Abstract
Northern wetlands make up a substantial terrestrial carbon sink and are often dominated by decay-resistant Sphagnum mosses. Recent studies have shown that planctomycetes appear to be involved in degradation of Sphagnum-derived debris. Novel trimethylornithine (TMO) lipids have recently been characterized as abundant lipids in various Sphagnum wetland planctomycete isolates, but their occurrence in the environment has not yet been confirmed. We applied a combined intact polar lipid (IPL) and molecular analysis of peat cores collected from two northern wetlands (Saxnäs Mosse [Sweden] and Obukhovskoye [Russia]) in order to investigate the preferred niche and abundance of TMO-producing planctomycetes. TMOs were present throughout the profiles of Sphagnum bogs, but their concentration peaked at the oxic/anoxic interface, which coincided with a maximum abundance of planctomycete-specific 16S rRNA gene sequences. The sequences detected at the oxic/anoxic interface were affiliated with the Isosphaera group, while sequences present in the anoxic peat layers were related to an uncultured planctomycete group. Pyrosequencing-based analysis identified Planctomycetes as the major bacterial group at the oxic/anoxic interface at the Obukhovskoye peat (54% of total 16S rRNA gene sequence reads), followed by Acidobacteria (19% reads), while in the Saxnäs Mosse peat, Acidobacteria were dominant (46%), and Planctomycetes contributed to 6% of the total reads. The detection of abundant TMO lipids in planctomycetes isolated from peat bogs and the lack of TMO production by cultures of acidobacteria suggest that planctomycetes are the producers of TMOs in peat bogs. The higher accumulation of TMOs at the oxic/anoxic interface and the change in the planctomycete community with depth suggest that these IPLs could be synthesized as a response to changing redox conditions at the oxic/anoxic interface.
INTRODUCTION
Peat-accumulating northern wetlands are important sinks for terrestrial carbon, making up one-third of the global soil organic carbon pool (1–4). Nutrient-poor and acidic conditions, as well as low temperatures and decay-resistant Sphagnum moss-dominated vegetation, result in low rates of microbial decomposition of plant debris and net carbon sequestration in these ecosystems (5–12). However, carbon respiration has been shown to accelerate in subsurface peat due to climate warming in the subarctic (13) and in simulations of climate warming (14). Additionally, the decomposition of organic matter in anoxic peat layers of northern wetlands is also a significant source of methane in the atmosphere (15–18). Permafrost melt has been shown to result in a net carbon release in northern tundra, including methane emission from thaw lakes (19–22). The microbial community responsible for decomposition of Sphagnum-derived litter is unique compared to other soil systems and is important to the global carbon cycle (23–25). Further study on the physiology of this microbial community is needed to understand how it will respond to changing environmental conditions.
Planctomycetes have recently been observed to be abundant microbes in Sphagnum-dominated northern wetlands and appear to play a role in Sphagnum degradation (24, 26, 27). All currently described peat-inhabiting planctomycetes have the ability to degrade various heteropolysaccharides (28–32), but the addition of available nitrogen to cellulose-amended Sphagnum peat resulted in a decrease in the relative abundance of planctomycetes compared to the total microbial community (25). In a 16S rRNA gene pyrosequencing survey of a northern acidic Sphagnum-dominated wetland, Serkebaeva et al. (33) observed that planctomycetes contribute to higher percentage of bacterial 16S rRNA gene reads in the anoxic subsurface peat layer than in the surface layer. These studies suggest that wetland-inhabiting planctomycetes preferentially occupy anoxic niches of the peat and are also more suited to nutrient-poor conditions, probably contributing to the final stages of plant litter decomposition. The functional role of planctomycetes in these ecosystems, however, remains poorly understood.
Characterizing the cell membranes of bacteria is important in understanding how they are adapted to their niches as their membranes come into contact with the environment (34, 35). Intact polar lipids (IPLs) are the building blocks of cell membranes, consisting of a polar head group connected to nonpolar core lipids. IPLs are thought to represent living biomass, and their molecular structures can be taxonomically and environmentally specific, making them useful biomarkers (36, 37). Novel trimethylornithine lipids (TMOs) (see Fig. S1 in the supplemental material) have recently been characterized as abundant lipids in various isolates of Sphagnum wetland planctomycetes (38). Like ornithine lipids (OLs), TMOs are composed of a core containing esterified normal and beta-hydroxy (βOH) fatty acid core lipids. The occurrence of TMOs in the environment has not yet been confirmed. Here, we applied a combined approach including IPL and molecular analysis of peat cores collected from two northern wetlands in order to investigate the preferred niche and abundance of TMO-producing planctomycetes and to shed light on their potential role in the microbial community of this ecosystem.
MATERIALS AND METHODS
Sample collection.
Acidic peat samples were collected from two Sphagnum-dominated ombrotrophic (receiving water and nutrients solely from atmospheric precipitation) peat bogs: Obukhovskoye bog in the Yaroslavl region, European north Russia (58°14′N, 38°12′E) (25, 33), which was sampled at five depth intervals (5 to 10 cm, 10 to 20 cm, 20 to 30 cm, 30 to 40 cm, and 40 to 50 cm), and Saxnäs Mosse raised bog near the village of Lidhult, southwestern Sweden (56°51′20.78″N, 13°27′39.62″E) (collected by Weijers et al. 39), which was sampled at 2-cm intervals throughout the 54-cm core (see Fig. S2 in the supplemental material). Sphagnum angustifolium and Sphagnum fuscum were the predominant vegetation species in Obukhovskoye bog (25), while Sphagnum magellanicum and Sphagnum papillosum were most abundant in the Saxnäs Mosse bog (39). The pH was 4.0 to 4.2 throughout the 50-cm-depth peat core from the Obukhovskoye bog. The pH level in the Saxnäs Mosse core was not recorded; however, ombrotrophic bogs in central and northern Sweden typically range between pH 3.7 and 4.2 (40). The Obukhovskoye core water table reached 15 cm at its highest point and was continuously water saturated and anoxic below 30 cm. The water table of the Saxnäs core ranged from 14 to 25 cm. The top 14 cm of the Saxnäs core consisted of nondecomposed vegetation followed by 13 cm of more decomposed material, and finally the remaining core consisted of highly decomposed peat. Peat samples were stored at −20°C until further analysis.
Reference culture of a peat-inhabiting planctomycete.
The Isosphaera-like bacterium strain PX4, which was isolated from just above the oxic/anoxic interface (15 to 20 cm) of the Obukhovskoye peat bog and is capable of growth under micro-oxic conditions (I. S. Kulichevskaya and S. N. Dedysh, unpublished data), was also analyzed to compare its IPL composition with that of environmental samples. For lipid analyses, strain PX4 was grown in medium M31, containing (g per liter of distilled water) the following: KH2PO4, 0.1; Hutner's basal salts (41), 20 ml; N-acetylglucosamine, 0.5; glucose, 0.5; and yeast extract, 0.1 (pH 5.8). Cultivation under fully oxic conditions was performed in 500-ml flasks containing 200 ml medium M31 with shaking at 120 rpm for 2 weeks at 20°C. Strain PX4 was then cultured in triplicate under oxic and micro-oxic conditions to observe potential responses in IPL composition. For cultivation under micro-oxic conditions, medium M31 was boiled for 10 min to remove oxygen. After that, hermetically closed 500-ml flasks were filled with 450 ml medium M31, inoculated with strain PX4, and incubated under static conditions for 2 weeks. The dissolved O2 concentration was measured in cultivation flasks prior to inoculation by using a sensION6 dissolved oxygen meter (Hach, USA). The respective dissolved oxygen concentrations were 7.0 and 1.5 mg O2 in “oxic” and “micro-oxic” flasks, respectively. Culture biomass was freeze-dried and stored at −20°C until further analysis.
IPL extraction and analysis.
Saxnäs Mosse peat samples were extracted and the IPLs were analyzed by Peterse et al. (42). Obukhovskoye peat samples and biomass of Isosphaera-like strain PX4 were freeze-dried and ground to a powder with a mortar and pestle prior to extraction. Lipids were extracted from the freeze-dried powdered peat by a modification of the method of Bligh and Dyer (43, 44). A solvent mixture (approximately 5 ml g−1 [dry weight] peat) of methanol (MeOH)-dichloromethane (DCM)-potassium phosphate buffer (2:1:0.8, vol/vol/vol) at pH 7.4 was added to ca. 0.3 to 1.3 g (dry weight) of peat in a centrifuge tube and placed in an ultrasonic bath for 10 min. The extraction was repeated twice more, and the extracts were combined for each sample. DCM and phosphate buffer were added to the combined extracts to yield a ratio of 1:1:0.9 (vol/vol/vol) and achieve separation of a DCM phase and an aqueous MeOH-phosphate buffer phase by centrifugation at 2,500 rpm for 2 min. The DCM phase, containing the IPLs, was pipetted off, passed over extracted cotton wool to remove any remaining particles, and collected in a glass tube. The aqueous phase was rinsed twice with DCM, and the rinses were also passed over extracted cotton wool and combined with the original DCM phase. The combined DCM phase and rinses were dried under an N2 flow and stored at −20°C until analysis.
Extracted IPLs from the Obukhovskoye core and Isosphaera-like strain PX4 were analyzed by high-performance liquid chromatography–electrospray ionization-ion trap mass spectrometry (HPLC-ESI/IT/MS) as described by Sturt et al. (36), with some modifications (38). An Agilent 1200 series high-performance liquid chromatograph (Agilent, San Jose, CA) with a thermostated autoinjector was coupled to a Thermo LTQ XL linear ion trap mass spectrometer with an Ion Max source and ESI probe (Thermo Scientific, Waltham, MA). Chromatographic separation was performed on a Lichrosphere diol column (250 mm by 2.1 mm; 5-μm particles) (Grace Alltech Associates Inc., Deerfield, IL). The MS scanning mass range of m/z 400 to 2,000 in positive-ion mode was used, followed by data-dependent dual-stage tandem MS (MS2), in which the four most abundant masses in the mass spectrum were fragmented successively. Each MS2 was followed by data-dependent triple-stage tandem MS (MS3), wherein the base peak of the MS2 spectrum was fragmented. IPL abundance was assessed by integrating the HPLC-ESI/IT/MS base peak chromatogram area per gram (dry weight) of peat. Performance of the HPLC-ESI/IT/MS was monitored by regular injections of platelet-activating factor (PAF) standard (1-O-hexadecyl-2-acetyl-sn-glycero-3-phosphocholine). The absolute amount of IPLs in the Obukhovskoye 30- to 40-cm peat was measured using the PAF internal standard and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-methyl external standard. Student t tests were performed using the GraphPad t test calculator (GraphPad Software, Inc. La Jolla, CA) in order to identify statistically significant differences in the fractional abundances of IPLs under different growth conditions; P values of <0.05 were considered statistically significant.
DNA extraction, PCR amplification, and phylogenetic analysis.
Peat samples collected at 10 to 12, 16 to 18, 22 to 24, 24 to 26, 28 to 30, and 40 to 42 cm of the Saxnäs Mosse bog were defrosted on ice prior to extraction, and the water content was removed by centrifugation at 4,000 × g for 10 min before proceeding with the DNA extraction (quantification values are given per gram [dry weight] as remaining drained wet-weight material from the extraction was later freeze-dried and the correction applied). Peat samples from the Obukhovskoye core at 5 to 10, 10 to 20, 20 to 30, 30 to 40, and 40 to 50 cm were extracted from freeze-dried material. DNA was extracted with the DNA PowerSoil isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) with a final volume of 60 μl. Integrity and concentration of the extracted DNA were tested by agarose gel electrophoresis and NanoDrop (Thermo Scientific, Waltham, MA) quantification. Amplification of the 16S rRNA gene fragment from members of the Planctomycetes was performed with the primer pair Pla352F/Pla920R (45) with DNA extracted from the Saxnäs Mosse peat collected at depths of 16 to 18, 22 to 24, and 40 to 42 cm. Total bacterial 16S rRNA gene amplification was performed with the 341F/907R primer pair (46, 47) with DNA extracted from the Saxnäs Mosse peat at a depth of 22 to 24 cm. The PCR mixture was as follows (final concentrations): 1× Q solution (PCR additive; Qiagen), 1× PCR buffer, bovine serum albumin (BSA) (200 μg ml−1), deoxynucleoside triphosphates (dNTPs) (20 μM), primers (0.2 pmol μl−1), MgCl2 (1.5 mM), and 1.25 U Taq polymerase (Qiagen, Valencia, CA, USA). PCR conditions for these amplifications were as follows: 95°C for 5 min; 30 to 35 cycles of 95°C for 1 min, the melting temperature (Tm) (see Table S1 in the supplemental material for details) for 1 min, and 72°C, for 1 min; and a final extension at 72°C for 5 min. PCR products were gel purified (QIAquick gel purification kit; Qiagen, Valencia, CA, USA), cloned using the TOPO-TA cloning kit from Invitrogen (Carlsbad, CA, USA), and transformed into Escherichia coli TOP10 cells following the manufacturer's recommendations. Recombinant clone plasmid DNAs were purified and sequenced by Baseclear (Leiden, The Netherlands). Sequences were analyzed for the presence of chimeras using the Bellerophon tool (http://greengenes.lbl.gov/). The phylogenetic affiliation of the partial planctomycete 16S rRNA gene sequences was compared to release 119 of the Silva NR SSU Ref database (http://www.arb-silva.de/) (48) using the ARB software package (49). Sequences were added to the reference tree supplied by the Silva database using the ARB Parsimony tool.
qPCR analysis.
Quantitative PCR (qPCR) analyses were performed on a Bio-Rad CFX96 real-time system/C1000 thermal cycler equipped with CFX Manager software. The copy numbers of total bacterial and planctomycete 16S rRNA genes were estimated by using the primers mentioned above. The qPCRs were performed in triplicate with standard curves from 100 to 107 molecules per microliter. Standard curves were generated as described before (50). For general bacterial and planctomycetes 16S rRNA gene quantification, 16S rRNA gene fragments cloned from the 22- to 24-cm peat were used as standards (accession numbers KP161600 for bacteria and KP161571 for planctomycetes). Gene copies were determined in triplicate on diluted DNA extract. The reaction mixture (25 μl) contained 1 U of Pico Maxx high-fidelity DNA polymerase (Stratagene, Agilent Technologies, Santa Clara, CA, USA), 2.5 μl of 10× Pico Maxx PCR buffer, 2.5 μl of 2.5 mmol liter−1 of each dNTP, 0.5 μl BSA (20 mg ml−1), 0.02 pmol μl−1 of primers, 10,000 times diluted SYBR green (Life Technologies, Carlsbad, CA, USA) (optimized concentration), 0.5 μl of 50 mmol liter−1 MgCl2, and ultrapure sterile water. All reactions were performed in iCycler iQ 96-well plates (Bio-Rad, Hercules, CA, USA) with optical tape (Bio-Rad). One microliter of diluted environmental DNA was added to 24 μl of mix in each well. The specificity of the reaction was tested with a gradient melting temperature assay. The cycling conditions for the qPCRs were as follows: 95°C for 4 min; 40 to 45 cycles of 95°C for 30 s, the Tm (see Table S1 in the supplemental material for details) for 40 s, and 72°C for 30 s, and a final extension at 80°C for 25 s. Specificity for qPCRs was tested by agarose gel electrophoresis and with a melting curve analysis (50 to 95°C, with a read every 0.5°C held for 1 s between each read). Efficiencies and r2 values of the qPCR analysis are specified in Table S1 in the supplemental material.
PCR amplicon library preparation for pyrosequencing and analysis.
PCRs were performed with the universal (Bacteria and Archaea) primers S-D-Arch-0519-a-S-15 (5′-CAG CMG CCG CGG TAA-3′) and S-D-Bact-785-a-A-21 (5′-GAC TAC HVG GGT ATC TAA TCC-3′) (51) adapted for pyrosequencing by the addition of sequencing adapters and multiplex identifier (MID) sequences. Each 30-μl PCR mixture comprised 5× Phusion HF buffer containing 1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 μM each primer, and 1 U Phusion high-fidelity DNA polymerase (Thermo Scientific, Pittsburgh, PA). The following PCR conditions were used: initial denaturation at 98°C for 30 s, followed by 25 cycles consisting of denaturation (98°C for 10 s), annealing (53°C for 20 s), and extension (72°C 30 s), and a final extension step at 72°C for 7 min. To minimize PCR bias, three individual reactions were performed per template. PCR products were pooled, loaded on a 0.8% agarose gel, and purified using the Qiagen QIAquick gel extraction kit (Qiagen, Germany). PCR products were quantified with the Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Life Technologies, The Netherlands). Equimolar concentrations of the barcoded PCR products were pooled and sequenced on the GS FLX Titanium platform (454 Life Sciences) by Macrogen Inc., South Korea. Samples were analyzed using the QIIME pipeline (52). Raw sequences were demultiplexed and then quality filtered with a minimum quality score of 25 and length between 250 and 350 bp and allowing a maximum of two errors in the barcode sequence. Taxonomy was assigned based on Blast and the SILVA database (48, 53). Representative operational taxonomic unit (OTU) sequences assigned to the planctomycetes were extracted through “classify.seqs” and “get.lineage” in mothur (54) by using the bacterial aligned sequence and taxonomy file from the SILVA SSURef database (v102), and then they were added to the guided tree of release 119 of the Silva NR SSU Ref database as specified above.
Accession numbers.
Partial planctomycete 16S rRNA gene sequence data have been deposited in the NCBI GenBank database under accession numbers KP161502 to KP161600. The pyrosequencing reads (raw data) have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA286090.
RESULTS
Distribution and abundance of IPLs in peat bogs.
The distribution of IPLs in the Saxnäs Mosse core was previously described by Peterse et al. (42). Briefly, in the surface layers of both peat bogs, betaine IPLs were highly abundant, along with phosphatidylcholine lipids (PCs) and followed by TMOs (Table 1). Betaine lipids became relatively less abundant starting at 16 to 18 cm in the Saxnäs Mosse core and at 10 to 20 cm in the Obukhovskoye core and continued to decline with increasing depth (Table 1). PCs were highly to moderately abundant from 2 to 20 cm in the Saxnäs Mosse bog, were more abundant from 20 to 32 cm, and fluctuated as moderate- to low-abundance IPLs down the rest of the core (Table 1). PCs were abundant throughout the Obukhovskoye bog. Phosphatidylethanolamines (PEs) and monomethylphosphatidylethanolamines (MMPEs) were low in abundance or not detected at middle and deep parts of both cores (Table 1).
TABLE 1.
Relative abundances of the most prevalent IPLs and the biopolymer polyhydroxybutyrate in the Saxnäs Mosse and Obukhovskoye bogs
| Bog | Depth (cm) | Relative abundancea |
|||||
|---|---|---|---|---|---|---|---|
| TMO | PC | Betaine | PHB | MMPE | PE | ||
| Saxnäs Mosse | 2–4 | 16.7 | 12.9 | 30.7 | 2.7 | ||
| 4–6 | 15.6 | 11.2 | 35.4 | 2.5 | |||
| 6–8 | 14.1 | 8.1 | 22.0 | 4.9 | |||
| 10–12 | 12.5 | 13.1 | 13.4 | 4.0 | 5.6 | ||
| 12–14 | 20.1 | 8.8 | 11.5 | 2.5 | 7.0 | ||
| 14–16 | 18.4 | 8.5 | 13.5 | 1.5 | 8.1 | ||
| 16–18 | 34.8 | 5.9 | 14.6 | 5.1 | |||
| 18–20 | 36.9 | 4.1 | 11.2 | ||||
| 20–22 | 35.7 | 10.9 | 3.7 | ||||
| 22–24 | 45.4 | 15.6 | 2.7 | ||||
| 24–26 | 50.6 | 15.9 | 1.2 | 1.6 | |||
| 30–32 | 30.2 | 10.2 | 1.5 | 3.0 | |||
| 32–34 | 12.4 | 2.7 | 1.4 | 3.1 | |||
| 34–36 | 4.8 | 1.0 | 1.3 | 3.6 | |||
| 36–38 | 5.2 | 0.4 | 1.2 | 4.2 | |||
| 42–44 | 5.1 | 12.5 | 3.2 | 7.5 | |||
| 44–46 | 3.5 | 10.0 | 1.4 | 6.6 | |||
| 46–48 | 2.6 | 7.7 | 0.5 | 7.7 | |||
| 48–50 | 3.7 | 10.6 | 1.3 | 5.7 | |||
| 50–52 | 2.5 | 9.1 | 1.7 | 6.5 | |||
| 52–54 | 3.3 | 9.5 | 1.4 | 6.9 | |||
| Obukhovskoye | 5–10 | 6.1 | 23.5 | 14.9 | 15.2 | 0.7 | 1.3 |
| 10–20 | 2.3 | 13.5 | 3.2 | 19.1 | 5.5 | 1.4 | |
| 20–30 | 1.9 | 16.1 | 2.2 | 6.2 | 5.7 | 2.0 | |
| 30–40 | 22.6 | 23.8 | 0.7 | 1.9 | |||
| 40–50 | 2.3 | 22.3 | |||||
Ionization and apparent abundance can differ between different types of IPLs, and thus the observed abundances are relative and not absolute. TMO, trimethylornithine; PC, phosphatidylcholine; PHB, polyhydroxybutyrate; MMPE, monomethyl phosphatidylethanolamine; PE, phosphatidylethanolamine. The IPL absolute abundances in the Obukhovskoye bog at 30 to 40 cm peat were 0.131, 0.139, 0.004, 0.022, and 0.010 μmol/g (dry weight) for TMO, PC, betaine, MMPE, and lyso-PC, respectively.
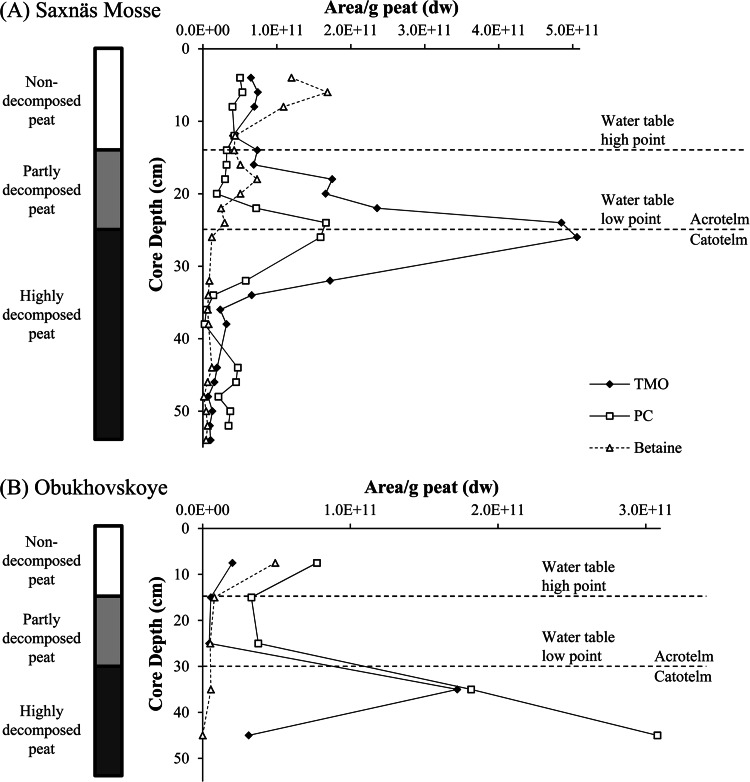
Trimethylornithine (TMO) IPLs were observed throughout both the Saxnäs Mosse and Obukhovskoye peat bogs in the oxic and anoxic layers (Fig. 1). The highest abundance of TMO IPLs was detected at the oxic/anoxic interface in both cores around and just below the water table low point (i.e., depths of approximately 25 and 30 cm, respectively) (Fig. 1). This was particularly evident in the Saxnäs Mosse bog given the higher sampling resolution (Fig. 1A). The oxic/anoxic interface is also the acrotelm (periodically water saturated oxic upper layer)/catotelm (continuously water saturated, anoxic lower layer) interface, where the transition from living vegetation (acrotelm) to dead plant material (catotelm) occurs. At these interface depths, TMOs are the most abundant IPL for the Saxnäs Mosse (24 to 26 cm) and second most abundant for the Obukhovskoye (30 to 40 cm) peat bogs, with PCs as the second most abundant IPL in Saxnäs Mosse interface peat and slightly more abundant than TMOs in Obukhovskoye interface peat (Table 1). The remaining IPLs identified at these depths were betaine and PE in the Saxnäs Mosse peat bog and betaine, MMPE, and lyso-PC in the Obukhovskoye peat bog (Fig. 2; Table 1).
FIG 1.
Relative abundances of trimethylornithine IPL (TMO), phosphatidylcholine IPL (PC), and betaine IPL based on HPLC-ESI/IT/MS chromatogram base peak area down core in Saxnäs Mosse peat (A) and Obukhovskoye peat (B). dw, dry weight.
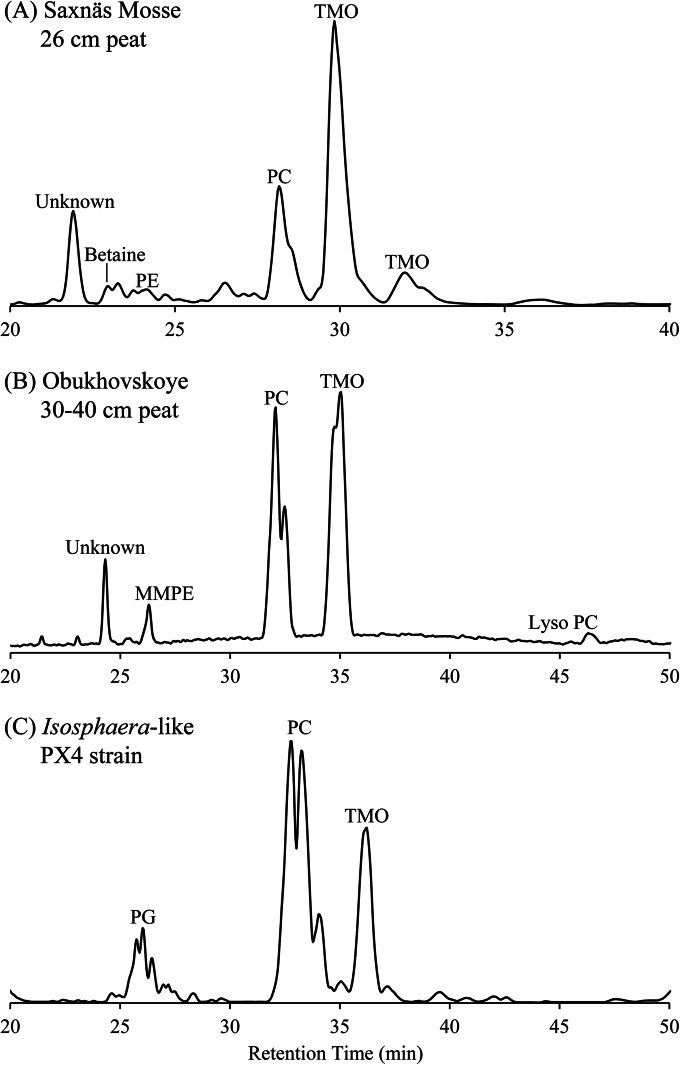
FIG 2.
HPLC-ESI/IT/MS base peak chromatogram of lipid extracts from Saxnäs Mosse peat at 24 to 26 cm (A), Obukhovskoye peat at 30 to 40 cm (B), and planctomycete Isosphaera-like strain PX4 isolated from Obukhovskoye bog (C). Retention times of IPLs in the various chromatograms are shifted due to slightly different chromatographic conditions used at the time of analysis. The unknown component was characterized by nonfragmentable m/z 680, 668, and 656 peaks and reported before by Peterse et al. (42). PC, phosphatidylcholine; TMO, trimethylornithine; MMPE, monomethylphosphatidylethanolamine; PG, phosphatidylglycerol.
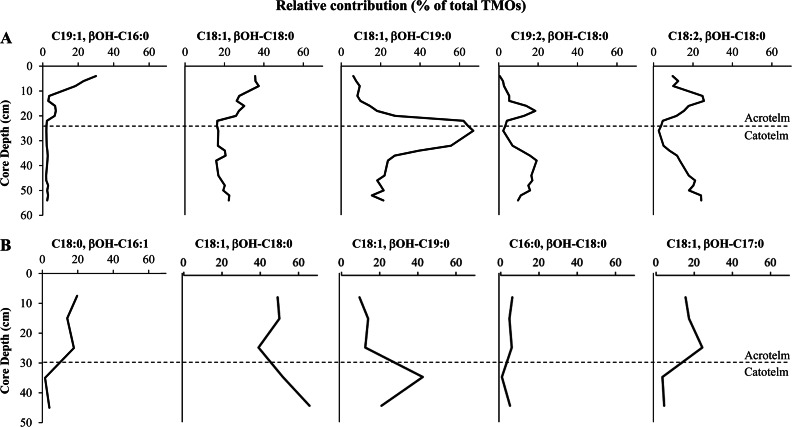
As the abundance of TMOs changed down core, the fatty acid composition of TMO lipids changed as well (Fig. 3). The most abundant TMO in the Saxnäs Mosse peat bog at the oxic/anoxic interface (where total TMOs peaked [Fig. 1]) contained predominantly (>60%) C18:1 and βOH-C19:0 fatty acids (Fig. 3A). The C18:1, βOH-C19:0 TMO also peaked (>40%) at the oxic/anoxic interface layer (30 to 40 cm) of the Obukhovskoye core, although not as highly as in the Saxnäs Mosse core (Fig. 3). One other individual TMO peaked near the oxic/anoxic interface of the Saxnäs Mosse core (C19:1, βOH-C19:0), but it made up only 5% of total TMOs at the interface depth. Two individual TMOs (core lipids C19:2, βOH-C18:0 and C18:2, βOH-C18:0) peaked above and below the oxic/anoxic interface of the Saxnäs Mosse core, and the most abundant individual TMOs in the near-surface layers (i.e., C19:1, βOH-C16:0 and C18:1, βOH-C18:0) declined with depth, followed by minor peaks around the oxic/anoxic interface. The C18:1, βOH-C18:0 TMO was the most consistently abundant from top to bottom in both cores, mostly ranging from 20 to 40% in the Saxnäs Mosse bog and from 39 to 65% in the Obukhovskoye bog. Other TMOs not described above followed similar trends in the Saxnäs Mosse core, with peaks above and below the oxic/anoxic interface and much lower contributions to total TMOs.
FIG 3.
Relative contributions (percentage of total TMO lipids) of the five most abundant TMO lipids down core in the Saxnäs Mosse (A) and Obukhovskoye (B) peat bogs.
IPLs of planctomycete strain Isosphaera sp. strain PX4.
The classes of IPLs identified in the Isosphaera-like strain PX4 lipid extract were similar to the IPLs identified at the oxic/anoxic interface in both peat bogs, particularly the Obukhovskoye bog (Fig. 2). PCs were the most abundant IPLs detected in strain PX4 lipid extract, followed by TMOs and phosphatidylglycerols (PGs) (Fig. 2C). By far the most abundant TMO core lipid fatty acids in the PX4 strain were C18:1, βOH-C18:0, which were also among the major TMO core lipids identified in both the Saxnäs Mosse and Obukhovskoye peat cores (Fig. 3). There was a statistically significant increase in the ratio of TMO/(PC + PG) IPLs in PX4 cultures grown under micro-oxic conditions compared to oxic conditions (Fig. 4A). There was also a statistically significant increase in the relative abundance of TMO-containing fatty acids C19:1 and βOH-C18:0 in biomass of strain PX4 grown under micro-oxic conditions compared to oxic conditions (Fig. 4B).
FIG 4.
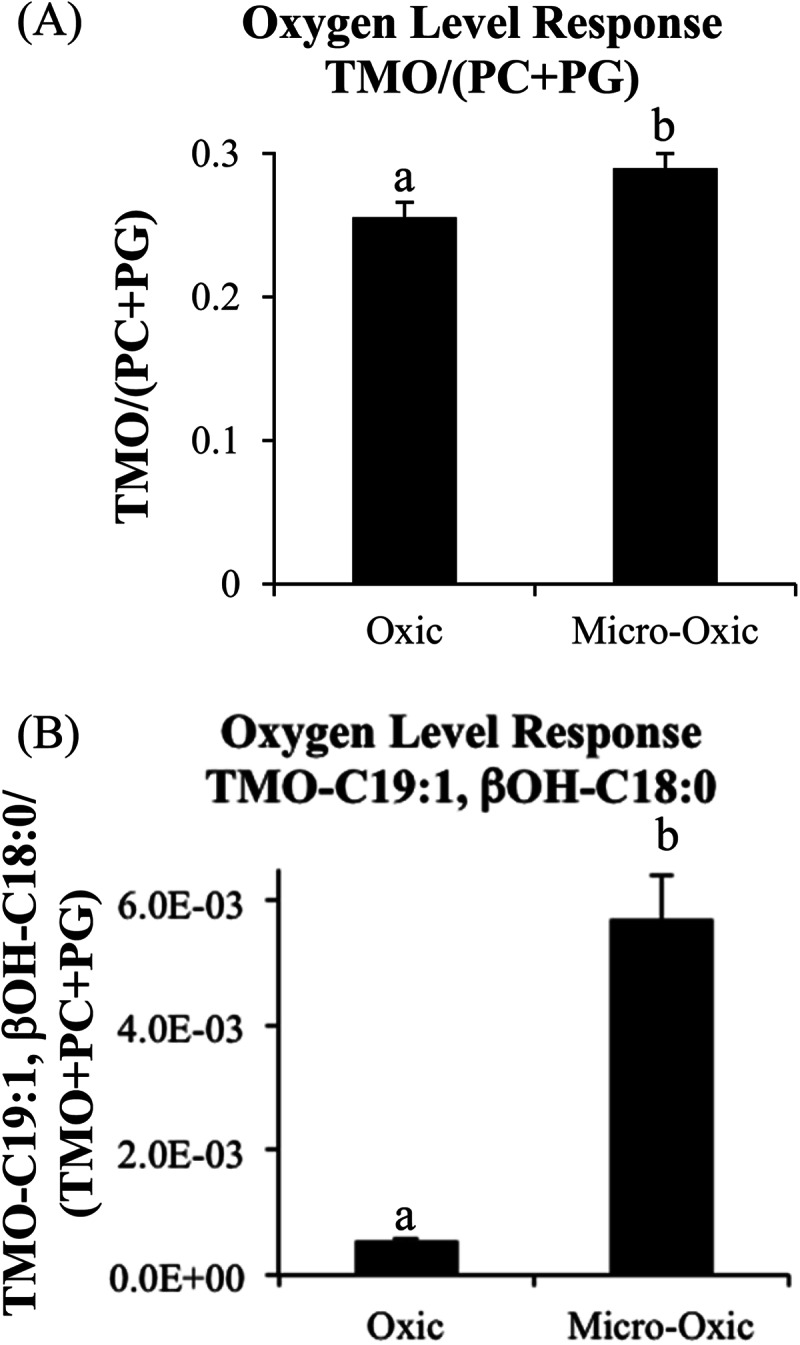
(A) Changes in Isosphaera-like strain PX4 IPL abundance when grown under oxic and micro-oxic conditions based on HPLC-ESI/IT/MS chromatogram base peak areas of TMO/(PC + PG) ratio (A) and TMO with fatty acids C19:1, βOH-C18:0/(total TMO + PC+ PG) (B). Differences between different growth conditions that are statistically significant by Student's t test are represented by letters (a and b) over each bar. TMO, trimethylornithine; PC, phosphatidylcholine; PG, phosphatidylglycerol.
Bacterial diversity determined by 16S rRNA gene amplicon pyrosequencing analysis.
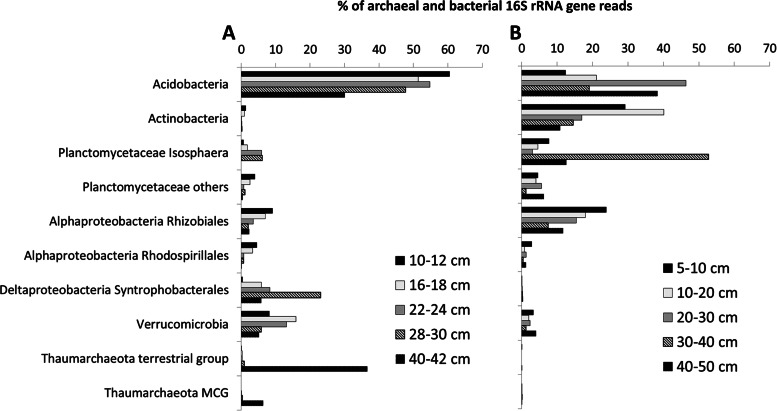
In order to determine the microbial diversity, partial 16S rRNA gene sequences were retrieved by pyrosequencing of the material sampled from depths of 10 to 12, 16 to 18, 22 to 24, 28 to 30, and 40 to 42 cm from the Saxnäs peat bog and from depths of 5 to 10, 10 to 20, 20 to 30, 30 to 40, and 40 to 50 cm from the Obukhovskoye core. In the Saxnäs peat bog at depths between 12 and 30 cm, bacterial 16S rRNA gene sequence reads comprised ca. 96% of the total reads, while in the deepest interval studied (40 to 42 cm), 52% of the reads were attributed to Bacteria and 48% to Archaea, specifically from the Thaumarchaeota terrestrial group (37%), and the miscellaneous Crenarchaeota group (8.5%) (Fig. 5A; see Table S2 in the supplemental material). Acidobacterial 16S rRNA gene sequences contributed on average 49% of the total reads at all depth intervals analyzed (Fig. 5A; see Table S2 in the supplemental material) and fell in Acidobacteria subgroups 1, 2, and 13 (see Fig. S3 in the supplemental material). Other bacterial 16S rRNA gene sequences attributed to Isosphaera-like planctomycetes, other planctomycetes, alphaproteobacterial families Rhizobiales and Rhodospirillales, deltaproteobacterial genus Syntrophobacter, and phylum Verrucomicrobia contributed on average 2 to 10% to the total reads (Fig. 5A; see Table S2 in the supplemental material). At the oxic/anoxic interface (22- to 24-cm depth) of the Saxnäs peat bog, total gene reads were distributed as follows: Acidobacteria, 55%; Planctomycetes Isosphaeraceae, 6%; Rhizobiales, 3.5%; Rhodospirillales, 0.7%; Synthrophobacter, 8%; and Verrucomicrobia, 13% (Fig. 5A; see Table S2 in the supplemental material).
FIG 5.
Percentages of total bacterial and archaeal reads attributed to different microbial groups detected in the Saxnäs Mosse peat bog at five different depths (10 to 42 cm) (A) and in the Obukhovskoye peat (five depths from 5 to 50 cm) (B) by 16S rRNA gene amplicon pyrosequencing analysis.
In the Obukhovskoye peat bog, Acidobacteria 16S rRNA gene reads contributed 27% of the total reads on average throughout the peat, followed by Actinobacteria reads (22%), Isosphaera-like planctomycetes (16%), Alphaproteobacteria Rhizobiales (15%), other planctomycetes (4%), and Verrucomicrobia (2%) (Fig. 5B; see Table S2 in the supplemental material). At the oxic/anoxic interface of the Obukhovskoye peat bog (maximum abundance of TMOs, 30- to 40-cm depth), a substantial change in the relative abundance of bacterial reads was observed with respect to the distribution above (20 to 30 cm) and below (40 to 50 cm) the interface, with the Isosphaera-like planctomycete 16S rRNA gene reads forming 53% of the total (Fig. 5B; see Table S2 in the supplemental material).
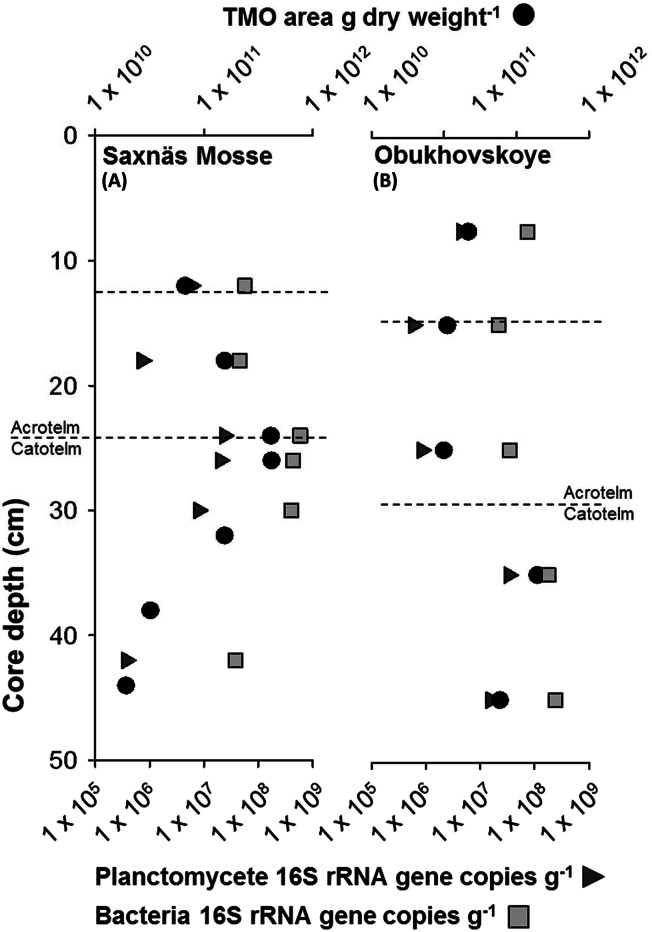
Planctomycete and total bacteria 16S rRNA gene abundances were quantified by quantitative PCR in seven peat intervals at depths between 10 and 42 cm in the Saxnäs Mosse core and in the 5 intervals between 5 and 50 cm in the Obukhovskoye peat bog. In the Saxnäs Mosse peat, the planctomycete 16S rRNA gene abundance ranged from 3.7 × 105 to 2.3 × 107 copies per gram (dry weight) of peat, with a maximum at the oxic/anoxic interface (average of 2.2 × 107 copies g−1) and a minimum at the deepest interval studied (i.e., 40 to 42 cm) (Fig. 6A). The bacterial 16S rRNA gene abundance was between 3.8 × 107 and 5.9 × 108 copies per gram of peat. Like in the case of the planctomycete 16S rRNA gene, the bacterial 16S rRNA gene abundance also showed maximum values at the oxic/anoxic interface (average of 5.1 × 108 copies g−1) and a minimum at the deepest interval studied (3.8 × 107 copies g−1) (Fig. 6A). In the Obukhovskoye peat, the planctomycete 16S rRNA gene abundance ranged from 6 × 105 copies g−1 (dry weight) at 10 to 20 cm to a maximum of 3 × 107 copies g−1 at the oxic/anoxic interface (maximum abundance of TMOs) at the 30- to 40-cm depth (Fig. 6B). The bacterial 16S rRNA gene abundance increased approximately 5-fold at 30 to 40 cm with respect to that at the uppermost layers. The maximum of bacterial 16S rRNA gene copies was detected between 40 and 50 cm (2.4 × 108 copies g−1 [dry weight]).
FIG 6.
Comparison of planctomycete and total bacterial 16S rRNA gene copy numbers per gram in comparison with total TMO abundance down core in the Saxnäs Mosse (A) and Obukhovskoye (B) peat bogs. Dashed lines indicate water table high and low points.
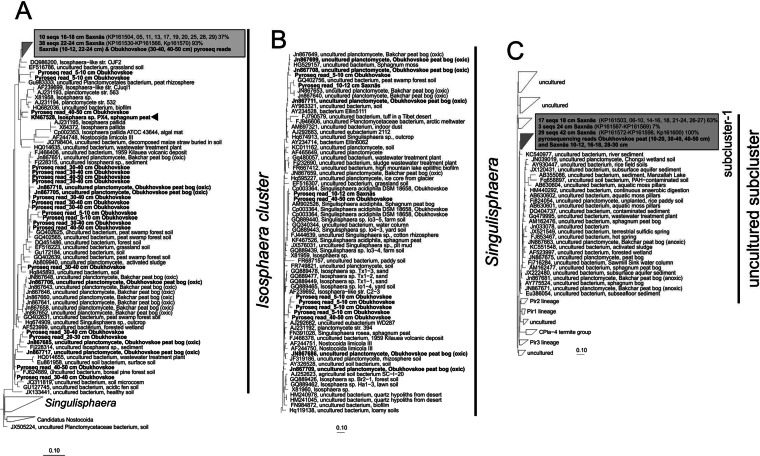
Planctomycete 16S rRNA gene fragments were amplified from the Saxnäs Mosse peat bog at intervals of 16 to 18 cm (acrotelm), 22 to 24 cm (oxic/anoxic interface), and 40 to 42 cm (catotelm) and cloned, and the obtained sequences were included in a phylogenetic tree together with the planctomycete reads retrieved by means of pyrosequencing analysis in both the Saxnäs Mosse and the Obukhovskoye peat bogs (Fig. 7). Approximately 93% (38 out of 41 clones) of the planctomycete sequences obtained from the Saxnäs Mosse peat oxic/anoxic interface belonged to the phylogenetic lineage defined by the genus Isosphaera (Fig. 7A). Some of the sequences included in this lineage have previously been retrieved from peat bogs, mainly from the oxic peat layer of the ombrotrophic bog Obukhovskoye (in bold, Fig. 7), and from the oligo-mesotrophic bog Bakchar (Fig. 7A) (27). The Isosphaera lineage also contained 37% of the sequences retrieved from the Saxnäs Mosse peat layer at 16 to 18 cm. On the other hand, all sequences obtained from the Saxnäs Mosse peat at 40 to 42 cm (n = 29 clones) fall in a subcluster (named here subcluster 1) (Fig. 7C) that is part of a lineage of uncultured planctomycete 16S rRNA gene sequences containing sequences previously retrieved from the anoxic peat layer of the Bakchar bog (27), in addition to many other environmental sequences (Fig. 7C). Approximately 63% of the sequences obtained from the Saxnäs Mosse peat at 16 to 18 cm and 7% of those obtained at 22 to 24 cm also group in this subcluster 1. Most of the representative reads of the pyrosequencing analysis from the Saxnäs Mosse peat layers at 10 to 12, 16 to 18, and 28 to 30 cm fell in subcluster 1 (Fig. 7C), while the reads from the 22- to 24-cm depth were part of the Isosphaera cluster (Fig. 7A). Pyrosequencing reads of the Obukhovskoye peat layers at 30 to 40 cm and 40 to 50 cm were also found in the Isosphaera cluster, while reads from the 5- to 10-cm sample were closely related to the Singulisphaera group, together with some reads of the Saxnäs Mosse peat sample at 10 to 12 cm and previously detected sequences from oxic parts of the Obukhovskoye and Bakchar peats. Pyrosequencing reads of the Obukhovskoye peat samples at 10 to 20, 30 to 40, and 40 to 50 cm also fall in subcluster 1 (Fig. 7C).
FIG 7.
Phylogenetic tree including the 16S rRNA gene sequences detected in the clone libraries of the Saxnäs Mosse peat bog at 16 to 18, 22 to 24, and 40 to 42 cm which are affiliated with planctomycete 16S rRNA gene sequences. Planctomycete 16S rRNA gene representative sequences obtained by pyrosequencing from the Saxnäs Mosse and the Obukhovskoye peat bog sediments are also included. (A) sequences closely related to the Isosphaera cluster; (B) sequences related to the Singulisphaera cluster; (C) sequences closely related to a cluster formed by uncultured planctomycetes. The scale bar indicates 0.10% estimated sequence divergence. Accession number of the sequences and percentages of sequences detected at a given depth by the clone libraries are indicated.
DISCUSSION
Abundance of Isosphaera-like planctomycetes at the oxic/anoxic interface of the Obukhovskoye and Saxnäs Mosse bogs.
A recent study estimated the bacterial diversity in the surface and subsurface layers of the acidic Sphagnum-dominated Obukhovskoye peat bog (33) and concluded that Acidobacteria, Proteobacteria, Actinobacteria, and Planctomycetes were the dominant phylum-level groups in both the oxic and anoxic zones of the peat. In our study, the percentage of Obukhovskoye peat bog reads attributed to Acidobacteria, Planctomycetes, Actinobacteria, and Rhizobiales were on average similar (15 to 27% of the total reads [see Table S2 in the supplemental material]), but it revealed that 16S rRNA reads attributed to Planctomycetes dominated at the oxic/anoxic interface (i.e., 53% of reads at 30 to 40 cm) (Fig. 5B). In the Saxnäs Mosse peat bog, we detected members of the Acidobacteria, Proteobacteria, Planctomycetes, and Verrucomicrobia as dominant groups, with Acidobacteria being represented on average by 50% of the total 16S rRNA gene reads (Fig. 5A). At this location, the percentage of 16S rRNA gene reads attributed to Planctomycetes was also highest at the oxic/anoxic interface (i.e., 7% of reads at 24 to 30 cm).
Total bacterial and planctomycete cell numbers were estimated, assuming that the average 16S rRNA copy number per bacterial cell is 3.6 (55), and that planctomycetes have an average of 2.5 copies of 16S rRNA gene per genome (56). Based on the copy numbers (Fig. 6), the maximum abundance of planctomycetes in the Saxnäs Mosse core was 2.2 × 107 cells per gram (dry weight) at the oxic/anoxic interface, with planctomycetes making up approximately 6% of the total bacterial cells. These values are comparable with those reported for diverse Sphagnum peat bogs in Russia by Ivanova and Dedysh (27) and in good agreement with the pyrosequencing data indicating that planctomycetes comprised 6% of the total reads at the oxic/anoxic interface. In the Obukhovskoye peat bog, the maximum abundance of planctomycetes at the 30- to 40-cm depth was estimated to be 1.3 × 107 cells per gram (dry weight) according to the same calculations, with planctomycetes accounting for 27% of the total bacterial cells.
Clone libraries and pyrosequencing indicated that at the oxic/anoxic interface of both the Saxnäs Mosse (24 to 26 cm) and the Obukhovskoye (30 to 40 cm) peat bogs there is also a marked change in the phylogenetic affiliation of the planctomycetes; i.e., almost all 16S rRNA gene reads and clone sequences were closely related to members of the Isosphaeraceae, whereas most other gene sequences recovered from different intervals were included in an uncultured planctomycete group (subcluster 1) (Fig. 7). Sequences of the Isosphaera group have previously been detected in both the oxic and anoxic parts of the Obukhovskoye peat bog (33). The only currently described member of the Isosphaera group (i.e., Isosphaera pallida) is aerobic, but the retrieval of environmental sequences from anoxic layers of the peat affiliated with this group suggests that other, uncultured Isosphaera species may be adapted to a microaerophilic or facultative anaerobic lifestyle. This would represent an advantage for this specialized planctomycete group to rapidly adapt to changing oxic/anoxic interfaces in peat bog systems. I. pallida is the type species of the genus Isosphaera (57, 58), but since I. pallida is a thermophilic planctomycete, it is deemed not to be relevant to northern wetlands. However, an Isosphaera-like bacterium, strain PX4, which was recently isolated from just above the oxic/anoxic interface of the Obukhovskoye peat bog and is capable of growth under micro-oxic conditions (Kulichevskaya and Dedysh, unpublished data), is phylogenetically related to the planctomycetes detected at the oxic/anoxic interface of the Saxnäs Mosse peat bog (Fig. 7A, triangle). Strain PX4 possesses a hydrolytic potential and is likely to be involved in the process of biopolymer degradation in peat (59).
TMO IPL production by Isosphaera-like planctomycetes.
TMO IPL abundance and planctomycete 16S rRNA sequences both peaked at the oxic/anoxic interface of the Obukhovskoye and Saxnäs Mosse peat bogs (Fig. 6). In addition, total bacterial abundance was also highest in those niches, suggesting that it is a hot spot for microbial activity, where planctomycetes play an important role. To date, planctomycetes are the only known TMO IPL producers in culture, including the species Singulisphaera acidiphila and S. rosea (38), which were isolated from Russian northern wetlands (29, 31) and are related to the Isosphaera group (Fig. 7). This suggests that the maximum TMO IPL abundance detected at the oxic/anoxic interfaces of two northern wetlands peat bogs may be attributed to Isosphaera-like planctomycetes thriving at the oxic/anoxic interface. Acidobacteria are more abundant at the oxic/anoxic interface of the Saxnäs Mosse peat bog (Fig. 5) and could potentially also be a source of TMO IPLs; however, previous studies analyzed the IPL composition of many acidobacterial species falling in subgroups 1, 3, 4, and 23 (60–62) and did not detect TMO IPLs. The 16S rRNA gene sequences retrieved in our analysis of the Saxnäs peat bog were closely related to Acidobacteria subgroups 1, 2, and 13 (see Fig. S3 in the supplemental material), and specifically, those included in subgroup 1 were closely related to previously tested strains with no TMO production capability. In addition, the percentage of reads attributed to acidobacteria decreased 2-fold (46 to 19%), and planctomycetes make up to 54% of the reads at the 30- to 40-cm depth in the Obukhovskoye peat, where maximum abundance of TMO was also detected. This evidence suggests that acidobacteria are not TMO producers but rather that Isosphaera-like planctomycetes are the most likely source of TMO lipids in this setting. This also applies to the Verrucomicrobia, which made up 13% of the total bacterial reads at the oxic/anoxic interface of the Saxnäs Mosse peat bog (Fig. 5A; see Table S2 in the supplemental material) but only 1.2% of the total reads in the Obukhovskoye peat at the peak of TMO depth. The Isosphaera-like strain PX4, which is closely related to the planctomycete 16S rRNA gene reads found at the oxic/anoxic interface of the Saxnäs Mosse and Obukhovskoye peats (Fig. 7A), contains TMO IPLs in high abundance (Fig. 2C), further supporting that the peak in TMO IPLs at the oxic/anoxic interfaces of the two peat bogs is due to the abundance of Isosphaera-like planctomycetes (Fig. 6).
Although TMO IPLs peak at the oxic/anoxic interface, these lipids can still be detected throughout the two peat bog cores (Fig. 1). Some of the most abundant Saxnäs Mosse and Obukhovskoye TMO core lipids (C19:1, βOH-C16:0; C18:1, βOH-C19:0; C19:2, βOH-C18:0; and C18:0, βOH-C16:1) have not yet been observed in planctomycete cultures and may be produced by uncultured species or result from adaptation to specific conditions in the peat. In addition, the most abundant TMO lipid at the oxic/anoxic interface of the Saxnäs Mosse bog (i.e., comprised of the C18:1 and βOH-C19:0 fatty acids) is likely derived from the Isosphaera-related species, since it clearly peaks at the oxic/anoxic interface of the peat (Fig. 3). Remarkably, the most abundant TMOs in northern wetland planctomycete species, including the Isosphaera-like strain PX4, do not contain the C18:1/βOH-C19:0 TMO in high abundance (Table 2). Apparently, the Isosphaera-like species thriving at the oxic/anoxic interfaces of the Saxnäs Mosse and Obukhovskoye peat bogs have different TMO compositions, which would be in line with the large variation in fatty acid composition of TMOs in planctomycetes (Table 2). Many of the other TMO lipids identified in the two peat cores (Fig. 3) have also been detected in cultured northern wetland planctomycetes (Table 2). Variations in the relative abundances of these TMOs (Fig. 3) are likely related to the changing composition of planctomycetes, which is evident from the genetic analyses (Fig. 7).
TABLE 2.
Relative abundance of the main TMO lipids of northern wetland planctomycetes
| Organism | Relative abundancea (% of total TMO abundance) of TMO with the indicated β-hydroxy fatty acid and regular fatty acid |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-16:0 |
β-17:0 |
β-18:0 |
β-20:0 |
|||||||||||||||||
| 14:0 | 16:0 | 16:1 | 16:0-OH | 18:0 | 18:1 | 18:2 | 20:1 | 16:1 | 18:1 | 16:0 | 16:1 | 16:2 | 18:1 | 18:2 | 19:0 | 19:1 | 20:1 | 18:1 | 20:1 | |
| Isosphaera-like PX4 strain | 3.8 | 6.8 | 81.9 | 2.2 | 5.3 | |||||||||||||||
| Singulisphaera acidiphila | 6.3 | 61.8 | 29.2 | 2.6 | ||||||||||||||||
| Singulisphaera rosea | 1.4 | 2.2 | 2.8 | 1.5 | 42.4 | 49.7 | ||||||||||||||
| Telmatocola sphagniphila | 20.9 | 12.1 | 23.0 | 20.2 | 9.9 | 13.8 | ||||||||||||||
| Gemmata-like SP5 strain | 2.6 | 26.8 | 5.8 | 6.7 | 47.1 | 8.5 | 2.5 | |||||||||||||
IPL and TMO abundances were assessed by integrating the HPLC-ESI/IT/MS base peak chromatogram area. Values for S. acidiphila, S. rosea, T. sphagniphila, and Gemmata-like SP5 strain TMO are from reference 38.
There are multiple lines of evidence supporting TMO production by members of the Isosphaera group, yet there is an apparent disproportionate contribution of TMOs to total IPLs versus the planctomycete-to-bacterium proportion at the oxic/anoxic interface of the Saxnäs Mosse peat (see Fig. S4 in the supplemental material). The relative abundance of TMOs makes up approximately 50% of total IPLs in the Saxnäs Mosse oxic/anoxic interface, but planctomycete cells accounted for only 6.6% of total bacterial cells at the same depth (24 to 26 cm [see Fig. S4 and Table S3 in the supplemental material; cell amounts are based on the 16S rRNA-to-cell conversion calculations described above]). Conversely, TMOs make up 22% of the total IPLs at the Obukhovskoye oxic/anoxic interface, and planctomycetes make up 27% of the total bacterial cells (see Fig. S4 and Table S3 in the supplemental material). The difference between the calculated percentage of planctomycete cells and the percentage of total bacterial 16S rRNA gene reads is probably due to differences in 16S rRNA gene copy number in the bacterial and planctomycete groups present at this depth at the two locations, inducing biases in the calculation of percentages of cells. In addition, we should also consider the possibility that PCR biases are introduced by the primers used for the quantification of 16S rRNA gene copies of bacteria and planctomycetes. The disproportionate amount of TMOs versus planctomycete cells at the Saxnäs Mosse oxic/anoxic interface (see Fig. S4 in the supplemental material) could be due to differences in the abundances of various microbial groups (Acidobacteria, Planctomycetaceae Isosphaera, etc.) in comparison with the Obukhovskoye bog (Fig. 5). Difficulty in extracting the membrane lipids of acidobacteria (60, 61), which represent 30 to 60% of all pyrosequencing reads in Saxnäs Mosse peat (Fig. 5A), could also result in underrepresentation of acidobacterial IPL contribution.
Purpose of TMO production.
The high relative abundances of TMOs and total bacterial cells at the oxic/anoxic interface suggests that there is some functional role of TMOs at this specific niche. Ornithine lipids (OLs) are relatively common among bacteria; approximately 50% of known bacterial species have the capability to produce ornithine lipids (63, 64). In certain bacteria, OLs can be produced in response to phosphorus limitation (65, 66) or modified in response to temperature or acid stress (67–69). TMOs are essentially modified OLs (38, 70); the addition of three methyl groups to the terminal nitrogen of TMOs results in a quaternary amine functional group, which is positively charged, making the lipid more polar and giving it a more cylindrical shape than conically shaped OLs, as observed in the methylation of conical PEs to yield cylindrical polar PCs. The increased relative abundance of TMOs compared to PCs and PGs and the increased relative abundance of TMOs with C19:1 and βOH-C18:0 fatty acids in PX4 strain cultures under micro-oxic growth conditions (Fig. 4) suggest that TMOs are used in response to low oxygen levels. As we hypothesized earlier (38), TMOs could be produced by northern wetland planctomycetes in order to provide greater membrane stability under rain-fed, acidic, low-nutrient conditions without using scarce phosphate. The high abundance of TMOs at the oxic/anoxic interface and increased relative TMO production in micro-oxic PX4 cultures indicate that there may be another niche-specific function of these lipids that is potentially linked to microaerophilic conditions and/or organic matter degradation.
Conclusions.
This study represents the first observation of TMOs in the environment. Initially discovered in northern wetland microbial isolates (38), it is now clear that TMOs constitute an important membrane lipid of microorganisms in northern European ombrotrophic bog ecosystems and possibly are an adaptation to the unique environmental conditions found at the oxic/anoxic interface. It still remains to be determined if these lipids are present in different types of peats or in other ecosystems. Future environmental and culture-based studies will be needed to tackle these questions.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Darwin Center for Biogeosciences grant number 142.16.3082. J.S.S.D. was supported by Gravitation grant SIAM (24002002) of the Netherlands Ministry of Education, Culture and Science and the Netherlands Science Foundation (NWO).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00324-15.
REFERENCES
- 1.Kivinen E, Pakarinen P. 1981. Geographical distribution of peat resources and major peatland complex types in the world. Ann Acad Sci Fenn 132:1–28. [Google Scholar]
- 2.Post WM, Emanuel WR, Zinke PJ, Stangenberger AG. 1982. Soil carbon pools and world life zones. Nature 298:156–159. doi: 10.1038/298156a0. [DOI] [Google Scholar]
- 3.Gorham E. 1991. Northern peatlands: role in carbon cycle and probable responses to climate warming. Ecol Appl 1:182–195. doi: 10.2307/1941811. [DOI] [PubMed] [Google Scholar]
- 4.Bain CG, Bonn A, Stoneman R, Chapman S, Coupar A, Evans M, Gearey B, Howat M, Joosten H, Keenleyside C, Labadz J, Lindsay R, Littlewood N, Lunt P, Miller CJ, Moxey A, Orr H, Reed M, Smith P, Swales V, Thompson DBA, Thompson PS, Van de Noort R, Wilson JD, Worrall F. 2011. IUCN UK Commission of Inquiry on Peatlands. IUCN UK Peatland Programme, Edinburgh, United Kingdom. [Google Scholar]
- 5.Clymo RS. 1965. Experiments on breakdown of Sphagnum in two bogs. J Ecol 53:747–758. doi: 10.2307/2257633. [DOI] [Google Scholar]
- 6.Moore PD, Bellamy DJ. 1974. Peatlands. Elek Science, London, United Kingdom. [Google Scholar]
- 7.Coulson JC, Butterfield J. 1978. An investigation of the biotic factors determining the rates of plant decomposition on blanket bog. J Ecol 66:631–650. doi: 10.2307/2259155. [DOI] [Google Scholar]
- 8.Clymo RS. 1984. The limits to peat bog growth. Philos Trans R Soc Lond B Biol Sci 303:605–654. doi: 10.1098/rstb.1984.0002. [DOI] [Google Scholar]
- 9.Johnson LC, Damman AWH. 1993. Decay and its regulation in Sphagnum peatlands. Adv Bryol 5:249–296. [Google Scholar]
- 10.Botch MS, Kobak KI, Vinson TS, Kolchugina TP. 1995. Carbon pools and accumulation in peatlands of the former Soviet Union. Global Biogeochem Cycles 9:37–46. doi: 10.1029/94GB03156. [DOI] [Google Scholar]
- 11.Verhoeven JTA, Liefveld WM. 1997. The ecological significance of organochemical compounds in Sphagnum. Acta Bot Neerl 46:117–130. [Google Scholar]
- 12.Aerts R, Wallén B, Malmer N, de Caluwe H. 2001. Nutritional constraints on Sphagnum—growth and potential decay in northern peatlands. J Ecol 89:292–299. doi: 10.1046/j.1365-2745.2001.00539.x. [DOI] [Google Scholar]
- 13.Dorrepaal E, Toet S, van Logtestijn RSP, Swart E, van de Weg MJ, Callaghan TV, Aerts R. 2009. Carbon respiration from subsurface peat accelerated by climate warming in the subarctic. Nature 460:616–619. doi: 10.1038/nature08216. [DOI] [Google Scholar]
- 14.Ise T, Dunn AL, Wofsy SC, Moorcroft PR. 2008. High sensitivity of peat decomposition to climate change through water-table feedback. Nat Geosci 1:763–766. doi: 10.1038/ngeo331. [DOI] [Google Scholar]
- 15.Matthews E, Fung I. 1987. Methane emissions from natural wetlands: global distribution, area, and environmental characteristics of sources. Global Biogeochem Cycles 1:61–86. doi: 10.1029/GB001i001p00061. [DOI] [Google Scholar]
- 16.Panikov NS. 1999. Fluxes of CO2 and CH4 in high latitude wetlands: measuring, modeling and predicting response to climate change. Polar Res 18:237–244. doi: 10.1111/j.1751-8369.1999.tb00299.x. [DOI] [Google Scholar]
- 17.Smith LC, MacDonald GM, Velichko AA, Beilman DW, Borisova OK, Frey KE, Kremenetski KV, Sheng Y. 2004. Siberian peatlands a net carbon sink and global methane source since the early Holocene. Science 303:353–356. doi: 10.1126/science.1090553. [DOI] [PubMed] [Google Scholar]
- 18.Yu ZC, Loisel J, Turetsky MR, Cai SS, Zhao Y, Frolking S, MacDonald GM, Bubier JL. 2013. Evidence for elevated emissions from high-latitude wetlands contributing to high atmospheric CH4 concentration in the early Holocene. Global Biogeochem Cycles 27:131–140. doi: 10.1002/gbc.20025. [DOI] [Google Scholar]
- 19.Zimov SA, Schuur EAG, Chapin FA. 2006. Permafrost and the global carbon budget. Science 312:1612–1613. doi: 10.1126/science.1128908. [DOI] [PubMed] [Google Scholar]
- 20.Walter KM, Zimov SA, Chanton JP, Verbyla D, Chapin FA. 2006. Methane bubbling from Siberian thaw lakes as a positive feedback to climate warming. Nature 443:71–75. doi: 10.1038/nature05040. [DOI] [PubMed] [Google Scholar]
- 21.Walter KM, Smith LC, Chapin FS. 2007. Methane bubbling from northern lakes: present and future contributions to the global methane budget. Philos Trans R Soc A 365:1657–1676. doi: 10.1098/rsta.2007.2036. [DOI] [PubMed] [Google Scholar]
- 22.Schuur EAG, Vogel JG, Crummer KG, Lee H, Sickman JO, Osterkamp TE. 2009. The effect of permafrost thaw on old carbon release and net carbon exchange from tundra. Nature 459:556–559. doi: 10.1038/nature08031. [DOI] [PubMed] [Google Scholar]
- 23.Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol 72:2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulichevskaya IS, Belova SE, Kevbrin VV, Dedysh SN, Zavarzin GA. 2007. Analysis of the bacterial community developing in the course of Sphagnum moss decomposition. Microbiologiia 76:702–710. [PubMed] [Google Scholar]
- 25.Pankratov TA, Ivanova AO, Dedysh SN, Liesack W. 2011. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ Microbiol 13:1800–1814. doi: 10.1111/j.1462-2920.2011.02491.x. [DOI] [PubMed] [Google Scholar]
- 26.Kulichevskaya IS, Pankratov TA, Dedysh SN. 2006. Detection of representatives of the Planctomycetes in Sphagnum peat bogs by molecular and cultivation approaches. Microbiology 75:389–396. [PubMed] [Google Scholar]
- 27.Ivanova AO, Dedysh SN. 2012. Abundance, diversity, and depth distribution of Planctomycetes in acidic northern wetlands. Front Microbiol 3:5. doi: 10.3389/fmicb.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulichevskaya IS, Ivanova AO, Belova SE, Baulina OI, Bodelier PLE, Rijpstra WIC, Sinninghe Damsté JS, Zavarzin GA, Dedysh SN. 2007. Schlesneria paludicola gen. nov, sp nov, the first acidophilic member of the order Planctomycetes from Sphagnum-dominated boreal wetlands Int J Syst Evol Microbiol 57:2680–2687. [DOI] [PubMed] [Google Scholar]
- 29.Kulichevskaya IS, Ivanova AO, Baulina OI, Bodelier PLE, Sinninghe Damsté JS, Dedysh SN. 2008. Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int J Syst Evol Microbiol 58:1186–1193. doi: 10.1099/ijs.0.65593-0. [DOI] [PubMed] [Google Scholar]
- 30.Kulichevskaya IS, Baulina OI, Bodelier PLE, Rijpstra WIC, Sinninghe Damsté JS, Dedysh SN. 2009. Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog. Int J Syst Evol Microbiol 59:357–364. doi: 10.1099/ijs.0.002378-0. [DOI] [PubMed] [Google Scholar]
- 31.Kulichevskaya IS, Detkova EN, Bodelier PLE, Rijpstra WIC, Sinninghe Damsté JS, Dedysh SN. 2012. Singulisphaera rosea sp. nov., a planctomycete from Sphagnum peat, and emended description of the genus Singulisphaera. Int J Syst Evol Microbiol 62:118–123. doi: 10.1099/ijs.0.025924-0. [DOI] [PubMed] [Google Scholar]
- 32.Kulichevskaya IS, Serkebaeva YM, Kim Y, Rijpstra WIC, Sinninghe Damsté JS, Liesack W, Dedysh SN. 2012. Telmatocola sphagniphila gen. nov, sp nov, a novel dendriform planctomycete from northern wetlands. Front Microbiol 3:146. doi: 10.3389/fmicb.2012.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serkebaeva YM, Kim Y, Liesack W, Dedysh SN. 2013. Pyrosequencing-based assessment of the bacteria diversity in surface and subsurface peat layers of a northern wetland, with focus on poorly studied phyla and candidate divisions. PLoS One 8:e63994. doi: 10.1371/journal.pone.0063994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper GM. 2000. The cell: a molecular approach, 2nd ed Sinauer Associates, Sunderland, MA. [Google Scholar]
- 35.Kujawinski EB. 2011. The impact of microbial metabolism on marine dissolved organic matter. Annu Rev Mar Sci 3:567–599. doi: 10.1146/annurev-marine-120308-081003. [DOI] [PubMed] [Google Scholar]
- 36.Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs KU. 2004. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry–new biomarkers for biogeochemistry and microbial ecology. Rapid Commin Mass Spectrom 18:617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- 37.Schubotz F, Wakeham SG, Lipp JS, Fredericks HF, Hinrichs KU. 2009. Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ Microbiol 11:2720–2734. doi: 10.1111/j.1462-2920.2009.01999.x. [DOI] [PubMed] [Google Scholar]
- 38.Moore EK, Hopmans EC, Rijpstra WIC, Villanueva L, Dedysh SN, Kulichevskaya IS, Wienk H, Schoutsen F, Sinninghe Damsté JS. 2013. Novel mono-, di-, and trimethylornithine membrane lipids in northern wetland planctomycetes. Appl Environ Microbiol 79:6874–6884. doi: 10.1128/AEM.02169-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weijers JWH, Schouten S, van der Linden M, van Geel B, Sinninghe Damsté JS. 2004. Water table related variations in the abundance of intact archaeal membrane lipids in a Swedish peat bog. FEMS Lett 239:51–56. doi: 10.1016/j.femsle.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Sjörs H, Bunnrsson U. 2002. Calcium and pH in north and central Swedish mire waters. J Ecol 90:650–657. doi: 10.1046/j.1365-2745.2002.00701.x. [DOI] [Google Scholar]
- 41.Staley JT, Fuerst JA, Giovannoni S, Schlesner H. 1992. The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata and Isosphaera, p 3710–3731. In Balows A, Truper H, Dworkin M, Harder W, Schleifer K-H (ed), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed Springer-Verlag, New York, NY. [Google Scholar]
- 42.Peterse F, Hopmans EC, Schouten S, Mets A, Rijpstra WIC, Sinninge Damsté JS. 2011. Identification and distribution of intact polar branched tetraether lipids in peat and soil. Org Geochem 42:1007–1015. doi: 10.1016/j.orggeochem.2011.07.006. [DOI] [Google Scholar]
- 43.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Canad J Biochem and Physiol 37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 44.Rütters H, Sass H, Cypionka H, Rullkötter J. 2002. Phospholipid analysis as a tool to study complex microbial communities in marine sediments. J Microbiol Methods 48:149–160. doi: 10.1016/S0167-7012(01)00319-0. [DOI] [PubMed] [Google Scholar]
- 45.Pollet T, Tadonleke RD, Humbert JF. 2011. Comparison of primer sets for the study of Planctomycetes communities in lentic freshwater ecosystems. Environ Microbiol Rep 3:254–261. doi: 10.1111/j.1758-2229.2010.00219.x. [DOI] [PubMed] [Google Scholar]
- 46.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muyzer G, Dewaal EC, Uitterlinden AG. 1993. Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S ribosomal RNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitcher A, Villanueva L, Hopmans EC, Schouten S, Reichart GJ, Sinninghe Damsté JS. 2011. Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone. ISME J 5:1896–1904. doi: 10.1038/ismej.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 54.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klappenbach JL, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res 29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Göker M, Cleland D, Saunders E. 2011. Complete genome sequence of Isosphaera pallida type strain (IS1BT). Stand Genomic Sci 4:63–71. doi: 10.4056/sigs.1533840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giovannoni SJ, Schabtach E, Castenholtz RW. 1987. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch Microbiol 147:276–284. doi: 10.1007/BF00463488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.International Committee on Systematic Bacteriology. 1995. Validation list no. 54. Int J Syst Bacteriol 45:619–620. doi: 10.1099/00207713-45-3-619. [DOI] [Google Scholar]
- 59.Naumoff DG, Ivanova AA, Dedysh SN. 2014. Phyolgeny of β-xylanases from Planctomycetes. Mol Microbiol 48:439–447. [PubMed] [Google Scholar]
- 60.Sinninghe Damsté JS, Rijpstra WIC, Hopmans EC, Weijers JWH, Foesel BU, Overmann J, Dedysh SN. 2011. 13,16-Dimethyl octacosanedioic acid (iso-diabolic acid), a common membrane-spanning lipid of Acidobacteria subdivisions 1 and 3. Appl Environ Microbiol 77:4147–4154. doi: 10.1128/AEM.00466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinninhe Damsté JS, Rijpstra WIC, Hopmans EC, Foesel BU, Wust PK, Overmann J, Tank M, Bryant DA, Dunfield PF, Houghton K, Stott MB. 2014. Ether- and ester-bound iso-diabolic acid and other lipids in members of Acidobacteria subdivision 4. Appl Environ Microbiol 80:5207–5218. doi: 10.1128/AEM.01066-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Losey NA, Stevenson BS, Busse HJ, Sinninghe Damsté JS, Rijpstra WIC, Rudd S, Lawson PA. 2013. Thermoanaerobaculum aquaticum gen. nov., sp nov., the first cultivated member of Acidobacterium subdivision 23, isolated from a hot spring. Int J Syst Evol Microbiol 63:4149–4157. doi: 10.1099/ijs.0.051425-0. [DOI] [PubMed] [Google Scholar]
- 63.Vences-Guzmán MA, Geiger O, Sohlenkamp C. 2012. Ornithine lipids and their structural modifications: from A to E and beyond. FEMS Microbiol Lett 335:1–10. doi: 10.1111/j.1574-6968.2012.02623.x. [DOI] [PubMed] [Google Scholar]
- 64.Vences-Guzmán MA, Guan Z, Escobedo-Hinojosa WI, Bermúdez-Barrientos JR, Geiger O, Sohlenkamp C. 7 August 2014. Discovery of a binfuctional acyltransferase responsible for ornithine lipid synthesis in Serratia proteamaculans. Environ Microbiol doi: 10.1111/1462-2920.12562. [DOI] [PubMed] [Google Scholar]
- 65.Weissenmayer B, Gao JL, Lopez-Lara IM, Geiger O. 2002. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol Microbiol 45:721–733. doi: 10.1046/j.1365-2958.2002.03043.x. [DOI] [PubMed] [Google Scholar]
- 66.Gao JL, Weissenmayer B, Taylor AM, Thomas-Oates J, Lopez-Lara IM, Geiger O. 2004. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol Microbiol 53:1757–1770. doi: 10.1111/j.1365-2958.2004.04240.x. [DOI] [PubMed] [Google Scholar]
- 67.Taylor CJ, Anderson AJ, Wilkinson SG. 1998. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology 144:1737–1745. doi: 10.1099/00221287-144-7-1737. [DOI] [PubMed] [Google Scholar]
- 68.Rojas-Jimenez K, Sohlenkamp C, Geiger O, Martinez-Romero E, Werner D, Vinuesa P. 2005. A CIC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol Plant Microbe Interact 18:1175–1185. doi: 10.1094/MPMI-18-1175. [DOI] [PubMed] [Google Scholar]
- 69.Vences-Guzman MA, Guan Z, Ormeno-Orillo E, Gonzalez-Silva N, Geiger O, Sohlenkamp C. 2011. Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol Microbiol 79:1496–1514. doi: 10.1111/j.1365-2958.2011.07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Escobedo-Hinojosa WI, Vences Guzmán MA, Schubotz F, Sandoval-Calderón M, Summons RE, López-Lara IM, Geiger O, Sohlenkamp C. 29 April 2015. OlsG (Sinac_1600) is an ornithine lipid N-methyltransferase from the planctomycete Singulisphaera acidiphila. J Biol Chem doi: 10.1074/jbc.M115.639575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.