Abstract
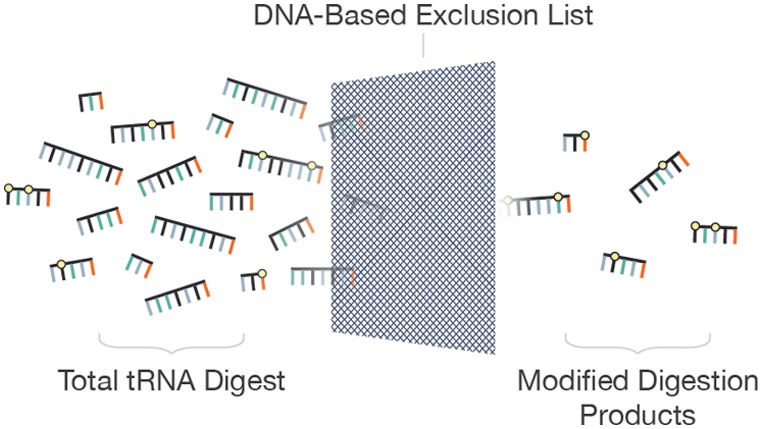
There has been a renewed appreciation for the dynamic nature of ribonucleic acid (RNA) modifications and for the impact of modified RNAs on organism health resulting in an increased emphasis on developing analytical methods capable of detecting modifications within specific RNA sequence contexts. Here we demonstrate that a DNA-based exclusion list enhances data dependent liquid chromatography tandem mass spectrometry (LC-MS/MS) detection of post-transcriptionally modified nucleosides within specific RNA sequences. This approach is possible because all post-transcriptional modifications of RNA, except pseudouridine, result in a mass increase in the canonical nucleoside undergoing chemical modification. Thus, DNA-based sequences reflect the state of the RNA prior to or in the absence of modification. The utility of this exclusion list strategy is demonstrated through the RNA modification mapping of total tRNAs from the bacteria Escherichia coli, Lactococcus lactis, and Streptomyces griseus. Creation of a DNA-based exclusion list is shown to consistently enhance the number of detected modified ribonuclease (RNase) digestion products by ∼20%. All modified RNase digestion products that were detected during standard data dependent acquisition (DDA) LC-MS/MS were also detected when the DNA-based exclusion list was used. Consequently, the increase in detected modified RNase digestion products is attributed to new experimental information only obtained when using the exclusion list. This exclusion list strategy should be broadly applicable to any class of RNA and improves the utility of mass spectrometry approaches for discovery-based analyses of RNA modifications, such as are required for studies of the epitranscriptome.
Transfer ribonucleic acids (tRNAs) deliver amino acids to the ribosome for the translation of messenger ribonucleic acids (mRNAs) into proteins. All tRNAs undergo post-transcriptional processing and modification,1 and those modifications have structural and functional significance. Post-transcriptionally modified nucleotides have a considerable influence on tertiary structure stabilization,2 the decoding process3 and aminoacyl tRNA synthetase recognition.4 To date, more than 100 different naturally occurring RNA modifications have been identified from all three domain of life.5
Recently, there has been a renewed appreciation for the dynamic nature of RNA modification and for the impact of modified RNAs on organism health.6−12 Thus, there has been an increased emphasis on developing analytical methods capable of detecting modifications within specific RNA sequence contexts. In addition to historical methods for the characterization of modified RNAs,13−15 approaches based on genomic sequencing technologies, for example, microarrays or RNA-seq,16−20 provide advantages related to sensitivity, broad applicability to sample types such as mRNA, and more readily available informatics tools to assist in processing large amounts of data. One trade-off is the limited number of modifications that can be characterized by such approaches.
Another popular approach for characterizing modified RNAs involves mass spectrometry.21−23 RNA modification mapping, as developed by McCloskey and co-workers,24−26 utilizes a base-specific ribonuclease (RNase) to cleave a larger RNA into smaller digestion products. These digestion products can be separated and analyzed using liquid chromatography tandem mass spectrometry (LC-MS/MS). Except for pseudouridine (Ψ), which is the isomer of uridine, all modified nucleotides are detected at a higher mass than the canonical nucleotide because of the chemical groups added during modification.27
Unlike genomic-based approaches, mass spectrometry approaches have more commonly been applied in a serial fashion (i.e., one RNA sequence at a time) and even in cases where multiplexing has been used (e.g., total census of modified nucleosides,28,29 comparative analysis of RNA digests (CARD)30,31), there are still limitations on the upper number of RNA sequences that are amenable to LC-MS/MS analysis. The challenge in the standard RNA modification mapping approach is seen by simply calculating the expected number of RNase digestion products from any given group of RNA sequences. For example, a recent publication demonstrating the mapping of modifications onto the total tRNA pool from Lactococcus lactis required the analysis of ∼200 RNase T1 digestion products that were detected during LC-MS/MS.32 Even then, multiple RNase digestions were required to obtain complete mapping coverage of the 40 unique tRNAs from this bacterium.
Because of the wealth of genomic information on RNAs,33 a unique aspect of RNA modification mapping is that complete RNA sequencing by mass spectrometry is not required—one simply seeks to identify those sequence locations that contain a modification. Presumably, by limiting MS/MS analysis to only RNase digestion products that contain one or more modified nucleotides, the information on modified RNase digestion products should increase thereby facilitating the parallel analysis of multiple modified RNAs.
One possible strategy for increasing the sample information generated during analysis is the use of an exclusion list with data dependent analysis (DDA) approaches.34 As previously implemented in proteomics, tryptic peptides that were detected from the previous run were added to the exclusion list for the following runs.35 By creating an exclusion list based on previously identified tryptic peptides, all subsequent analyses enabled deeper coverage of the sample leading to an overall increase in protein identifications.
Here we describe a different implementation of an exclusion list approach. As genomic information already defines the RNA sequence of interest and as all post-transcriptional modifications except pseudouridine will result in a mass increase to the canonical nucleotide being modified, it follows that the mass of unmodified RNase-generated digestion products are predictable a priori and can be used to populate an exclusion list. Post-transcriptionally modified digestion products will appear at a different (higher) mass value than those predicted by transcribing the known gene sequences. Theoretically with such an exclusion list, only modified digestion products will trigger data dependent collision-induced dissociation (CID) to yield MS/MS spectra. The anticipated advantages include an improvement in the MS/MS coverage of modified digestion products, a reduction in the number of MS/MS spectra that must be interpreted during LC-MS/MS, and the capacity to handle more complex mixtures of RNAs. As demonstrated here, this exclusion list strategy does not require knowledge from previous MS/MS analyses nor prior knowledge of RNA modification status for implementation. This DNA-based exclusion list strategy provides an unbiased approach for generating LC-MS/MS data from modified RNAs with deeper sequence coverage than is obtained by standard DDA methods.
Experimental Section
Material
Streptomyces griseus ATCC 13350 was obtained from American Type Culture Collection (ATCC, Manassas, VA). Lactococcus lactisssp. cremoris MG 1363 was a gift from Prof. Bert Poolman. Escherichia coli total tRNA, lysozyme chloride from chicken egg white, phenol, chloroform, RNase T1, 1,1,1,3,3,3 hexafluoro-2-propanol (HFIP) and TriReagent were obtained from Sigma-Aldrich (St. Louis, MO). Triethylamine (TEA) was purchased from ThermoFisher Scientific (Waltham, MA). HPLC grade methanol and water (Burdick & Jackson Kalamazoo, MI) was used during chromatography.
Sample Preparation
S. griseus and L. lactis were cultured as described.32,36 Transfer RNAs were isolated using lysozyme buffer and TriReagent, followed by phenol-chloroform precipitation and ion exchange column purification.32S. griseus tRNAs were digested to nucleosides using Nuclease P1, snake venom phosphodiesterase, and Antarctic phosphatase as described.37 For RNA modification mapping, eight micrograms of purified total tRNAs were added to 400 U of RNase T1 in 4 μL of 220 mM ammonium acetate buffer. The mixture was incubated at 40 °C for 2 h.
Liquid Chromatography Tandem Mass Spectrometry
Analysis of the S. griseus total tRNA nucleoside digests were done using a Hitachi D-7000 HPLC equipped with a UV detector. Nucleosides were separated on a Supelcosil LC-18S column, 5 μm, 2.1 mm × 250 mm (Supelco), with mobile phase A (MPA) of 5 mM ammonium acetate, pH 5.3, and mobile phase B (MPB) of 40% aqueous acetonitrile at a flow rate of 300 μL min–1. The column eluent was split immediately post column, 1/3 to a Thermo Scientific (Waltham, MA) LTQ-XL linear ion trap mass spectrometer equipped with an ion max electrospray source and 2/3 to the UV detector set at 260 nm.
Mass spectra were recorded in the positive ion mode over an m/z range of 105–800 with a capillary temperature of 275 °C, spray voltage of 4.0 kV and sheath gas, auxiliary gas and sweep gas of 35, 20, and 20 arbitrary units, respectively. Data dependent MS/MS of each of the two most abundant ions were recorded throughout the LC/MS run.
RNase T1 digestion products were separated on an Xbridge C18 column, 3.5 μm, 1 mm × 150 mm (Waters) with MPA of 400 mM HFIP, 16 mM TEA in water, pH 7 and MPB of 50% MPA and 50% methanol at a flow rate 30 μL min–1. Digestion products were eluted using a gradient from 5%B to 99%B in 49 min, followed by 99% B for 5 min.
RNase T1 digestion products were analyzed using the Thermo LTQ-XL linear ion trap mass spectrometer. Electrospray ionization mass spectra were recorded in negative polarity at a capillary temperature of 275 °C, spray voltage of 3.7 kV, and 35, 14, and 10 arbitrary flow units of sheath, auxiliary and sweep gas, respectively. Each analysis segment contains a full scan from m/z 650 to 1800 (scan event 1) followed by four data-dependent scans triggered by the four most abundant precursors from scan event 1 (scan events 2–5). The maximum injection period for MS/MS was fixed at 250 ms with an intensity threshold of 500 counts. Samples were analyzed with the same MS/MS parameters for both standard DDA and exclusion list methods. During analysis using an exclusion list, the LC-MS/MS run was divided into six time segments with excluded m/z values added to the appropriate segment based on the elution time of the RNase T1 digestion product. Each ion selected for CID was analyzed for up to 8 scans before it was added to a dynamic exclusion list for 45 s for both data dependent approaches.
Data Analysis
All instrument control and data processing steps were performed with Xcaliber software. The tRNA genomic sequences were obtained from the Genomic tRNA Database (http://gtrnadb.ucsc.edu/).33E. coli tRNA sequences with post-transcriptional modifications were obtained from Modomics (http://modomics.genesilico.pl/).5 Two approaches were used to analyze the LC-MS/MS data. Manual annotation of MS/MS spectra was performed by calculating expected m/z values of all RNase T1 digestion products based on either genomic tRNA sequences or the known post-transcriptionally modified E. coli tRNA sequences using the MongoOligo calculator (http://mods.rna.albany.edu/masspec/Mongo-Oligo). Automated annotation was conducted using RoboOligo38 and in-house software. The criterion for detection of a particular RNase T1 digestion product based on MS/MS analysis was at least 80% of the expected c- and y-type ions were found during spectral annotation,32 except for 7-methylguanosine containing digestion products, which fragment uniquely.39
Results and Discussion
In the standard DDA-based LC-MS/MS RNA modification mapping approach, all precursors with an ion abundance above the user-defined threshold are candidates for CID analysis. However, as the goal of RNA modification mapping experiments is to place modified nucleosides in the correct RNA sequence context, RNase digestion products that do not contain a modified nucleoside can be considered to be information-deficient oligonucleotides. Moreover, as unmodified oligonucleotides are typically much more abundant than digestion products containing modifications,24 the standard DDA approach can be inefficient for detecting modified digestion products unless a large number of DDA scans are available during analysis. To overcome these limitations, the use of DNA-based exclusion lists was investigated. These static exclusion lists are generated based on the known sequences of the RNAs being analyzed.
Exclusion List Generation
It has been shown that standard DDA creates sampling bias against low abundance precursor ions relative to other sample components.35 To determine whether an exclusion list strategy would increase the information obtained during LC-MS/MS analysis of modified oligonucleotides, this strategy was first evaluated using E. coli total tRNAs. The modification status of these tRNAs is well characterized through a number of independent analyses.5 Moreover, E. coli total tRNA has been used previously in developing new analytical mass spectrometry approaches for RNA modification mapping of complex mixtures.30,31,40,41 Using the existing annotated E. coli tRNA sequences,5,42 a total of 73 unique RNase T1 digestion products containing at least one post-transcriptionally modified nucleoside (excluding pseudouridine) are expected.
To create the exclusion list, the genomic tRNA (tDNA) sequences for E. coli were in silico digested with RNase T1 to generate a catalog of all possible unmodified digestion products. The appropriate m/z values including multiply charged ions for each unmodified digestion product were then determined. For E. coli, the 47 unique tDNA sequences yielded 215 predicted RNase T1 digestion products. Reducing these digestion products to those of unique mass resulted in 122 digestion products, leading finally to 191 unique m/z values to populate the exclusion list.
To minimize exclusion list interferences initially observed during an HPLC elution time-independent exclusion strategy (data not shown), these m/z values were separated into five groups based on the known elution times of the digestion products. These time periods (1–30, 30–35, 35–40, 40–45, and 45–50 min) were determined empirically to best minimize overlap and match the known length-dependence of oligonucleotide separations seen when using ion pairing reversed phase HPLC.43
During initial studies on this exclusion list approach, it was discovered that sodium adducts could trigger data dependent CID (data not shown). In addition to the predicted molecular ions for each RNase T1 digestion product, the mass of a sodium atom was added to each of the unique m/z values calculated above to minimize DDA of such adducts. While potassium adducts were also found to interfere in select instances, the potential overlaps between unmodified digestion products with potassium and modified sequences were determined to be significant enough to warrant not adding potassium adducts to the exclusion list. Thus, starting with tDNA sequences, a final time-dependent exclusion list for molecular ions (multiply charged when oligomer length ≥4) and sodium adducts was used for the RNA modification mapping of E. coli and all other bacteria investigated here.
Exclusion List DDA Approach for Total tRNA Modification Mapping
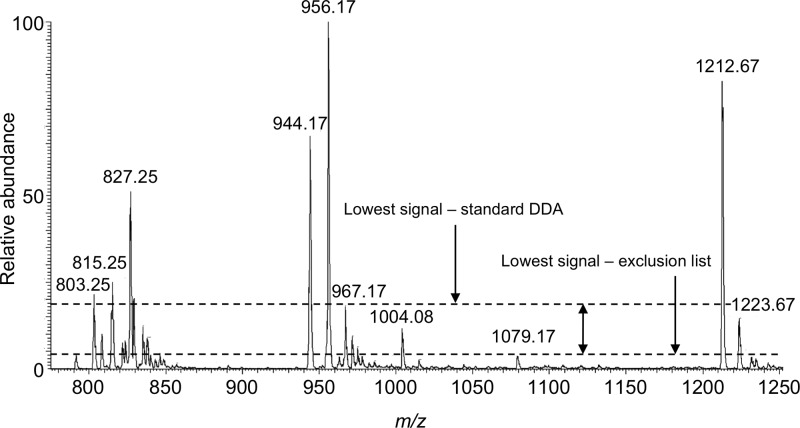
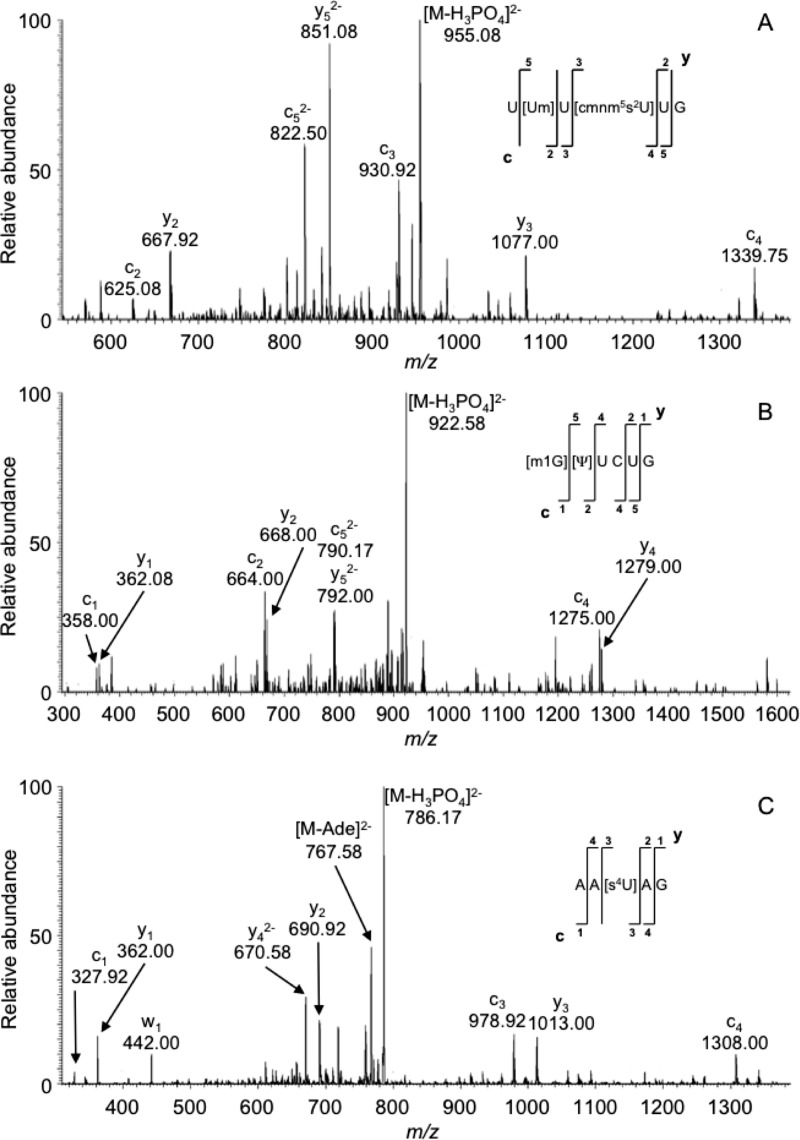
After finalizing the approach for creating an exclusion list, RNase T1 digestion products from E. coli total tRNA were analyzed. A representative example of the gains available by an exclusion list approach is illustrated in Figure 1. More than 20 m/z values are seen in the mass spectrum associated with an LC elution period from 38.83–39.04 min. During standard DDA, the least abundant m/z value that triggered CID was m/z 967.17. In contrast, using the E. coli tDNA sequences to construct an exclusion list resulted in an improvement in the dynamic range of the MS/MS analysis, with the least abundant m/z value that now triggered CID being m/z 1079.17. This dynamic range improvement resulted in the detection and MS/MS analysis of three post-transcriptionally modified digestion products: AA[s4U]AG (m/z 835.17 Figure 2A), [m1G]UUCUG (m/z 971.67 Figure 2B), and U[Um]U[cmnm5s2U]UG (m/z 1004.08 Figure 2C). Significantly, even multiple replicate analyses during standard DDA failed to trigger MS/MS of these three modified oligonucleotides.
Figure 1.
Mass spectrum arising from the LC-MS/MS analysis of RNase T1 digestion of E. coli total tRNAs. The mass spectral data corresponds to ions eluting from 38.83–39.04 min. As noted, the dynamic range for data dependent MS/MS increases when a tDNA-based exclusion list is implemented.
Figure 2.
MS/MS spectra corresponding to previously unreported post-transcriptionally modified RNase T1 digestion products from E. coli total tRNAs. (A) Annotated mass spectrum of digestion product with precursor m/z of 1004.34. (B) Annotated mass spectrum of digestion product with precursor m/z of 971.67. (C) Annotated mass spectrum of digestion product with precursor m/z of 835.2. Modified nucleosides: cmnm5s2U: 5-carboxymethylaminomethyl-2′-O-methyluridine; s4U: 4-thiouridine; others defined in Table 4.
Given the potential improvements seen by increasing the dynamic range for MS/MS analysis of RNase digestion products containing post-transcriptionally modified nucleosides, the next goal was to determine the effectiveness of this strategy for RNA modification mapping across the entire total tRNA sample. A standard DDA-based LC-MS/MS analysis of RNase T1 digestion products from E. coli total tRNAs resulted in the detection of 50–51 digestion products across three replicate analyses (Table 1). Of the total, 49 digestion products were detected consistently in all three analyses. When the exclusion list approach was used, an additional 8–10 expected digestion products were detected (Table 1). During these replicate exclusion list analyses, 59 digestion products were detected consistently. Thus, the use of an exclusion list increased the number of detected modified digestion products by ∼20%. Importantly, the use of an exclusion list retained detection of all modified RNase T1 digestion products detected by standard DDA, thus the 20% improvement can be attributed to previously undetected digestion products.
Table 1. Number of RNase T1 Digestion Products Identified (and Percentage out of 73 Predicted) from E. coli Total tRNA via Replicate Runs of Standard DDA-Based LC-MS/MS and LC-MS/MS Using an Exclusion List.
| method | replicate 1 | replicate 2 | replicate 3 |
|---|---|---|---|
| standard DDA | 51 (70%) | 50 (68%) | 51 (70%) |
| exclusion list | 59 (81%) | 60 (82%) | 60 (82%) |
| % increase | 16% | 20% | 18% |
Despite this 20% increase, 13 predicted digestion products were not detected even using the exclusion list approach. To understand why, these digestion products were examined in greater detail (Table 2). It was found that six predicted digestion products were not detected at the MS stage. For half of those not detected (DDAUG, UU[cmo5U]G and ACU[cmo5U]G), an m/z signal and corresponding MS/MS data for a hyper- or hypomodified variant were detected during both standard DDA and exclusion list analysis. Detection of these variants suggests a difference exists in the modification states of these tRNAs for the sample analyzed as compared to those already catalogued in the modified tRNA databases.5,42 Importantly, their absence was not attributed to any deficiency in the exclusion list strategy.
Table 2. Predicted E. coli RNase T1 Digestion Products and the Source tRNAs for Each That Were Not Detected by LC-MS/MS with Either a Standard DDA Strategy or the Exclusion List Strategya.
| RNase T1 digestion product | tRNA | explanation |
|---|---|---|
| DDAUG | Cys (GCA) | detected as DUAUG |
| UU[cmo5U]G | Pro (UGG) | detected as [Um]U[cmo5U]G |
| ACU[cmo5U]G | Thr (UGU) | detected as A[Cm]U[cmo5U]G |
| CCCCUCCU[t6A]AG | Arg (CCU) | low abundance tRNA |
| ACU[k2C]AU[t6A]A[Ψ]CG | Ile (CAU) | low ionization efficiency |
| AC[s2C]U[mnm5U]CU[t6A]AG | Arg (UCU) | low abundance tRNA |
| CCCUU[s4U]AG | Ile (CAU) | interference |
| AADC[Gm]G | Leu (UAA) | interference |
| AAADC[Gm]G | Leu (CAA) | interference |
| CC[s2C]UCCG | Arg (CCG) | present but not selected for MS/MS |
| [m7G]UCUCAGa | Arg (CCG) | present but not selected for MS/MS |
| [m7G][acp3U]CACAGa | Met (CAU) | present but not selected for MS/MS |
| CCU[mnm5U]CCAAGa | Gly (UCC) | present but not selected for MS/MS |
Detected using an enhanced exclusion list (see text for details).
The other three predicted digestion products not detected at the MS stage (CCCCUCCU[t6A]AG, ACU[k2C]AU[t6A]A[Ψ]CG, and AC[s2C]U[mnm5U]CU[t6A]AG) were not found in a hyper- or hypomodified variant. Their absence could be attributed to an absence of the predicted tRNA in the analyzed sample, to the tRNA being present at levels below the limits of detection of the LC-MS system used, or to low ionization efficiency of the particular RNase T1 digestion product. To determine whether the expected tRNA was present in the sample, other signature digestion products40 associated with these three tRNAs (tRNA-Arg (CCU), tRNA-Ile (CAU) and tRNA-Arg (UCU)) were searched for within the LC-MS/MS data set. Low abundance unmodified signature digestion products for tRNA-Arg (CCU) and tRNA-Arg (UCU) were found within the exclusion list data set (MS only) supporting the explanation that these tRNAs are present but at amounts approaching the limit of detection by our LC-MS/MS system. In contrast, two high abundance signature digestion products for tRNA-Ile (CAU) were detected. As these signature digestion products are modified, they also generated MS/MS data during the exclusion list analysis. Detection of these modified signature digestion products suggests that the undetected modified digestion product for tRNA-Ile (CAU), ACU[k2C]AU[t6A]AUCG, has an ionization efficiency much lower than those of other coeluting digestion products thereby limiting detection of this expected product.
The identification of three of the remaining undetected E. coli total tRNA RNase T1 digestion products was hindered by interferences with very similar digestion products. For example, the coeluting digestion products CCCUU[s4U]AG and CCCCU[s4U]AG diverge by 1 Da because of a C to U difference in the two sequences. Other coeluting digestion products of closely related sequence and mass include AADC[Gm]G/AADD[Gm]G and AAADC[Gm]G/AAADD[Gm]G. While the mass difference for both pairs is 3 Da (C → D), the resulting MS/MS spectra cannot readily differentiate components of dissimilar ion abundance. In these cases, only the higher abundance digestion product was considered to be identified and included in the calculated values shown in Table 1.
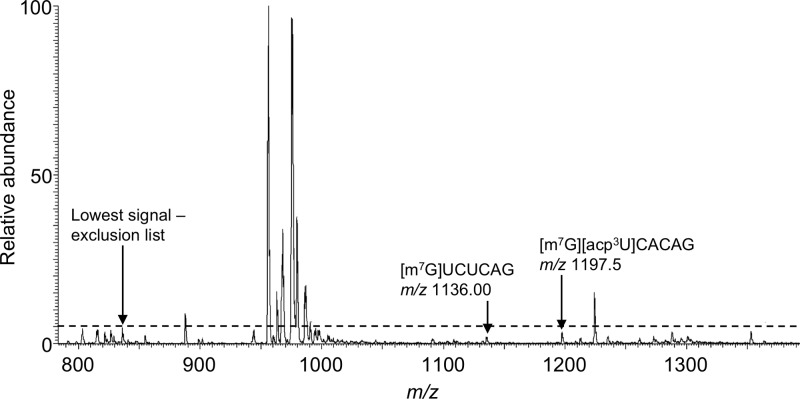
Finally, four predicted modified digestion products were found to be present within the MS data but did not trigger data dependent CID for MS/MS with either approach. Thus, these four digestion products were available for detection but were missed even during the exclusion list approach due to a limited number of data dependent events per analysis segment. A representative example is seen in Figure 3. Improvements in chromatography or an increase in the number of data dependent events might address the MS/MS detection of such low abundance digestion products.
Figure 3.
Mass spectrum arising from the LC-MS/MS analysis of RNase T1 digestion of E. coli total tRNAs. The mass spectral data corresponds to ions eluting from 39.70–39.81 min. In this instance, even the implementation of the tDNA-based exclusion list does not enable detection of two low abundance modified RNase T1 digestion products. Modified nucleosides: acp3U: 3-(3-amino-3-carboxypropyl)uridine; others defined in Table 4.
Alternatively, as was done in a proteomics implementation of the exclusion list strategy,35 one could analyze the same sample a second time wherein the original exclusion list is enhanced with the m/z values of modified digestion products detected in the initial LC-MS/MS analysis. Such an approach was also tested here by creating an enhanced exclusion list containing all unmodified digestion products plus m/z values of modified digestion products detected by the exclusion list approach. When this enhanced static exclusion list was created and applied to the analysis of E. coli tRNAs, three of the four predicted modified digestion products that were present in the MS data but did not initially trigger MS/MS could now be detected in the MS/MS data. Only CC[s2C]UCCG from tRNA-Arg(CCG) failed to trigger data dependent CID when using the enhanced exclusion list.
Exclusion List Improvements in Total tRNA Modification Mapping
An added benefit of using the exclusion list strategy on this well-characterized sample was the detection of four previously unidentified RNase T1 digestion products with modifications. The four modified oligonucleotides are (6)AA[s4U]AG(10) from tRNA-Gly (GCC), (31)AUU[cmo5U]AG(36) from tRNA-Leu (UAG), (36)U[t6A]AUCAG(42) from tRNA-Thr (UGU) and (2)UCCCCU[s4U]CG(10) from tRNA-Glu (UUC). All four post-transcriptionally modified oligonucleotides were detected solely during the exclusion list approach and were detected reproducibly in each of the three replicate analyses. As these four modified digestion products were previously unreported, they are not included in the calculated values within Table 1.
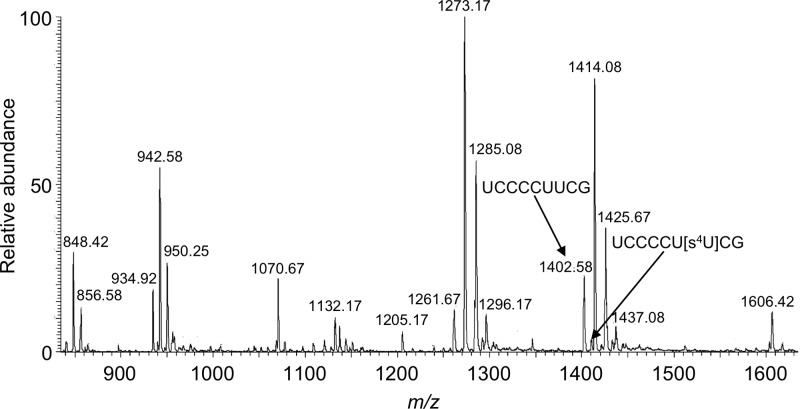
To determine whether these previously unreported sites of modification arise within a hypomodified context, the unmodified versions of each digestion product were sought within the data. The only hypomodified RNase T1 digestion product found corresponded to (2)UCCCCUUCG(10) from tRNA-Glu (UUC). This digestion product was detected at high abundance with the modified counterpart present at <15% of the unmodified variant (Figure 4), consistent with hypomodification of 4-thiouridine at position 8 within this tRNA.
Figure 4.
Mass spectrum arising from the LC-MS/MS analysis of RNase T1 digestion of E. coli total tRNAs. The mass spectral data corresponds to ions eluting from 42.27–42.57 min. Both an unmodified (m/z 1402.58) and hypomodified (m/z 1410.08) RNase T1 digestion product from E. coli tRNA-Thr (UGU) can be identified in this spectrum.
Taken together, these data demonstrate that the use of a tDNA-based exclusion list for tRNA modification mapping is significantly more effective than the standard DDA approach. Of the 73 predicted modified RNase T1 digestion products from E. coli, only three previously known digestion products present in the sample could not be detected by the strategy implemented here. Moreover, the dynamic range improvements achieved during MS/MS enabled the detection of four previously unknown modified digestion products. As implementation of an exclusion list does not compromise the data attainable by the standard DDA approach, this strategy should prove to be useful for characterizing complex mixtures of modified oligonucleotides.
Lactococcus lactis Total tRNAs
Buoyed by the improvements in RNA modification mapping using an exclusion list strategy with the known tRNAs from E. coli, the same strategy was then applied to L. lactis total tRNAs. Recently we reported initial results on the tRNA modification profiles for L. lactis, which were obtained using multiple RNases and a standard DDA-based LC-MS/MS strategy.32 From those initial results, a total of 58 unique RNase T1 digestion products containing at least one post-transcriptionally modified nucleoside (pseudouridine excepted) are expected.
As noted in Table 3, 47–48 modified digestion products were identified using the exclusion list, which was nearly a 20% improvement over the standard DDA approach. As with the E. coli analyses, all of the digestion products detected by standard DDA were again detected when using the exclusion list strategy. However, a significant fraction (20/58 = 34%) of the predicted RNase T1 digestion products32 were not found in the exclusion list MS/MS data. A closer examination of the data revealed the low coverage of expected modified digestion products can be explained by three factors: (1) no MS signal was detected for the predicted modified digestion product; (2) coelution of closely related modified sequences that cannot be unambiguously identified based on MS/MS results; and (3) previously unreported modifications yield new (unpredicted) digestion products. Updating the L. lactis tRNA sequences with modifications detected in this work and recomputing the expected digestion products and their detection reduces the number of undetected yet predicted digestion products to 20%. This value is more consistent with that seen during the E. coli analyses, where 13 of the 73 predicted digestion products were not detected in the exclusion list MS/MS data.
Table 3. Number of RNase T1 Digestion Products Identified from L. lactis Total tRNA via Replicate Runs of Standard DDA-Based LC-MS/MS and LC-MS/MS using an Exclusion List.
| method | replicate 1 | replicate 2 | replicate 3 |
|---|---|---|---|
| standard DDA | 40 | 39 | 41 |
| exclusion list | 47 | 47 | 48 |
| % increase | 18% | 20% | 17% |
Streptomyces griseus Total tRNA
While E. coli total tRNAs were used to develop this exclusion list strategy and L. lactis total tRNAs served as a previously evaluated data set to confirm the general characteristics of this approach, a more fruitful application would be for RNA modification mapping of bacteria that are presently uncharacterized. Thus, total tRNAs from S. griseus, a Gram-positive bacterium with high GC content, were chosen. The modified nucleosides in S. griseus and their placement on S. griseus tRNA sequences have not been reported previously, thus this organism should be a reasonable test of this exclusion list approach. To facilitate characterization of the RNase T1 digestion products, first a census of modifications was obtained from the LC-MS/MS analysis of a nucleoside digest.24 Seventeen unique modified nucleosides were identified from S. griseus tRNAs (Table 4). In comparison to Bacillus subtilis, another Gram-positive firmicute, S. griseus was found to lack mo5U, cmnm5U, cmnm5s2U, k2C, Q, m5U, and s4U. The first five of these modifications are located at position 34 of anticodon loop.5 The absence of m5U (rT) was intriguing, as this modification is typically present as part of the conserved TΨC loop in tRNAs.5 4-Thiouridine (s4U) is commonly found at position 8 of tRNAs.5 Overall, S. griseus tRNAs appear to be less extensively modified than other bacterial tRNAs. Thus, the expectation is that fewer modified RNase T1 digestion products will be present.
Table 4. Nucleosides from S. griseus Total tRNAs Detected Using UV-LC-MS/MS.
| RT (min) | MH+ | nucleoside |
|---|---|---|
| 4.09 | 247 | D, dihydrouridine |
| 4.46 | 245 | Ψ, pseudouridine |
| 12.74 | 258 | Cm, 2′-O-methylcytidine |
| 20.34 | 259 | Um, 2′-O-methyluridine |
| 20.97 | 269 | I, inosine |
| 21.91 | 282 | m1A, 1-methyladenosine |
| 23.49 | 298 | m7G, 7-methylguanosine |
| 28.90 | 298 | m1G, 1-methylguanosine |
| 30.51 | 298 | Gm, 2′-O-methylguanosine |
| 32.53 | 413 | t6A, N6-threonylcarbamoyladenosine |
| 34.23 | 298 | m2G, N2-methylguanosine |
| 37.83 | 282 | Am, 2′-O-methyladenosine |
| 39.45 | 282 | m2A/m6A, 2-methyladenosine/N6-methyladenosine |
| 45.46 | 352 | io6A, N6-(cis hydroxyisopentenyl)adenosine |
| 51.65 | 382 | ms2i6A, 2-methylthio-N6-isopentenyladenosine |
| 51.70 | 398 | ms2io6A, 2-methylthio-N6-(cis hydroxyisopentenyl)adenosine |
| 52.75 | 336 | i6A, N6-isopentenyladenosine |
As before, the RNase T1 digestion products from S. griseus total tRNA were analyzed using both the standard DDA and exclusion list approaches (Table 5). Compared to the information obtained during a standard DDA analysis, the exclusion list approach yielded 18% more post-transcriptionally modified digestion products. As before, all modified digestion products detected by standard DDA were also seen in the exclusion list data set, thus the increase in detected modified digestion products is new information from the experimental analysis. These data confirm the gains achieved by an exclusion list strategy are consistent regardless of the bacterium studied with ∼20% more digestion products being identified by MS/MS with the exclusion list. While a significant number of the modified digestion products can be mapped back onto a unique genomic tRNA sequence, complete modification mapping and unambiguous sequence assignment requires complementary data from additional RNase digestions.32
Table 5. Number of RNase T1 Digestion Products Identified from S. griseus Total tRNA via Replicate Runs of Standard DDA-Based LC-MS/MS and LC-MS/MS Using an Exclusion List.
| method | replicate 1 | replicate 2 | replicate 3 |
|---|---|---|---|
| standard DDA | 40 | 39 | 39 |
| exclusion list | 47 | 46 | 46 |
| % increase | 18% | 18% | 18% |
Data Analysis
The previous results demonstrate that use of a tDNA-based exclusion list leads to a valuable increase in the number of modified digestion products detected by LC-MS/MS when analyzing total tRNAs from bacteria. Although it was initially assumed that the exclusion list would reduce the number of MS/MS spectra that must be interpreted for RNA modification mapping, experimentally it was found that there were minimal differences in the number of MS/MS spectra generated in each case. During standard DDA, 6766 MS/MS scan events from L. lactis and 8290 MS/MS scan events from S. griseus, on average, were generated. Use of the exclusion list approach found that 5753 MS/MS scan events from L. lactis and 7910 MS/MS scan events from S. griseus, on average, were generated.
While the number of MS/MS scan events was similar in both approaches, a significant difference was found when these spectra were annotated. On average, 143 and 162 RNase T1 digestion products from L. lactis and S. griseus, respectively, could be annotated from the MS/MS data generated by the standard DDA approach. Those numbers decrease to 65 annotated digestion products from L. lactis and 82 annotated digestion products from S. griseus, on average, during the exclusion list analyses. This reduction in the number of annotated MS/MS spectra arises as a significant fraction of the MS/MS scans during exclusion list analysis were from low abundance m/z values not associated with tRNA digestion products. Thus, with the exceptions already noted earlier, the exclusion list strategy enables a thorough analysis of all m/z values that arise from sample-related oligonucleotides. Although not investigated here, it is anticipated that a higher DDA threshold would reduce the number of MS/MS scan events further when the exclusion list strategy is implemented.
One limitation of an RNA modification mapping approach that uses LC-MS/MS is the inability to assign specific modified nucleoside identities in the MS/MS spectrum, where isomeric modifications could exist. For example, modified RNase T1 digestion products found to contain a methylated guanosine in the S. griseus data could arise from four possibilities: m1G, m2G, m7G, or Gm (Table 4). While one can take advantage of unique MS/MS fragmentation for some modifications, such as m7G,39 in general one must rely upon known homology in tRNA modification sites or conduct more detailed sample fractionation and analysis.32 It is anticipated that the deeper coverage in modification mapping data that is facilitated by this exclusion list approach may increase the likelihood that such isomeric possibilities are encountered.
Conclusions
Extending RNA modification mapping studies into mixtures of RNAs is challenging because of the more abundant population of unmodified digestion products in the sample. By creating an exclusion list built from the known DNA sequences of the RNA samples of interest, MS/MS cycles can be focused on digestion products containing one or more modified nucleosides. This approach provides deeper LC-MS/MS coverage of modified digestion products present in the sample enabling detection of previously unreported sequence locations of modified nucleosides. While only illustrated using complex RNase T1 digestion products from bacterial total tRNA samples, the approach should be applicable to other RNases and other RNA classes. Moreover, this concept should also facilitate sequence-based placement of DNA lesions, which also result in a mass change of the oligonucleotide and occur at relatively low levels within the sample, as well as the characterization of low-level impurities within synthetic oligonucleotide samples.44
While this exclusion list strategy enhances the information that can be obtained during sample analysis, complete and unambiguous RNA modification mapping still requires the use of multiple RNases to provide appropriate sequence coverage.32 Moreover, multiple RNases are required as closely related coeluting modified digestion products can be challenging to identify even when using this exclusion list strategy. To minimize such effects, a time-dependent exclusion list was used in this work, although improved chromatographic methods are an alternative that could also be explored.
Although species-specific exclusion lists were generated in this work to demonstrate proof-of-concept, this approach should be readily extendable to a universal exclusion list that would exclude any possible particular RNase digestion product combination of base compositions. Such a universal exclusion list would facilitate application of RNA modification mapping into samples that are composed of more than one type of RNA or that arise from more than one organism, such as samples isolated from the microbiome. Moreover, a universal exclusion list approach should improve the capabilities of LC-MS/MS for the discovery-based characterization of dynamic changes in RNA modification status allowing this technique to be used in large-scale studies of the epitranscriptome.
Acknowledgments
The authors thank Ningxi Yu for assistance with spectral annotation and Prof. Bert Poolman (University of Groningen) for the gift of Lactococcus lactis. Financial support of this work was provided by the National Institutes of Health (GM58843) and the University of Cincinnati.
The authors declare no competing financial interest.
References
- Jackman J. E.; Alfonzo J. D. Wiley Interdiscip Rev. RNA 2013, 4, 35–48 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P.; Sprinzl M.; Watanabe K.; Albani M.; Kersten H. Nucleic Acids Res. 1979, 6, 1571–1581 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris P. F. Nucleic Acids Res. 2004, 32, 223–238 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T.; Miyauchi K. FEBS Lett. 2010, 584, 272–277 10.1016/j.febslet.2009.11.085. [DOI] [PubMed] [Google Scholar]
- Machnicka M. A.; Milanowska K.; Osman Oglou O.; Purta E.; Kurkowska M.; Olchowik A.; Januszewski W.; Kalinowski S.; Dunin-Horkawicz S.; Rother K. M.; Helm M.; Bujnicki J. M.; Grosjean H. Nucleic Acids Res. 2013, 41, D262–D267 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S.; Ignatova Z. Nat. Rev. Genet. 2015, 16, 98–112 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- Klungland A.; Dahl J. A. Curr. Opin. Genet. Dev. 2014, 26, 47–52 10.1016/j.gde.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Meyer K. D.; Jaffrey S. R. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y.; Chen-Kiang S.; Mason C. E. RNA Biol. 2013, 10, 342–346 10.4161/rna.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.; Bernstein D. A.; Mumbach M. R.; Jovanovic M.; Herbst R. H.; León-Ricardo B. X.; Engreitz J. M.; Guttman M.; Satija R.; Lander E. S.; Fink G.; Regev A. Cell 2014, 159, 148–162 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. G.; Batlle E.; Ribas de Pouplana L. Trends Mol. Med. 2014, 20, 306–314 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Carlile T. M.; Rojas-Duran M. F.; Zinshteyn B.; Shin H.; Bartoli K. M.; Gilbert W. V. Nature 2014, 515, 143–146 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.; Nilsson L.; Kurland C. G. J. Mol. Biol. 1996, 260, 649–663 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M.; Gomes S.; Geslain R.; Dai Q.; Rosner M. R.; Pan T. Nucleic Acids Res. 2009, 37, 7268–7280 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M.; Fu Y.; Pavon-Eternod M.; He C.; Pan T. RNA 2010, 16, 1317–1327 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinlay A.; Gerard W.; Fields S. BioTechniques 2012, 52, 109–111 10.2144/000113801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y.; Lyko F.; Helm M. Nucleic Acids Res. 2010, 38, 1415–1430 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. X.; Yi C.; He C. Nat. Biotechnol. 2012, 30, 1107–1116 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M.; Pollex T.; Hanna K.; Lyko F. Nucleic Acids Res. 2009, 37, e12. 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattenoz P. B.; Taft R. J.; Westhof E.; Mattick J. S. RNA 2013, 19, 257–270 10.1261/rna.036202.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giessing A. M. B.; Kirpekar F. J. Proteomics 2012, 75, 3434–3449 10.1016/j.jprot.2012.01.032. [DOI] [PubMed] [Google Scholar]
- Taucher M.; Breuker K. Angew. Chem., Int. Ed. 2012, 51, 11289–11292 10.1002/anie.201206232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston K. W.; Limbach P. A. RNA Biol. 2014, 11, 1568–1585 10.4161/15476286.2014.992280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak J. A.; Pomerantz S. C.; Crain P. F.; McCloskey J. A. Nucleic Acids Res. 1993, 21, 4577–4585 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg R.; Urban C.; Reuner B.; Scharff P.; Pomerantz S. C.; McCloskey J. A.; Gross H. J. Nucleic Acids Res. 1993, 21, 5583–5588 10.1093/nar/21.24.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T. M.; Nair V.; Guymon R.; Pomerantz S. C.; Crain P. F.; Davis D. R.; McCloskey J. A. Nucleic Acids Symp. Ser. 2004, 48, 263–264 10.1093/nass/48.1.263. [DOI] [PubMed] [Google Scholar]
- Helm M.; Alfonzo J. D. Chem. Biol. 2014, 21, 174–185 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. T.; Dyavaiah M.; DeMott M. S.; Taghizadeh K.; Dedon P. C.; Begley T. J. PLoS Genet. 2010, 6, e1001247. 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D.; Chan C. T. Y.; Gu C.; Lim K. S.; Chionh Y. H.; McBee M. E.; Russell B. S.; Babu I. R.; Begley T. J.; Dedon P. C. Nat. Protoc. 2014, 9, 828–841 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Limbach P. A. Anal. Chem. 2012, 84, 8607–8613 10.1021/ac301638c. [DOI] [PubMed] [Google Scholar]
- Li S.; Limbach P. A. Analyst 2013, 138, 1386–1394 10.1039/c2an36515d. [DOI] [PubMed] [Google Scholar]
- Puri P.; Wetzel C.; Saffert P.; Gaston K. W.; Russell S. P.; Cordero Varela J. A.; van der Vlies P.; Zhang G.; Limbach P. A.; Ignatova Z.; Poolman B. Mol. Microbiol. 2014, 93, 944–956 10.1111/mmi.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. P.; Lowe T. M. Nucleic Acids Res. 2009, 37, D93–D97 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen P.; Spicer V.; Rydzak T.; Sparling R.; Levin D.; Wilkins J. A.; Krokhin O. Proteomics 2012, 12, 1160–1169 10.1002/pmic.201100425. [DOI] [PubMed] [Google Scholar]
- Rudomin E. L.; Carr S. A.; Jaffe J. D. J. Proteom Res. 2009, 8, 3154–3160 10.1021/pr801017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd M. D.; Kharel M. K.; Bosserman M. A.; Rohr J. In Current Protocols in Microbiology; John Wiley & Sons, Inc.: Hoboken, NJ, 2005. [Google Scholar]
- Russell S. P.; Limbach P. A. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2013, 923–924, 74–82 10.1016/j.jchromb.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample P. J.; Gaston K. W.; Alfonzo J. D.; Limbach P. A. Nucleic Acids Res. 2015, 43, e64. 10.1093/nar/gkv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. Y.; Javid B.; Addepalli B.; Piszczek G.; Strader M. B.; Limbach P. A.; Barry C. E. 3rd. Antimicrob. Agents Chemother. 2013, 57, 6311–6318 10.1128/AAC.00905-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.; Limbach P. A. RNA 2007, 13, 295–303 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel C.; Limbach P. A. J. Proteomics 2012, 75, 3450–3464 10.1016/j.jprot.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jühling F.; Mörl M.; Hartmann R. K.; Sprinzl M.; Stadler P. F.; Pütz J. Nucleic Acids Res. 2009, 37, D159–D162 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apffel A.; Chakel J. A.; Fischer S.; Lichtenwalter K.; Hancock W. S. Anal. Chem. 1997, 69, 1320–1325 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- Nikcevic I.; Wyrzykiewicz T. K.; Limbach P. A. Int. J. Mass Spectrom. 2011, 304, 98–104 10.1016/j.ijms.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]