstrate that each member of the ayrRABC operon is required to tolerate SPase inhibition.
Derepression of ayrRABC bypasses the requirement for SPase.
Because derepression of ayrRABC in S. aureus confers high-level resistance to SPase inhibition, we speculated that it may render SPase nonessential. Using the same chromosomal integration approach, we attempted to delete the SPase gene spsB in ARC0001 and its parental wild-type strain, N315. Not surprisingly, attempts to delete spsB in the wild-type strain resulted in the recovery of the wild-type sequence. In contrast, the ARC0001∆spsB strain was viable. In addition to confirming the absence of spsB at its chromosomal locus via sequencing, we demonstrated that spsB was not present elsewhere in the genome of ARC0001∆spsB via PCR and RT-PCR of genomic DNA and RNA, respectively, using primers internal to the spsB gene (see Table S4 in the supplemental material).
Deletion of spsB was also verified phenotypically via an examination of the susceptibility of N315 and ARC0001∆spsB to gentamicin and the β-lactam antibiotics cefoxitin, oxacillin, and penicillin G. Previously, we and others demonstrated that the activity of these antibiotics is uniquely synergistic with that of the arylomycins (13, 20). As controls, we also examined susceptibilities to CCCP, vancomycin, daptomycin, erythromycin, trimethoprim, and tetracycline. For each control antibiotic tested, the observed susceptibilities of N315 were virtually identical to those observed for N315 and ARC0001 (15). However, the susceptibility of ARC0001∆spsB to gentamicin and each β-lactam was 8- to 64-fold greater than that of N315 or ARC0001 (see Table S5 in the supplemental material), consistent with the absence of SPase activity.
The activity of the ayrRABC complements SPase deletion.
We next evaluated protein secretion in N315, ARC0001, and ARC0001∆spsB using one-dimensional (1D) SDS-PAGE (Fig. 2). Identical patterns of secretion were observed for the wild type and ARC0001, which suggests that ARC0001 still employs SPase even though the ayrRABC operon is derepressed. In contrast, a small but clearly significant alteration in the pattern of secretion was apparent for ARC0001∆spsB. To confirm that this altered pattern of secretion was the result of the loss of SPase activity, secretion was examined in the presence of arylomycin M131 (4× MIC). As expected, wild-type cells showed drastically reduced levels of secretion in the presence of the arylomycin. However, treatment of ARC0001 cells with the arylomycin did not reduce secretion and resulted in a secretion pattern indistinguishable from that of ARC0001∆spsB, clearly revealing that the altered pattern does indeed result from the absence of SPase activity. Moreover, the pattern and quantity of secreted proteins observed with ARC0001∆spsB were unchanged by the addition of the arylomycin, confirming that secretion did not depend on SPase function.
FIG 2 .
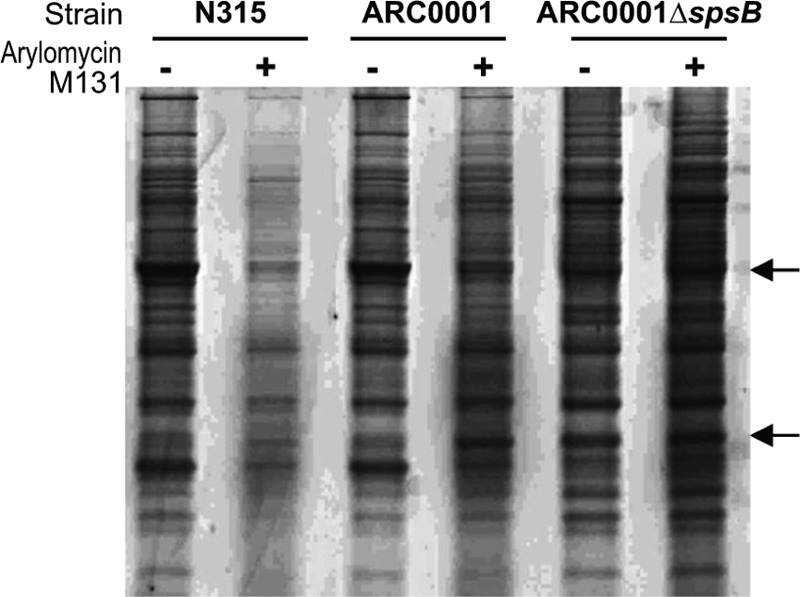
The extracellular proteomes of N315, ARC0001, and ARC0001∆spsB in the absence and presence of arylomycin M131. To minimize cell lysis, strains were grown until wild-type N315 reached an OD590 of 4.0, corresponding to late exponential growth. Strains were then balanced to an OD590 of 4.0, pelleted by centrifugation, and washed twice with medium before resuspension in fresh medium. Strains were then grown in the presence or absence of 4× MIC of arylomycin M131 (N315) for 20 min. Cells were pelleted and the supernatant was collected. Proteins in the supernatant were precipitated overnight with 10% (wt/vol) TCA, washed with acetone, resuspended in 1× loading dye, and visualized by SDS-PAGE. Arrows denote bands where significant differences are observed in the extracellular proteomes of wild-type N315 and ARC0001∆spsB.
The secreted proteomes of ARC0001∆spsB and N315 were next compared in more detail by isolating proteins from the medium, digesting with trypsin, using reductive dimethylation (ReDiMe) to label the wild-type fragments with heavy isotopes and label the ARC0001∆spsB fragments with light isotopes, and then analyzing the combined samples via multidimensional mass spectrometry (MudPIT) (21, 22). In total, tryptic peptides corresponding to 38 proteins that are encoded with signal peptides were identified (Table 1), which is consistent with previous studies of the S. aureus secretome (6). The quantity of secreted proteins was then determined from the ratio of parent ion peak areas, which demonstrated that while tryptic peptides for the same proteins were detected from each strain, several were detected at different levels (Table 1). Sixteen of the 38 secreted proteins were detected in both strains at similar levels (within 2-fold), while 20 were detected at 2- to 8-fold-reduced levels (for example, the proteases staphylokinase and SspP, the complement inhibitor Sak, and a hypothetical surface protein SA2285). Two proteins (SA0663 and SdrE) were detected at 6- to 8-fold-elevated levels in the ∆spsB strain, while PrsA was detected at a 16-fold-elevated level. PrsA is a peptidyl-prolyl isomerase involved in protein folding of secreted proteins in the extracellular environment (23), and we previously demonstrated that it is upregulated at the transcriptional level by SPase inhibition. Examination of signal sequences revealed no obvious correlation with observed differences in secretion.
TABLE 1 .
Comparison of the secreted proteomes of wild-type N315 and ARC0001∆spsB
| Locus | Name | Description | Fold changea | SPase signal sequenceb |
|---|---|---|---|---|
| SA2285 | Putative surface protein | 0.13 ± 0.01 | MRDKKGPVNKRVDFLSNKLNKYSIRKFTVGTASILIGSLMYLGTQQEAEA | |
| SA1758 | Sak | Staphylokinase | 0.17 ± 0.01 | MLKRSLLFLTVLLLLFSFSSITNEVSA |
| SA1754 | Scn | Staphylococcal complement inhibitor | 0.26 ± 0.10 | MKIRKSILAGTLAIVLASPLVTNLDKNEAQA |
| SA0620 | Peptidase M23 | 0.28 ± 0.03 | MKKLAFAITATSGAAAFLTHHDAQA | |
| SA1813 | Leukocidin/hemolysin toxin family | 0.30 ± 0.04 | MKNKKRVLIASSLSCAILLLSAATTQANSAHK | |
| SA1268 | EbhB | Extracellular matrix-binding protein | 0.32 ± 0.05 | MNYRDKIQKFSIRKYTVGTFSTVIATLVFLGFNTSQAHA |
| SA2431 | IsaB | Immunodominant staphylococcal antigen B | 0.34 ± 0.04 | MNKTSKVCVAATLALGTLIGVTVVENSAPTSKQAQA |
| SA2097 | Secretory antigen precursor | 0.34 ± 0.08 | MKKLVTATTLTAGIGTALVGQAHHADA | |
| SA1725 | SspP | Staphopain A | 0.36 ± 0.08 | MKRNFPKLIALSLIFSLSVTPIANA |
| SA0107 | Spa | Immunoglobulin G-binding protein A | 0.37 ± 0.06 | MKKKNIYSIRKLGVGIASVTLGTLLISGGVTPAANA |
| SA0587 | PsaA | Metal-binding protein PsaA | 0.38 ± 0.04 | MKKLVPLLLALLLLVAACGTGGKQS |
| SA0914 | Chitinase | 0.38 ± 0.06 | MNKLLQSLSALGVSATLVTPNLNADA | |
| SA2074 | ModA | Probable molybdate-binding protein | 0.40 ± 0.08 | MKMKRFIAIVMALFLVLAGCSNS |
| SA0222 | Coa | Staphylocoagulase precursor | 0.45 ± 0.16 | MKKQIISLGALAVASSLFTWDNKADA |
| SA2208 | HlgC | Gamma-hemolysin component C | 0.45 ± 0.05 | MLKNKILATTLSVSLLAPLANPLLENAKA |
| SA1761 | Sep | Enterotoxin P | 0.47 ± 0.16 | MSKIKKTTFILLSFIALTLITSPFVNCSEK |
| SA1755 | Chp | Chemotaxis inhibitory protein | 0.47 ± 0.10 | MKKKLATTVLALSFLTAGISTHHHSAKA |
| SA0905 | Atl | Bifunctional autolysin | 0.47 ± 0.08 | MAKKFNYKLPSMVALTLVGSAVTAHQVQA |
| SA2356 | IsaA | Probable transglycosylase IsaA | 0.51 ± 0.05 | MKKTIMASSLAVALGVTGYAAGTGHQAHA |
| SA2206 | Sbi | Immunoglobulin-binding protein Sbi | 0.52 ± 0.04 | MKNKYISKLLVGAATITLATMISNGEAKA |
| SA0570 | Hypothetical | 0.55 ± 0.04 | MKKLLTASIIACSVVMGVGLVNTSAEA | |
| SA1964 | FmtB | Methicillin resistance protein | 0.56 ± 0.11 | MNLFRQQKFSIRKFNVGIFSALIATVTFISTNPTTASA |
| SA0265 | LytM | Glycyl-glycine endopeptidase | 0.57 ± 0.03 | MKKLTAAAIATMGFATFTMAHQADA |
| SA2353 | SsaA1 | Staphylococcal secretory antigen SsaA1 | 0.61 ± 0.12 | MKKIVTATIATAGLATIAFAGHDAQA |
| SA2093 | SsaA2 | Staphylococcal secretory antigen ssaA2 | 0.61 ± 0.05 | MKKIATATIATAGFATIAIASGNQAHA |
| SA1000 | Fibrinogen-binding protein | 0.62 ± 0.20 | MKKNFIGKSILSIAAISLTVSTFAGESHA | |
| SAP010 | BlaZ | Beta-lactamase | 0.63 ± 0.10 | MKKLIFLIVIALVLSACNSNSSHA |
| SA1003 | Fibrinogen-binding protein | 0.63 ± 1.09 | MKNKLIAKSLLTIAAIGITTTTIASTADA | |
| SA0394 | Hypothetical | 0.66 ± 0.05 | MRENFKLRKMKVGLVSVAITMLYIMTNGQAEA | |
| SA2437 | N-Acetylmuramoyl-l-alanine amidase | 0.74 ± 0.07 | MPKNKILIYLLSTTLVLPTLVSPTAYA | |
| SA0744 | Emp | Extracellular matrix protein-binding protein | 0.82 ± 0.21 | MKKKLLVLTMSTLFATQLINSNHAKA |
| SA0393 | Set15 | Exotoxin 15 | 0.88 ± 0.05 | MKLKNIAKASLALGILTTGMITTTAQPVKA |
| SA0520 | SdrD | Serine-aspartate repeat-containing protein D | 0.90 ± 0.29 | MLNRENKTAITRKGMVSNRLNKFSIRKYTVGTASILVGTTLIFGLGNQEAKA |
| SA1751 | Map | Truncated map-w protein | 1.15 ± 0.21 | MKFKSLITTTLALGVIASTGANFNTNEASA |
| SA0908 | Hypothetical | 1.47 ± 0.42 | MNKFLKYFLILLALVLIVVPIVFATLLFKTSQDA | |
| SA0710 | Peptidase M23 | 1.58 ± 0.16 | MKKTLTVTVSSVLAFLALNNAAHA | |
| SA0663 | Hypothetical | 6.31 ± 5.41 | MNTKYFLAVGAVASVLTLGACGNSNS | |
| SA0521 | SdrE | Serine-aspartate repeat-containing protein E | 8.13 ± 5.15 | MINRDNKKAITKKGMISNRLNKFSIRKYTVGTASILVGTTLIFGLGNQEAKA |
| SA1659 | PrsA | Peptidylprolyl isomerase | 16.53 ± 6.0 | MKMINKLIVPVTASALLLGACGASA |
Reported as the average for all detected tryptic peptides for a given protein. Data are averages and standard errors of the means for three independent biological samples.
Underlined amino acids correspond to tryptic peptides detected in ARC0001∆spsB which contain part of the SPase signal sequence.
As expected for secretion mediated by SPase, none of the tryptic peptides isolated from the wild-type sample contained any part of the signal peptide; however, peptide fragments for eight of the proteins were found with N termini that precisely matched the predicted SPase cleavage site (see Fig. S3 in the supplemental material). In contrast, with ARC0001∆spsB, tryptic peptides containing portions of the N-terminal leader peptides of 12 of the 38 proteins were detected (Table 1; also, see Fig. S3 in the supplemental material). These data suggest that AyrABC mediates secretion of the same proteins as does SPase but either via cleavage at a more N-terminal site or perhaps by extricating the entire intact preprotein from the cytoplasmic membrane.
Conclusions.
SPase functions at the terminal step of the general secretory pathway by releasing translocated proteins from the cytoplasmic membrane at a defined cleavage site. We have provided evidence of an inducible alternative terminal step in S. aureus encoded by the ayrRABC operon, which under normal conditions is repressed by AyrR but which is derepressed by SPase inhibition. The natural physiological role of the pathway may be to support SPase function at times when elevated secretion is required and the capacity of SPase has been exceeded. Alternatively, it may have evolved to facilitate survival in the presence of an SPase inhibitor. The latter possibility is consistent with the fact that SPase is the only component of the general secretory pathway that is exposed on the outer surface of the cytoplasmic membrane, making it uniquely susceptible to extracellular inhibitors. The evolution of four different families of arylomycins (10) further suggests that that SPase inhibition may have represented a significant selection pressure, as does the existence of multiple, redundant SPases in many Gram-positive bacteria.
Hints as to the mechanism by which AyrABC mediates the release of proteins from their attachment to the cell may be gleaned from an analysis of the individual genes. ayrB and ayrC encode an ABC transporter. It is possible that the ABC transporter functions as part of the translocation process, perhaps augmenting or even replacing other components of the general secretory pathway. However, bacterial ABC transporters have been identified that extract lipophilic moieties from the cytoplasmic membrane (24), and AyrBC may function after Sec translocation to extract intact preproteins or proteins after intramembrane cleavage. The role of AyrA, which encodes a membrane protein of unknown function, is even less clear, but it is interesting to speculate that it could be a protease responsible for intramembrane cleavage, if one is required, which is consistent with its predicted membrane localization. It is also interesting that ayrR homologs in other species appear to control the regulation of proteases (25, 26). A more complete understanding of the pathway awaits detailed analysis of these proteins as well as the signal responsible for derepression of the operon, which is in progress. Undoubtedly, understanding the mechanistic details of this alternate terminal step of the Sec pathway will lead to a greater understanding of secretion and the response to secretion stress.
SUPPLEMENTAL MATERIAL
Alignment of RNA-Seq reads to the S. aureus NCTC8325 genome sequence. The ayrRABC operon corresponds to the region from position 343,418 to 345,827 in the NCTC8325 chromosome. The major transcriptional start site corresponds to coordinate 434,400, which is 18 nt upstream of the GTG start codon. A small number of reads corresponded to coordinate 343,248 suggesting a secondary minor promoter region further upstream. Overlapping RNA reads were detected for the entire ayrRABC locus, suggesting that they are cotranscribed. Download
Solid-state synthesis of AyrR. The entire AyrR protein was synthesized using standard methods for Boc solid-state peptide synthesis employed in our laboratory (R. Adhikary, J. Zimmermann, J. Liu, P. E. Dawson, F. E. Romesberg, J. Phys. Chem. B 117:13082–13089, 2013). Briefly, the peptide was synthesized on 0.4 mmol of p-methyl-benzhydrylamine HCl (MBHA) resin using 2.2 mmol of protected amino acid, 4 ml 0.5 M of the peptide coupling reagent HCTU (2.0 mmol), and 700 μl N,N-diisopropylethylamine (DIEA) (~4.1 mmol), with a coupling time of 20 min. The first amino acid, Boc-Lys(Fmoc)-OH, was coupled onto the MBHA resin, and the Fmoc group was removed by reaction with 20% piperidine-dimethylformamide (vol/vol); Boc groups were deprotected with neat trifluoroacetic acid (TFA) for 2 min. An extra lysine residue was added at the C terminus of AyrR to conjugate biotin with the side chain of lysine. The biotinyl-N-hydroxysuccinimide ester was generated by first dissolving 2 mmol N-hydroxysuccinimide and 2 mmol biotin in 5 ml dimethyl sulfoxide (DMSO) and then adding 2 mmol N,Nʹ-diisopropyl carbodiimide. After 30 min, the mixture was added to the resin and coupled for 1 h, followed by the addition of 2 mmol DIEA, which was coupled for an additional hour. The complete coupling was confirmed by a ninhydrin test. The peptide was cleaved from the dry resin support using anhydrous HF with 10% (vol/vol) anisole for 1 h at 0°C. The resulting polypeptide was purified via reverse-phase high-performance liquid chromatography (HPLC) using a linear gradient of solvent A (0.1% [vol/vol] aqueous TFA) and solvent B (90% [vol/vol] acetonitrile, 10% [vol/vol] water, 0.1% [vol/vol] TFA). Purity was assessed by analytical HPLC (A) and mass spectrometry (B and C). The observed mass of the pure biotinylated peptide is 8,027.0 Da (calculated: 8,027.3 Da); shown are the mass spectrum (B) and reconstructed mass spectrum (C). Download
A representation of tryptic peptides detected from the secreted proteome. A schematic of a secreted protein depicting its signal peptide (grey) and mature protein (white) with the location of 3 tryptic peptides is shown: (1) tryptic peptides identified in the ARC0001∆spsB strain which contain amino acids located in the signal peptide and which overlap the SPase cleavage site; the SPase cleavage site is denoted by a dash in the amino acid sequence; (2) tryptic peptides found in wild-type N315 that correspond to the N terminus of the mature SPase processed secreted protein; and (3) representative tryptic peptides from the mature protein. Below the schematic are parent ion chromatograms of three representative proteins (immunoglobulin G-binding protein A [Spa SA0107], the putative surface protein SA2285, and the staphylococcal complement inhibitor SA1754), detected from the secreted proteome of wild-type (blue trace) or ARC0001∆spsB (red trace) S. aureus. Download
Primers used in this study.
MICs and frequencies of resistance to arylomycin M131.
ayrR mutations that evolve to arylomycin M131 in N315∆SA0336.
Amplification of spsB from RNA and genomic DNA.
MICs of N315, ARC0001, and ARC0001∆spsB
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (AI-109809). A.C. was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Citation Craney A, Dix MM, Adhikary R, Cravatt BF, Romesberg FE. 2015. An alternative terminal step of the general secretory pathway in Staphylococcus aureus. mBio 6(4):e01178-15. doi:10.1128/mBio.01178-15.
REFERENCES
- 1.Blobel G, Sabatini DD. 1971. Ribosome-membrane interaction in eukaryotic cells, p 193–195. In Manson LA (ed), Biomembranes. Springer, New York, NY. [Google Scholar]
- 2.Milstein C, Brownlee GG, Harrison TM, Mathews MB. 1972. A possible precursor of immunoglobulin light chains. Nat New Biol 239:117–120. doi: 10.1038/newbio239117a0. [DOI] [PubMed] [Google Scholar]
- 3.Du Plessis DJ, Nouwen N, Driessen AJ. 2011. The Sec translocase. Biochim Biophys Acta 1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Rusch SL, Kendall DA. 2007. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 46:9665–9673. doi: 10.1021/bi7010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driessen AJ, Nouwen N. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 6.Schallenberger MA, Niessen S, Shao C, Fowler BJ, Romesberg FE. 2012. Type I signal peptidase and protein secretion in Staphylococcus aureus. J Bacteriol 194:2677–2686. doi: 10.1128/JB.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schimana J, Gebhardt K, Holtzel A, Schmid DG, Sussmuth R, Muller J, Pukall R, Fiedler HP. 2002. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by streptomyces sp. Tü6075. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot (Tokyo) 55:565–570. [DOI] [PubMed] [Google Scholar]
- 8.Roberts TC, Smith PA, Cirz RT, Romesberg FE. 2007. Structural and initial biological analysis of synthetic arylomycin A2. J Am Chem Soc 129:15830–15838. doi: 10.1021/ja073340u. [DOI] [PubMed] [Google Scholar]
- 9.Smith PA, Roberts TC, Romesberg FE. 2010. Broad spectrum antibiotic activity of the arylomycin natural products is masked by natural target mutations. Chem Biol 17:1223–1231. doi: 10.1016/j.chembiol.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan YX, Romesberg FE. 2012. Latent antibiotics and the potential of the arylomycins for broad-spectrum antibacterial activity. Med Chem Comm 3:916–925. doi: 10.1039/c2md20043k. [DOI] [Google Scholar]
- 11.Roberts TC, Schallenberger MA, Liu J, Smith PA, Romesberg FE. 2011. Initial efforts toward the optimization of arylomycins for antibiotic activity. J Med Chem 54:4954–4963. doi: 10.1021/jm1016126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Smith PA, Steed DB, Romesberg F. 2013. Efforts toward broadening the spectrum of arylomycin antibiotic activity. Bioorg Med Chem Lett 23:5654–5659. doi: 10.1016/j.bmcl.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Therien AG, Huber JL, Wilson KE, Beaulieu P, Caron A, Claveau D, Deschamps K, Donald RG, Galgoci AM, Gallant M, Gu X, Kevin NJ, Lafleur J, Leavitt PS, Lebeau-Jacob C, Lee SS, Lin MM, Michels AA, Ogawa AM, Painter RE, Parish CA, Park YW, Benton-Perdomo L, Petcu M, Phillips JW, Powles MA, Skorey KI, Tam J, Tan CM, Young K, Wong S, Waddell ST, Miesel L. 2012. Broadening the spectrum of beta-lactam antibiotics through inhibition of signal peptidase type I. Antimicrob Agents Chemother 56:4662–4670. doi: 10.1128/AAC.00726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powers ME, Smith PA, Roberts TC, Fowler BJ, King CC, Trauger SA, Siuzdak G, Romesberg FE. 2011. Type I signal peptidase and protein secretion in Staphylococcus epidermidis. J Bacteriol 193:340–348. doi: 10.1128/JB.01052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craney A, Romesberg FE. 2015. A putative Cro-like repressor contributes to arylomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 59:3066–3074. doi: 10.1128/AAC.04597-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffman JL, Brennan RG. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr Opin Struct Biol 12:98–106. doi: 10.1016/S0959-440X(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 17.Sarai A, Takeda Y. 1989. Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc Natl Acad Sci U S A 86:6513–6517. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ptashne M. 1986. A genetic switch: gene control and phage λ, 3rd ed. Cell Press and Blackwell Scientific Publications, Cambridge, MA. [Google Scholar]
- 19.Mesak LR, Yim G, Davies J. 2009. Improved lux reporters for use in Staphylococcus aureus. Plasmid 61:182–187. doi: 10.1016/j.plasmid.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Smith PA, Powers ME, Roberts TC, Romesberg FE. 2011. In vitro activities of arylomycin natural-product antibiotics against Staphylococcus epidermidis and other coagulase-negative staphylococci. Antimicrob Agents Chemother 55:1130–1134. doi: 10.1128/AAC.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Inloes JM, Hsu KL, Dix MM, Viader A, Masuda K, Takei T, Wood MR, Cravatt BF. 2014. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci U S A 111:14924–14929. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyyryläinen HL, Sarvas M, Kontinen VP. 2005. Transcriptome analysis of the secretion stress response of Bacillus subtilis. Appl Microbiol Biotechnol 67:389–396. doi: 10.1007/s00253-005-1898-1. [DOI] [PubMed] [Google Scholar]
- 24.Narita S. 2011. ABC transporters involved in the biogenesis of the outer membrane in gram-negative bacteria. Biosci Biotechnol Biochem 75:1044–1054. doi: 10.1271/bbb.110115. [DOI] [PubMed] [Google Scholar]
- 25.Biswas S, Biswas I. 2014. A conserved streptococcal membrane protein, LsrS, exhibits a receptor-like function for lantibiotics. J Bacteriol 196:1578–1587. doi: 10.1128/JB.00028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, Xu Y, Sitkiewicz I, Ma Y, Wang X, Yestrepsky BD, Huang Y, Lapadatescu MC, Larsen MJ, Larsen SD, Musser JM, Ginsburg D. 2012. Inhibitor of streptokinase gene expression improves survival after group A streptococcus infection in mice. Proc Natl Acad Sci U S A 109:3469–3474. doi: 10.1073/pnas.1201031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of RNA-Seq reads to the S. aureus NCTC8325 genome sequence. The ayrRABC operon corresponds to the region from position 343,418 to 345,827 in the NCTC8325 chromosome. The major transcriptional start site corresponds to coordinate 434,400, which is 18 nt upstream of the GTG start codon. A small number of reads corresponded to coordinate 343,248 suggesting a secondary minor promoter region further upstream. Overlapping RNA reads were detected for the entire ayrRABC locus, suggesting that they are cotranscribed. Download
Solid-state synthesis of AyrR. The entire AyrR protein was synthesized using standard methods for Boc solid-state peptide synthesis employed in our laboratory (R. Adhikary, J. Zimmermann, J. Liu, P. E. Dawson, F. E. Romesberg, J. Phys. Chem. B 117:13082–13089, 2013). Briefly, the peptide was synthesized on 0.4 mmol of p-methyl-benzhydrylamine HCl (MBHA) resin using 2.2 mmol of protected amino acid, 4 ml 0.5 M of the peptide coupling reagent HCTU (2.0 mmol), and 700 μl N,N-diisopropylethylamine (DIEA) (~4.1 mmol), with a coupling time of 20 min. The first amino acid, Boc-Lys(Fmoc)-OH, was coupled onto the MBHA resin, and the Fmoc group was removed by reaction with 20% piperidine-dimethylformamide (vol/vol); Boc groups were deprotected with neat trifluoroacetic acid (TFA) for 2 min. An extra lysine residue was added at the C terminus of AyrR to conjugate biotin with the side chain of lysine. The biotinyl-N-hydroxysuccinimide ester was generated by first dissolving 2 mmol N-hydroxysuccinimide and 2 mmol biotin in 5 ml dimethyl sulfoxide (DMSO) and then adding 2 mmol N,Nʹ-diisopropyl carbodiimide. After 30 min, the mixture was added to the resin and coupled for 1 h, followed by the addition of 2 mmol DIEA, which was coupled for an additional hour. The complete coupling was confirmed by a ninhydrin test. The peptide was cleaved from the dry resin support using anhydrous HF with 10% (vol/vol) anisole for 1 h at 0°C. The resulting polypeptide was purified via reverse-phase high-performance liquid chromatography (HPLC) using a linear gradient of solvent A (0.1% [vol/vol] aqueous TFA) and solvent B (90% [vol/vol] acetonitrile, 10% [vol/vol] water, 0.1% [vol/vol] TFA). Purity was assessed by analytical HPLC (A) and mass spectrometry (B and C). The observed mass of the pure biotinylated peptide is 8,027.0 Da (calculated: 8,027.3 Da); shown are the mass spectrum (B) and reconstructed mass spectrum (C). Download
A representation of tryptic peptides detected from the secreted proteome. A schematic of a secreted protein depicting its signal peptide (grey) and mature protein (white) with the location of 3 tryptic peptides is shown: (1) tryptic peptides identified in the ARC0001∆spsB strain which contain amino acids located in the signal peptide and which overlap the SPase cleavage site; the SPase cleavage site is denoted by a dash in the amino acid sequence; (2) tryptic peptides found in wild-type N315 that correspond to the N terminus of the mature SPase processed secreted protein; and (3) representative tryptic peptides from the mature protein. Below the schematic are parent ion chromatograms of three representative proteins (immunoglobulin G-binding protein A [Spa SA0107], the putative surface protein SA2285, and the staphylococcal complement inhibitor SA1754), detected from the secreted proteome of wild-type (blue trace) or ARC0001∆spsB (red trace) S. aureus. Download
Primers used in this study.
MICs and frequencies of resistance to arylomycin M131.
ayrR mutations that evolve to arylomycin M131 in N315∆SA0336.
Amplification of spsB from RNA and genomic DNA.
MICs of N315, ARC0001, and ARC0001∆spsB