Abstract
Drinking water assessments use a variety of microbial, physical, and chemical indicators to evaluate water treatment efficiency and product water quality. However, these indicators do not allow the complex biological communities, which can adversely impact the performance of drinking water distribution systems (DWDSs), to be characterized. Entire bacterial communities can be studied quickly and inexpensively using targeted metagenomic amplicon sequencing. Here, amplicon sequencing of the 16S rRNA gene region was performed alongside traditional water quality measures to assess the health, quality, and efficiency of two distinct, full-scale DWDSs: (i) a linear DWDS supplied with unfiltered water subjected to basic disinfection before distribution and (ii) a complex, branching DWDS treated by a four-stage water treatment plant (WTP) prior to disinfection and distribution. In both DWDSs bacterial communities differed significantly after disinfection, demonstrating the effectiveness of both treatment regimes. However, bacterial repopulation occurred further along in the DWDSs, and some end-user samples were more similar to the source water than to the postdisinfection water. Three sample locations appeared to be nitrified, displaying elevated nitrate levels and decreased ammonia levels, and nitrifying bacterial species, such as Nitrospira, were detected. Burkholderiales were abundant in samples containing large amounts of monochloramine, indicating resistance to disinfection. Genera known to contain pathogenic and fecal-associated species were also identified in several locations. From this study, we conclude that metagenomic amplicon sequencing is an informative method to support current compliance-based methods and can be used to reveal bacterial community interactions with the chemical and physical properties of DWDSs.
INTRODUCTION
Transmission of pathogens via contaminated water is a significant cause of illness worldwide. It has been estimated that one-third of gastrointestinal illnesses are caused by contaminated drinking water (1), and 4% of all deaths worldwide are due to polluted drinking water and poor sanitation (2). In developed nations water quality assessments and treatment facilities have been introduced to reduce microbial contamination, resulting in a significant reduction in drinking water-related illnesses and deaths. Water treatment commonly involves the reduction of organics and other contaminants via coagulation and sedimentation, separation of any remaining solids via filtration, and finally disinfection via chemical oxidants or ultraviolet (UV) radiation. The addition of chemical oxidants such as chlorine and monochloramine is the most common method of drinking water disinfection (3). The level of treatment required varies from system to system, with some drinking water distribution systems (DWDSs) receiving only one or two levels of treatment, while others require multiple treatments to create water suitable for end use.
Currently, most drinking water quality assessments do not directly taxonomically identify the many multifarious bacterial taxa present in drinking water systems. Instead, a variety of biological, physical, and chemical indicators are used to assess the efficiency of water treatment processes and the quality of the product water. These indicators quantify bacterial cells using flow cytometry or heterotrophic plate counts and measure concentrations of nutrients, such as ammonia, organic carbon, nitrates, and nitrites. Quantitative PCR (qPCR) can be used to directly quantify specific bacterial taxa (4), such as fecal indicators or known pathogens, but this does not allow measurements of whole-community diversity. Conversely, denaturing gradient gel electrophoresis (DGGE) can be used to estimate the biodiversity of bacterial communities (5), but individual taxa cannot be identified without additional expensive DNA sequencing.
Recently, metagenomic amplicon sequencing has emerged as a promising technique for characterizing complex biological communities as multiple taxa within communities deriving from multiple samples can be sequenced in a relatively rapid and inexpensive manner. Metagenomics has been successfully used to investigate bacterial communities present in end-use tap water, experimental drinking water systems, and bacterial biofilms within pipes (6–10). However, very few studies have explored bacterial communities throughout full-scale DWDSs or investigated community variations between different systems. In the present study, we characterize changes in bacterial community structure at different locations along two full-scale DWDSs and also determine the viability of metagenomic sequencing as a monitoring tool for assessing system efficiency and water quality. We also measured various water quality parameters at the same sample locations to validate the metagenomic findings. Using these approaches, bacterial diversity, water quality, and treatment efficiency were assessed at multiple sites along two distinct DWDSs: (i) a linear DWDS in Western Australia (WA) that undergoes monochloramine disinfection before distribution and (ii) a complex, branching DWDS in South Australia (SA) that is treated by a four-stage water treatment plant (WTP) prior to monochloramine disinfection and distribution. From this metagenomic data set, we aim to guide future management and water quality monitoring strategies and potentially reduce associated human health risks by extending the current knowledge of bacterial communities in DWDSs.
MATERIALS AND METHODS
Site descriptions. (i) WA DWDS.
The WA DWDS was chosen for this study because it is a simple, linear system with minimal catchment runoff present in the source water. A large proportion of the catchment area is state-owned and -managed forest, and the remaining land is primarily low-intensity agricultural land, such as grazing pastures. The water is a blend of groundwater and desalinated seawater and is supplemented from the metropolitan system. The source water undergoes disinfection with monochloramine before distribution. Areas local to the reservoir are supplied via several large storage tanks (Fig. 1).
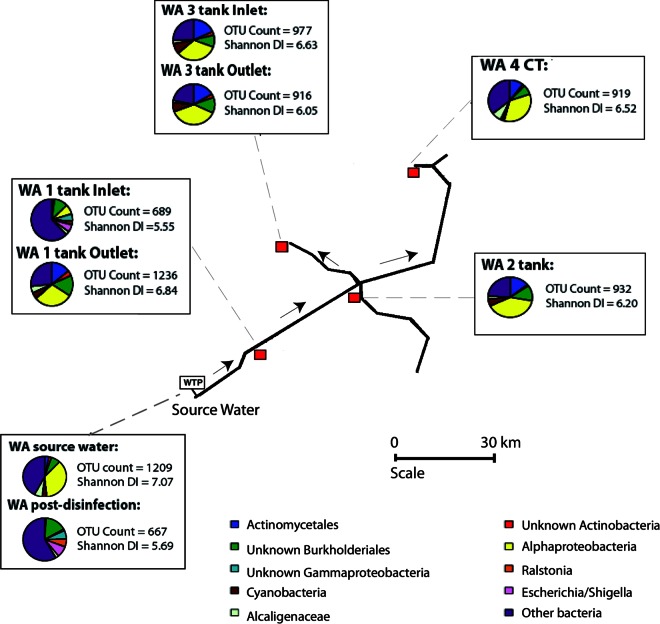
FIG 1.
A diagram of the bacterial diversity within the WA DWDS, including pie charts that depict the 10 most prolific bacterial taxa. WTP, water treatment plant (monochloramine dosing only); CT, customer tap. Red squares indicate sample collection locations. The WA reservoir is fed with a mixture of groundwater and desalinated water and has no physical filtration system. The WA WTP utilizes monochloramine. DI, diversity index.
(ii) SA DWDS.
The SA DWDS is a larger, more complex system with multiple distinct distribution branches (Fig. 2). The source water is supplied from river water with the catchment area passing through state-owned forests and agricultural areas that produce wool, cotton, wheat, cattle, pigs, dairy, rice, wine, fruit, and vegetables. The source water is treated within a four-stage water treatment plant (WTP), with stages consisting of coagulation, flocculation, sedimentation, and filtration, followed by UV disinfection and monochloramine disinfection before distribution.
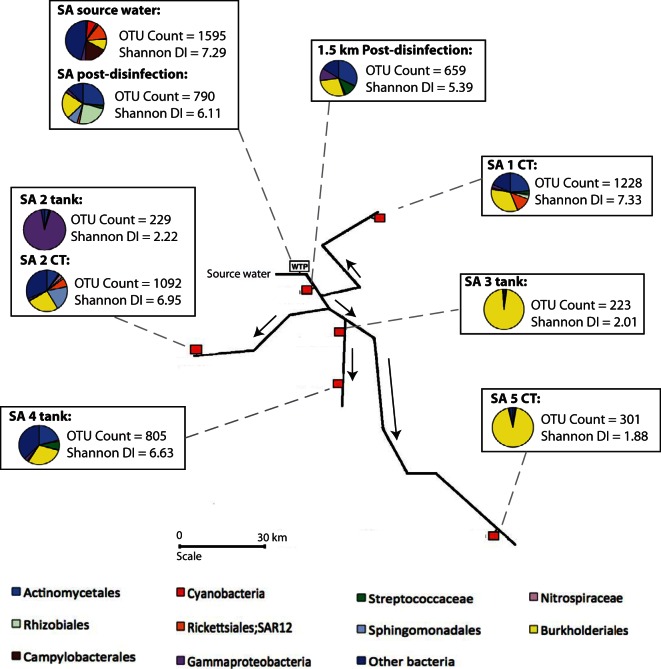
FIG 2.
A diagram of the bacterial diversity within the SA DWDS, including pie charts that depict the 11 most prolific bacterial taxa. WTP, water treatment plant; CT, customer tap. Red squares indicate sample collection points. The SA system utilizes river water as the source and uses a four-stage treatment process, followed by ultraviolet (UV) and monochloramine disinfection.
Sample collection.
For each system, 600-ml water samples were collected from the source water, from within the treatment plant (WTP), and also at each sample point indicated in Fig. 1 and 2. Sample locations covered the expanse of the distribution systems from the source water through to various township storage tanks and customer taps (CTs). For three of the end-user locations, samples were collected from within the township storage tank (tank) and also from a customer tap (CT) after the water had left the tank (tank outlet tap or a nearby CT) to assess whether bacterial diversity was affected by tank storage. In the SA system, additional WTP and source water samples were collected 2 weeks later to determine if bacterial communities are subject to short-term temporal fluctuations. Samples were collected in sterile bottles, immediately placed on ice, and transported to dedicated laboratory facilities, where they were stored at 4°C and processed within 24 h of collection.
Water quality analyses.
Traditional, compliance-based water quality analyses, such as nutrient and chemical analyses, flow cytometry, and quantitative PCR amplification of nitrification-associated genes, were conducted for each of the samples. Classical chemical indicators for nitrification, such as free ammonia, nitrate, nitrite, and monochloramine levels, were measured using standard protocols (11). Flow cytometry (FCM), in combination with a BacLight bacterial viability kit (Molecular Probes, USA), was utilized to enumerate the bacterial cells present in the water, as previously described (12). For the FCM analyses, duplicate 500-μl aliquots were subsampled from each of the 600-ml samples, with one aliquot stained with SYTO9 to stain both intact (live) and membrane-damaged (dead) cells while the other aliquot was stained with both SYTO9 and propidium iodide (which stains only membrane-damaged cells). FCM enumeration using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) was performed separately to count the total cell population (stained with SYTO9 alone) and live cells (the population of cells stained with SYTO9 but not propidium iodide). In order to calculate cell densities, sample tubes were weighed pre- and postanalysis to allow calculation of the volume analyzed. The resulting FCM data were analyzed using CellQuest software (Becton Dickinson, USA). The percent viable cells (Table 1) was calculated as follows: live cell count per milliliter/total cell count per milliliter × 100.
TABLE 1.
Sample names and descriptions of water quality for each system
| System and sample | Sample description | pH | Turbidity (NTU)a | DOC (mg/liter)b | NH2Cl (mg/liter)c | Nitrite as N (mg/liter) | Nitrate as N (mg/liter) | AOB presenced | AOA presenced | % viable cellse | Total no. of cells/ml |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WA system | |||||||||||
| Source water | Raw, untreated inlet | 7.6 | 0.49 | 1.70 | <0.1 | <0.003 | 0.049 | − | +++ | 58.8 | 1.66E+06 |
| Postdisinfection | Posttreatment | 8.2 | 0.83 | 2.10 | 3.53 | 0.004 | 0.036 | − | + | 3.4 | 2.72E+06 |
| WA-1 inlet | Storage tank inlet | 8.3 | 0.85 | 2.20 | 3.59 | 0.007 | 0.036 | + | ++ | 3.9 | 2.64E+06 |
| WA-1 outlet | Storage tank outlet | 8.2 | 0.74 | 2.20 | 3.34 | 0.004 | 0.045 | + | +++ | 3.9 | 2.76E+06 |
| WA-2 tank | Storage tank | 8.2 | 1.01 | 2.50 | 3.11 | 0.004 | 0.042 | + | ++ | 4.5 | 2.78E+06 |
| WA-3 inlet | Storage tank inlet | 8.4 | 0.35 | 2.20 | 2.84 | 0.005 | 0.047 | + | ++ | 2.7 | 2.89E+06 |
| WA-3 outlet | Storage tank outlet | 8.4 | 0.33 | 2.30 | 2.52 | 0.006 | 0.053 | ++ | ++ | 3.8 | 2.00E+06 |
| WA-4 CT | Customer tap | 8.3 | 0.24 | 2.10 | 2.46 | 0.413 | 0.303 | +++ | + | 3.3 | 2.52E+06 |
| SA system | |||||||||||
| Source water | Raw, untreated inlet | 7.4 | 77.0 | 12.6 | <0.1 | 0.012 | 0.184 | +++ | +++ | 58.1 | 1.98E+07 |
| Postdisinfection | Posttreatment | 8.4 | 0.40 | 5.40 | 4.7 | 0.013 | 0.253 | + | +++ | 13.6 | 4.83E+05 |
| 1.5 km postdisinfection | 1.5-km point postdisinfection | 8.2 | 0.20 | 5.20 | 3.6 | 0.009 | 0.269 | + | ++ | 4.9 | 3.08E+05 |
| SA-1 CT | Customer tap | 8.9 | 0.10 | 5.00 | 1.0 | 0.059 | 0.600 | + | +++ | 11.5 | 1.24E+05 |
| SA-2 tank | Storage tank | 8.3 | 0.20 | 5.50 | 1.4 | 0.054 | 0.488 | + | ++ | 5.1 | 2.14E+05 |
| SA-2 CT | Customer tap | 7.9 | 0.20 | 4.90 | <0.1 | 0.009 | 1.150 | ++ | ++ | 50.0 | 6.90E+05 |
| SA-3 tank | Storage tank | 8.5 | 0.60 | 5.80 | 2.4 | 0.021 | 0.338 | + | − | 2.3 | 3.59E+05 |
| SA-4 tank | Storage tank | 8.5 | 8.00 | 5.50 | 2.4 | 0.026 | 0.396 | + | +++ | 5.3 | 3.23E+05 |
| SA-5 CT | Customer tap | 9.0 | 0.30 | 4.10 | 2.1 | 0.022 | 0.464 | ++ | +++ | 51.6 | 3.80E+04 |
NTU, nephelometric turbidity unit.
DOC, dissolved organic carbon.
Monochloramine.
The presence of ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (A) was semiquantified using the height of the DNA melting peaks. The standard was nominally scored as high (+++), with weak peaks scored as +, peaks between the weak and control levels as ++, and negative PCR results as −.
See Materials and Methods for the method of calculation.
Basic qPCR assays were used for the semiquantification of the number of ammonia-oxidizing bacteria (AOB) and archaea (AOA) in each sample using the DNA extracts, described below. The assays used primers for AOA and AOB as previously described (13), except that the PCR reagents were replaced with Fast Start Essential Probe Master Mix (Roche) with the addition of 3.3 μM SYTO9 (final concentration; Molecular Probes). The amplifications were conducted on a Rotor-Gene 6000 HRM (high-resolution melt) system (Qiagen) using the recommended cycling conditions, followed by DNA melting curve analysis on the HRM channel from 70°C to 95°C in 0.2°C increments. The first-derivative melting profiles were visualized with the digital filter set to “none.” The presence of nonspecific amplicon prevented conventional quantification using the amplification curves. Instead, the presence of AOA/AOB was semiquantified using the height of the DNA melting peaks relative to the peak in the positive-control reaction containing 104 copies of the target fragment.
Metagenomic sequencing. (i) DNA extraction.
Within 24 h of collection, three 50-ml subsamples from each of the 600-ml water samples were centrifuged at 4,000 rpm for 30 min to pellet the intact bacterial cells, and the supernatant was discarded. The pellet from each of these subsamples was resuspended in 180 μl of sterile nuclease-free water (Sigma-Aldrich, Castle Hill, Australia) and transferred to a MoBio Ultraclean Soil DNA Isolation Kit (MoBio Laboratories, Inc., Solana Beach, CA). DNA was extracted alongside extraction blanks consisting of 180 μl of sterile nuclease water, according to the manufacturer's recommendations. Once the extractions were complete, DNA yield was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and the replicate subsamples were combined to reduce sequencing costs, resulting in 10 samples for the SA system and 8 samples for the WA system (Table 1).
(ii) PCR amplification and NGS.
Amplicon libraries were prepared in dedicated sterile PCR hoods. Prior to the addition of GoTaq, the PCR master mix was subjected to ultraviolet radiation for 15 min to reduce contamination from laboratory bacteria (14). Sample DNA was added in a second sterile hood in a separate room. A two-step nested-PCR approach was adopted, first amplifying a long DNA fragment prior to amplifying a short fragment appropriate for amplicon-based next-generation sequencing (NGS). This was done to minimize the contribution of highly degraded, uninformative, and contaminant DNA (15). A real-time quantitative PCR (RT-qPCR) platform was used for the PCR amplification step in order to enable comparison of DNA melt curves and to determine if samples were contaminated by DNA present in reagents and plasticware. In the first RT-qPCR, universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACCTTGTTACGACTT-3′) were used to amplify a 1,500-bp fragment within the 16S rRNA gene (12). Reaction mixtures of 25 μl were prepared in quadruplicate and pooled post-PCR to reduce PCR bias (16). Each reaction mixture contained 5 μl of DNA extraction template, 5% dimethyl sulfoxide (DMSO), 3.3 μM SYTO9 dye, 0.4 U μl−1 of GoTaq (Promega), a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), a 0.5 μM concentration of each primer, 1× PCR buffer, 2.5 mM MgCl2, and 4.4 μl of nuclease-free water. Reaction mixtures were amplified using the following parameters: 95°C for 3 min, followed by 35 cycles at 94°C for 30 s, 50°C for 60 s, and 72°C for 120 s. In the second RT-qPCR, a smaller 176-bp region from within the 1,500-bp PCR product was amplified using bar-coded Ion Torrent fusion primers: 341F (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGxxxxxxxCCTACGGGAGGCAGCAG-3′; x represents the bar code) and 518R (5′-CCTCTCTATGGGCAGTCGGTGATATTACCGCGGCTGCTGG-3′). Reaction mixtures were prepared as described above. The second round of amplification was performed with the following parameters: 95°C for 3 min, followed by 35 cycles at 95°C for 20 s, 65°C for 20 s, and 72°C for 30 s. PCR products were purified (Ampure; Agencourt Bioscience), quantified, and pooled to equimolar concentrations before being sequenced on an Ion Torrent Personal Genome Machine using an Ion PGM 200 sequencing kit (Life Technologies, Carlsbad, CA, USA).
Data analysis.
Reads that contained zero mismatches within the bar code and primer sequence were demultiplexed into their respective samples, and bar codes and primer sequences were trimmed using CutAdapt, version 1.1 (17). Reads were then filtered for quality (Phred threshold of 20 for 90% of sequence base pairs) and length (>100 bp) using the FASTX-Toolkit (version 0.0.13 [http://hannonlab.cshl.edu/fastx_toolkit]). The resulting files were converted to a QIIME software-compatible fna (fast nucleic acid) file (script available from http://www.u.arizona.edu/∼gwatts/azcc/QIIMEfastaFormatter.pl) and imported into QIIME (18).
Within QIIME, sequences were clustered to 97% similarity to form operational taxonomic units (OTUs) and aligned using Pynast. Chimeric reads were detected and removed using ChimeraSlayer (19). The standard default threshold (0.8) was applied to assign taxonomy using the Greengenes database (version gg_12_8; [http://greengenes.lbl.gov]). Contaminant OTUs identified in the extraction blanks were removed from the samples. All samples were rarefied at 8,904 sequences (i.e., the lowest number of reads present within a single sample) to ensure an even read coverage for each sample and to minimize skewed diversity estimates. Diversity estimates were obtained for each sample by calculating Chao species richness and Shannon-Weiner diversity indices. The number of operational taxonomic units (OTUs) per sample was also measured. Sequences were also reclustered at 98% similarity, and taxonomy was reassigned to improve phylogenetic resolution for some closely related species. Further, sequences from the specific genera of interest were extracted from the sequence files and then blasted against the NCBI database (accessed 17 October 2014) to allow further phylogenetic discrimination between closely related taxa. Exploratory and statistical analyses of the data were carried out using QIIME, PASW Statistics, version 18 (SPSS, Inc., Chicago, IL), and Explicet (20). Jackknifed trees using the unweighted-pair group method using average linkages (UPGMA) based on weighted hierarchal clustering (6,670 replicate sequences; E value, 10) were constructed to visualize sample similarity based on phylogenetic distance between taxa (UniFrac software) (21) and were viewed using FigTree, version 1.4, software (tree.bio.ed.ac.uk/software). Heat maps were constructed within Explicet to visualize dominant taxa (>0.1%) for each sample and determine dissimilarity between samples.
Microarray data accession number.
Raw data files were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA287069.
RESULTS
Treatment effectiveness.
To assess the efficiency of DWDS treatments, bacterial communities pre- and posttreatment were compared. Metagenomic amplicon sequencing revealed that the bacterial communities were significantly altered after treatment in both systems (P = < 0.001; Mann-Whitney U test), demonstrating the impact that current water treatment technologies can have on bacterial communities. The source water samples had the highest bacterial diversity in both systems (Fig. 1 and 2), and corresponding FCM data revealed that both systems' source water had similar proportions of viable (active) bacterial cells (approximately 58%). In contrast, the first samples collected posttreatment had a significantly lower OTU count (P = < 0.001 for both systems; Mann-Whitney U test) and a reduced percentage of viable cells (3.4% for the WA system and 13.6% for the SA system). Although the percentage of viable cells decreased in the WA system posttreatment, the FCM data revealed that the total number of bacterial cells remained the same as that of the source water, whereas the water treatment in the SA system reduced the total bacterial cell count by almost 2 orders of magnitude (Table 1). The difference in total cell counts postdisinfection is likely due to the four-stage treatment process within the SA system that removes both the live and inactive (deceased) bacterial cells from the water, whereas in the WA system the inactive cells remain in the water. The SA system's source water had a marginally higher diversity index and OTU count than that of the WA system (Shannon diversity index, 7.29 and 7.07 for SA and WA, respectively; OTU count, 1,596 and 1,209 for SA and WA, respectively), and postdisinfection the SA system still had a higher diversity index and OTU count than the WA system, which is likely due to the higher starting diversity and OTU counts (Fig. 1 and 2).
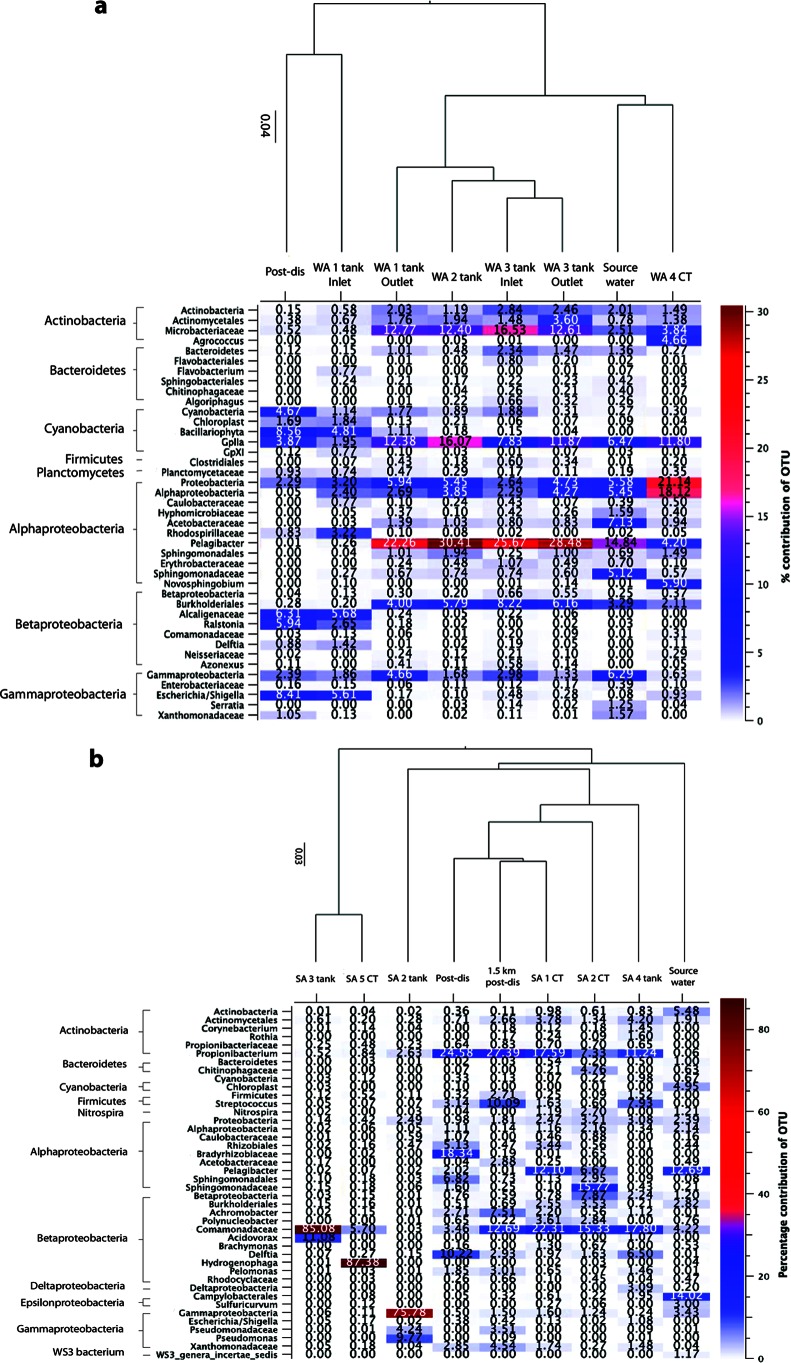
In both systems, the source water was dominated by unknown bacterial taxa (both systems, 27%), emphasizing the limitations of current knowledge (Fig. 3). The community structures in the WA and SA source water samples were different from one another, with some taxa present in one system but undetectable in the other. For example, bacteria in the order Campylobacterales were highly abundant in the SA source water (∼14%) but were negligible in the WA source water (<1%). Similarly, the proportions of Acetobacteraceae and Sphingomonadales in the WA source water were relatively high (∼7% and ∼6%, respectively) compared to those in the SA source water (<1% for both). In the future, this type of information could lead to more effective treatment strategies, customized for the unique diversity present in different source waters. Postdisinfection, the bacterial community structure in the two systems also differed; however, these differences were much less pronounced. In the SA postdisinfection sample, there was a relatively high proportion of Sphingomonadales (8%), Rhizobiales (22%), and Streptococcus (3%), which were negligible in the WA system's postdisinfection sample. Instead, cyanobacteria dominated the WA postdisinfection sample, whereas in the SA postdisinfection sample cyanobacteria were present only in low abundances (<1%). The high abundance of cyanobacteria in the WA postdisinfection sample suggests that chloramination alone is not efficient at destroying robust cyanobacterial cells.
FIG 3.
Heatmaps depict the bacterial taxa that contributed to >0.01% of the total diversity in at least one sample, and cluster dendrograms estimate sample similarity based on weighted UniFrac comparisons of bacterial communities for those samples for the WA DWDS (a) and SA DWDS (b).
In the SA system, additional WTP and source water samples were collected 2 weeks after the initial sample collections to examine whether bacterial communities are subject to short-term temporal fluctuations (see Fig. SM-2 in the supplemental material). Interestingly, this basic temporal study carried out within the SA DWDS revealed that the source water samples collected 2 weeks apart had significantly different bacterial communities (P = 0.01, UniFrac Monte-Carlo significance test), but the treated water samples did not (P = 0.72, UniFrac Monte-Carlo significance test). These data suggest that treatments are consistent and that disinfection selects for the same taxa irrespective of the starting diversity in the source water. Further, this result also suggests that temporal snapshots of community diversity may not be adequate for analysis of highly diverse source waters.
Perhaps surprisingly, the second sampling point postdisinfection (at 1.5 km postdisinfection) in the SA system had a lower diversity index and OTU count than the primary postdisinfection sample point (Fig. 2). This demonstrates that continued exposure to the residual disinfectant has a persistent effect on the bacterial taxa present and can further reduce bacterial diversity. The FCM viable cell counts (Table 1) support this as the viable cell count decreased from 13.6% in the primary postdisinfection location to 4.9% at the second postdisinfection location. The viable cell counts in the WA system suggest that the disinfection is achieving maximum inactivation at the initial dose point as there is no further reduction in the proportion of viable cells after this point, with viable cell counts remaining stable at around 3.5%. This may be due to the lower dissolved organic carbon (DOC) amount and different bacterial diversity within the WA source water, enabling the disinfectant to have a more rapid impact on the bacterial communities.
Nitrification.
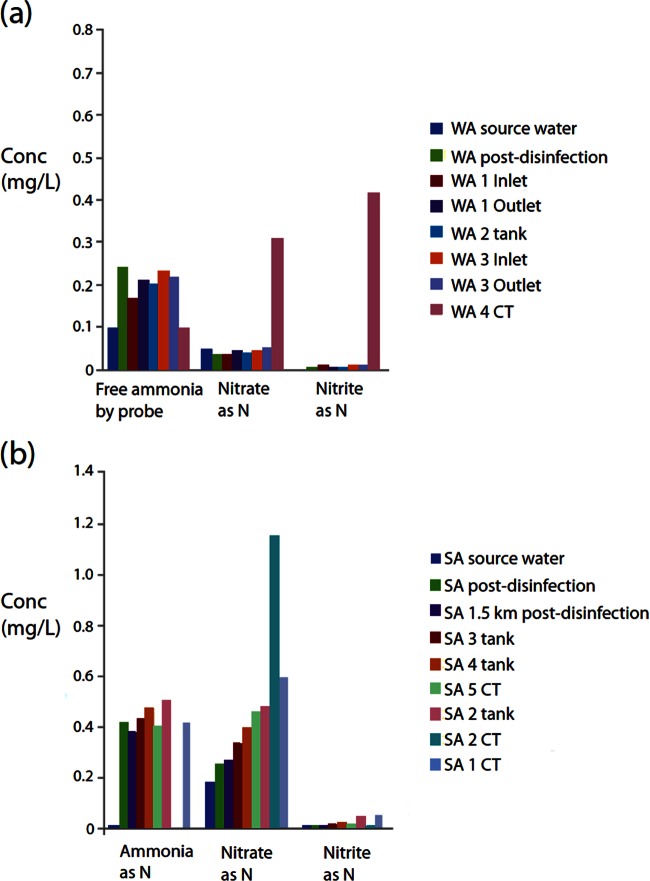
Nitrification is a major issue for chloraminated DWDSs as it leads to eventual decay of the disinfectant, which in turn can result in bacterial regrowth. Water quality analyses of the two DWDSs revealed that some sites may be subject to differing stages of nitrification. The CT sample at SA site 2 (SA-2 CT) had elevated nitrate and reduced nitrite with no residual ammonia or chloramine, suggesting that this site was completely nitrified (Fig. 4). The WA-4 CT sample had elevated amounts of nitrite and nitrate and reduced ammonia and chloramine, suggesting that nitrification was occurring at this site but had not progressed enough to completely remove the disinfectant. The SA-1 CT sample had slightly increased nitrate and nitrite, decreased chloramine, and an increased percentage of viable cells compared to other samples in the SA DWDS, suggesting that the site could be in the early stages of nitrification and regrowth.
FIG 4.
Nutrient concentrations for both the WA DWDS (a) and SA DWDS (b). As water quality indices were measured by different water corporations, the protocols used differ slightly, with free ammonia measured for the WA DWDS and ammonia as nitrogen measured for the SA DWDS.
Metagenomic community analysis detected the nitrifying bacteria Nitrospira in SA-1 CT (1.2%), SA-2 CT (2.6%), and WA-4 CT (0.55%) samples. With the exception of the source water, the proportion of Nitrospira was either below 1% or completely undetected for all other SA samples and below 0.05% or undetected for all other WA samples. The nitrifying OTUs of Nitrospira and Nitrobacter were the only nitrifying genera detected by the metagenomic analyses and were a minor component of the community present. However, the FCM data indicated there were approximately 690,000 total cells ml−1 in the SA-2 CT sample (50% of which were viable), 2,520,000 total cells ml−1 in WA-4 CT (3.3% were viable), and 124,000 total cells ml−1 in the SA-1 CT sample (11.5% were viable). Using these numbers, we can estimate that there are 9,000 live Nitrospira cells ml−1 in the SA-2 CT sample, 415 live cells ml−1 in the WA-4 CT sample, and 170 live cells ml−1 in the SA-1 CT sample. However, it is possible that additional, unknown nitrifying taxa are present in the samples but are not classified in genetic databases, which prevents their detection when meta-bar coding is used.
The qPCR results for ammonia-oxidizing genes suggest that ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) were present throughout both of the DWDSs (Table 1). This contrasts with the metagenomic data, which detected only nitrifying species at locations displaying chemical signs of nitrification and did not detect known ammonia oxidizers at any location. The qPCR results indicated that AOA were present at the WA-4 CT in lower numbers than AOB, whereas both AOA and AOB were present in moderate numbers at the SA-2 CT location. The SA-1 CT location appeared to be dominated by AOA. These results indicate that biological nitrification within DWDSs is a complex process and may be caused by different taxonomic groups at different locations throughout a DWDS, highlighting the limitations associated with using only one particular method to detect nitrification in DWDSs.
The unexpected low diversity within the SA-2 tank and the extremely high diversity in the nearby SA-2 CT site, as well as a sharp increase in cell counts and viable cells, support the suggestion that nitrification and bacterial regrowth (due to the loss of residual disinfectant) are occurring rapidly between these two sample points. The unusual Gammaproteobacteria-dominated community (75%) in the SA-2 tank, combined with the nitrified SA-2 CT sample further along the DWDS, may indicate that these Gammaproteobacteria predispose DWDSs to nitrification. The monochloramine concentrations support this theory as monochloramine had completely decayed at the SA-2 CT location (Table 1). However, the concentration of ammonia in the SA-2 tank was not depleted, and therefore it is unlikely that the unknown Gammaproteobacteria are ammonia oxidizers. BLAST results of individually extracted Gammaproteobacteria sequences from the SA-2 tank (see Table SM-5 in the supplemental material) showed that the sequences were a close match to Pseudomonas, a group that contains strains previously isolated from a nitrifying inoculum (Pseudomonas peli).
Chloramine-resistant taxa.
The concentration of monochloramine varied throughout the systems (Table 1). As would be expected, the source water samples had the least amount of monochloramine present, and the postdisinfection samples closest to the WTP had the highest. However, the monochloramine levels did not decay evenly in accordance with distance from the WTP, suggesting that additional biological factors were affecting the monochloramine concentrations. Previous studies have suggested that taxa abundant in locations with high disinfectant concentrations are likely to be disinfection-resistant or -tolerant bacterial groups (22). The relatively low bacterial diversity and high abundance of specific dominant groups in samples containing the highest concentrations of monochloramine suggest that these bacterial groups are monochloramine-resistant taxa. Burkholderiales were found in relatively high proportions in the postdisinfection samples of both systems, demonstrating that this bacterial group is able to tolerate high levels of disinfectant (Fig. 1, 2, and 3). The Burkholderiales taxa differed slightly between the two systems (Fig. 3), with the SA postdisinfection sample dominated by Achromobacter (3%), Delftia (10%), Pelomonas (2%), and an unknown member of the Comamonadaceae (3%), whereas the WA postdisinfection sample contained mostly Alcaligenaceae (6%) and Ralstonia (6%). Burkholderiales also dominated (>85%) two of the end-use locations within the SA DWDS: the SA-3 tank and SA-5 CT site (Fig. 3b). Closer inspection revealed that the Burkholderiales taxa in the SA-3 tank sample belonged to the Comamonadaceae family and that the SA-5 CT sample contained predominantly Hydrogenophaga, a yellow-pigmented, hydrogen-oxidizing bacterium. These two end-use samples had unusually low diversity indexes and OTU counts compared to those of all other samples (Mann-Whitney U; P = <0.001) and therefore could indicate locations where monochloramine-resistant taxa are outcompeting other bacterial taxa for resources. These taxa could be targets for future microbiological studies as the tanks were relatively unusual and may provide useful insights into bacterial community dynamics within DWDSs that utilize monochloramine. The order Sphingomonadales was also identified in relatively high proportions in the SA postdisinfection sample but not in the WA postdisinfection sample. Nevertheless, the relatively high abundance of this order in samples with large amounts of monochloramine warrants further investigation in future studies.
Pathogen detection.
The OTU tables for each system were screened for common waterborne pathogens to determine if metagenomics is a feasible tool for pathogen identification in DWDSs. Several genera known to contain pathogenic species were identified in the systems; however, in most cases the 16S gene fragments did not allow species- or strain-level resolution of the taxa. Therefore, we utilized an approach similar to current compliance methods, where potential coliform microorganisms are identified as indicators of contamination even if specific species or strains cannot be resolved (23). The coliform bacterial genera Escherichia and Shigella were identified in relatively large proportions in the WA postdisinfection sample (8%) and in the WA-1 inlet sample (5%) but were negligible in the SA postdisinfection sample. This suggests that the filtration steps used in the SA WTP are crucial for complete removal of fecal coliform cells. However, as the FCM analyses revealed that the WA disinfection step reduces the percentage of viable bacterial cells to just 3.4%, it is likely that the Escherichia and Shigella taxa detected by metagenomic sequencing are intact but nonviable cells (i.e., they are effectively neutralized, but the cells have not yet degraded). In an attempt to discriminate between Escherichia and Shigella, all associated sequences were extracted from the data set and blasted separately in NCBI. Results indicated that the closest matches to the taxa found in the WA system were Escherichia coli, Escherichia fergusonii, Shigella dysenteriae, and Shigella sonnei, some of which are human pathogens (see Table SM-5 in the supplemental material). Other taxa known to contain pathogenic species were also identified throughout the systems. For example, Burkholderia, a genus known to contain pathogenic species, was identified in the SA-3 tank while other samples contained the genera Campylobacter (SA-1 CT), Mycobacterium (SA source water, SA postdisinfection, SA-1 CT, SA-3 tank, and SA-5 CT), and Legionella (SA source water). In the WA DWDS, Leptospira (WA-4 CT), Mycobacterium (WA-3 and WA-4 CT), Clostridium (WA-3 inlet), and Legionella (WA source water, WA postdisinfection, WA-4 CT, and WA-3 inlet and outlet) were identified.
DISCUSSION
Bacterial communities in drinking water systems are complex, and although current monitoring methods are useful to quantify total and live bacterial cells and detect specific taxa, they do not allow the intricate and complex nature of these diverse communities to be effectively evaluated. In this study, the addition of metagenomic analyses to currently used compliance-based methods revealed complex interactions between biological communities and various DWDS parameters, such as water treatment, monochloramine tolerance, nitrification, and tank storage effects. To summarize, the water treatment regimes within both the WA and SA systems were initially highly effective at reducing bacterial community diversity and cell counts. However, some sample locations at a further distance from the WTPs displayed bacterial regrowth and increases in diversity. The data also revealed that nitrification was occurring at multiple locations at various degrees of severity and that monochloramine-tolerant bacterial groups, including Burkholderiales and Sphingomonadales, were present in some locations. The metagenomic analyses successfully identified genera associated with fecal coliforms and pathogens; however, the universality and small size of the gene fragments used in this study prevented specific pathogens from being identified.
In this study, nitrification was found to be occurring in both systems further along the DWDS. Nitrification refers to the biological oxidation of ammonia to nitrite, commonly conducted by the bacterial genus Nitrosomonas (24), followed by the oxidation of nitrite to nitrate, usually by Nitrobacter or Nitrospira (25). Free ammonia is often present in DWDSs as a consequence of the process used to generate chloramine. Nitrospira was detected in WA-4 and SA-2 CT sites, and water quality analyses revealed little to no ammonia or monochloramine at these locations and elevated levels of nitrate. Nitrospira has been considered the most dominant and ubiquitous genus among the nitrifying bacteria (26) and was only one of two nitrifying bacterial genera detected by the metagenomic analysis. However, it is possible that there were other nitrifying bacteria present that have not been classified in genetic databases. The qPCR results indicated that there were AOA- and AOB-associated genes present throughout the systems. However, the metagenomic data detected nitrifiers only when various stages of nitrification were also confirmed by the water quality data. The discrepancy between these results could be due to a number of different reasons; for example, ammonia oxidizers may have been at low abundance, and their DNA signals may have been obscured by more highly abundant taxa, preventing detection via metagenomic sequencing. In contrast, it could be that the highly specific nature of the qPCR is detecting AOA and AOB even when the abundances of those taxa are below effectual levels, which would explain their consistent presence throughout the systems. Alternatively, this result may indicate that ammonia-oxidizing genes are present in other bacterial species that may be taxonomically unknown at present. In addition to detecting nitrifying bacteria, the metagenomic data revealed that the end-user locations WA-4 and SA-2 CT had OTU counts, biodiversity measurements, and OTU proportions more similar to those of the source water than to those of the postdisinfection samples, suggesting that bacterial population regrowth during nitrification was occurring at these sites. The detection of nitrifying species in DWDS locations in the early stages of nitrification could be used as a preventative measure, identifying sections of a DWDS that may require remedial action, such as flushing or additional disinfection, to avoid potential future, more severe nitrification episodes.
Taxa identified in samples with large amounts of disinfectant (monochloramine) are likely to be species that are resistant to or tolerant of disinfectants. For example, the Burkholderiales, which are known to contain strains resistant to antibiotics (27), were identified in high percentages in the postdisinfection samples in both systems. The sensitivity of Burkholderia to chlorine (28) and monochloramine (29) is known to vary considerably, with some isolates more tolerant than others. Previous research has revealed that cocultures of the pathogenic Burkholderia species Burkholderia pseudomallei with the ubiquitous freshwater amoeba Acanthamoeba astronyxis greatly enhances the survival of B. pseudomallei in the presence of monochloramine, with 100 times more monochloramine required to maintain disinfectant efficacy when the amoeba was present (29). This is due to the ability of Acanthamoeba cysts to resist common disinfectants, such as monochloramine and chlorine, and their ability to bear phagocytosis-resistant bacteria within, thus protecting the bacteria from water treatments (30). While these particular species may not have been present in these systems, it may be the case that similar interkingdom interactions can occur for other Burkholderiales species. As eukaryotes were not sequenced in this study, it is not known what amoebae were present in the SA and WA systems. Further, other studies found that viable B. pseudomallei cells can be recovered from water containing up to 1,000 ppm of free chlorine and after the disinfectant had previously successfully reduced cells to compliance levels, suggesting that species affected by initial disinfection could recover to harmful levels later in the DWDS (31). Burkholderiales have also previously been identified in drinking water pipe biofilms (32), which may potentially serve as reservoirs to seed cells back into the water postdisinfection. This provides a further possible explanation for the relatively high abundance of Burkholderiales in the postdisinfection samples as biofilm microorganisms are known to be more resistant to drinking water disinfection than free-living microorganisms (33). Again, although these previous studies have focused on specific pathogenic strains of Burkholderiales, it is possible that other species within this order are able to resist disinfection in the same way. In addition to Burkholderiales, Sphingomonadales are also known to contain chlorine-resistant species (34), are found in biofilms (32), and were detected in relatively high proportions in postdisinfection samples. Therefore, it is plausible that the Sphingomonas identified in this study could also be resistant to disinfectants, explaining its high proportions in samples with elevated amounts of monochloramine.
Bacterial genera known to contain notable waterborne pathogens were identified at some locations along the DWDSs. Clostridium, Campylobacter, Corynebacterium, Escherichia coli, Mycobacterium, Legionella, Burkholderia, and Leptospira were all identified in various samples throughout the systems, some close to the consumer end-use locations. As mentioned previously, the Burkholderia genus contains pathogenic species that have been shown to resist disinfectants and antibiotics. Burkholderia species are of particular public health significance in warmer climates (35), with outbreaks of the waterborne disease melioidosis sporadically occurring in South America, Asia, and Australia (36). Campylobacter was present in relatively high abundance in the SA source water. Campylobacter is a known fecal-associated bacteria that usually requires a host to survive and grow (37); the presence of this taxon in the source water combined with its failure to reappear further along the DWDS postdisinfection supports this concept. However, while Campylobacter can survive only a few days without a host in water above 15°C, it can survive for many weeks in a viable but nonculturable state in water at 4°C (37); therefore, its presence may be of importance during colder winter months. Cyanobacteria were found in high abundances in the WA postdisinfection sample, likely because chloramine disinfection alone is not capable of destroying filamentous or colonial cells or because akinetes, which are environmentally resistant, dormant cells with thick walls, were present (38). Some cyanobacterial species are of public health significance due to the production of toxins, while others can cause the deterioration of the esthetic quality of the water due to the production of secondary metabolites that produce an earthy, musty taste and odor (39). Despite the presence of these genera, it must be stressed that the metagenomic data presented here do not confirm the presence of specific pathogenic strains or species and also do not provide information on the viability or infectivity of the taxa detected, making it difficult to quantitatively assess any public health significance. Even so, the ability to identify multiple genera in a large number of samples rapidly and inexpensively demonstrates the usefulness of a metagenomic monitoring approach for screening DWDSs for potential pathogens and other organisms detrimental to water quality. In addition, increased phylogenetic resolution may be obtainable by targeting loci with greater specificity to target groups, and the inclusion of eukaryotic analyses might reveal important community relationships, such as the symbiotic relationship between Acanthamoeba and Burkholderia discussed above.
In the future, metagenomics should be used alongside current assessment methods to generate databases that contain information about bacterial taxa typically found in drinking water systems. Such databases would eventually be able to provide information about which taxa indicate a healthy, high-quality DWDS and may also help diagnose problems within systems, enabling an appropriate solution to be achieved more rapidly. However, there are limitations associated with metagenomic approaches for assessing DWDS health and quality. For example, metagenomics alone does not provide information on the amount of total or viable cells in the system; therefore, intact organisms that are no longer alive and active may be detected (as was seen in the SA postdisinfection sample). Further, the choice of loci is critical and should be dependent upon the proposed research question. Nonetheless, this study demonstrates that metagenomics used in collaboration with other techniques, such as FCM analyses and nutrient measures, can provide highly useful information about the health status and efficiency of particular DWDSs, and future use of metagenomics will foster a more detailed picture of bacterial community dynamics within DWDSs.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Australian Research Council grants LP110100459 and LP0991985.
We gratefully acknowledge the assistance of Renae Philips, Melody Lau, and the Water Treatment and Distribution Research technical team and the support of Ralph Henderson from the Water Corporation, Western Australia.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01297-15.
REFERENCES
- 1.Hunter PR. 1997. Waterborne disease: epidemiology and ecology. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 2.Prüss A, Kay D, Fewtrell L, Bartram J. 2002. Estimating the burden of disease due to water, sanitation and hygiene at a global level. Environ Health Perspect 110:537–542. doi: 10.1289/ehp.02110537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Environmental Protection Agency. 1999. EPA guidance manual: alternative disinfectants and oxidants, EPA 815-R-99-014. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 4.Bej AK, Steffan RJ, DiCesare J, Haff L, Atlas RM. 1990. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol 56:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holinger EP, Ross KA, Robertson CE, Stevens MJ, Harris JK, Pace NR. 2014. Molecular analysis of point-of-use municipal drinking water microbiology. Water Res 49:225–235. doi: 10.1016/j.watres.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 7.Douterelo I, Sharpe RL, Boxall JB. 2013. Influence of hydraulic regimes on bacterial community structure and composition in an experimental drinking water distribution system. Water Res 47:503–516. doi: 10.1016/j.watres.2012.09.053. [DOI] [PubMed] [Google Scholar]
- 8.Bal Krishna KC, Sathasivan A, Ginige MP. 2013. Microbiol community changes with decaying chloramine residuals in a lab-scale system. Water Res 47:4666–4679. doi: 10.1016/j.watres.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Alvarez V, Revetta RP, Santo Domingo JW. 2012. Metagenomic analyses of drinking water receiving different disinfection treatments. Appl Environ Microbiol 78:6095–6102. doi: 10.1128/AEM.01018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu WT. 2012. Microbial community dynamics of an urban drinking water distribution system subjected to phases of chloramination and chlorination treatments. Appl Environ Microbiol 78:7856–7865. doi: 10.1128/AEM.01892-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed American Public Health Association, Washington, DC. [Google Scholar]
- 12.Hoefel D, Monis PT, Grooby WL, Andrews S, Saint CP. 2005. Profiling bacterial survival through a water treatment process and subsequent distribution system. J Appl Microbiol 99:175–186. doi: 10.1111/j.1365-2672.2005.02573.x. [DOI] [PubMed] [Google Scholar]
- 13.van der Wielen PW, Voost S, van der Kooij D. 2009. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl Environ Microbiol 75:4687–4695. doi: 10.1128/AEM.00387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamariz J, Voynarovska K, Prinz M, Caragine T. 2006. The application of ultraviolet irradiation to exogenous sources of DNA in plasticware and water for the amplification of low copy number DNA. J Forensic Sci 51:790–794. doi: 10.1111/j.1556-4029.2006.00172.x. [DOI] [PubMed] [Google Scholar]
- 15.Champlot S, Berthelot C, Pruvost M, Bennett EA, Grange T, Geigl E. 2010. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS One 5:e13042. doi: 10.1371/journal.pone.0013042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polz MF, Cavanaugh CM. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M. 2012. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, Feazel LM, Park K, Pace NR, Frank DN. 2013. Explicet: graphical user interface software for metadata-driven management, analysis, and visualization of microbiome data. Bioinformatics 29:3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridgway HF, Olson BH. 1982. Chlorine resistance patterns of bacteria from two drinking water distribution systems. Appl Environ Microbiol 44:972–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashbolt NJ, Grabow WOK, Snozzi M. 2001. Indicators of microbial water quality, p 289–316. In Fewtrell L, Bartram J (ed), Water quality: guidelines, standards, health risk assessment and management for water-related infectious disease. IWA Press, London, United Kingdom. [Google Scholar]
- 24.Koops HP, Bottcher B, Moller UC, Pommerening-Roser A, Stehr G. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol 137:1689–1699. doi: 10.1099/00221287-137-7-1689. [DOI] [Google Scholar]
- 25.Nogueria R, Melo LF. 2006. Competition between Nitrospira spp. and Nitrobacter spp. in nitrite-oxidising bioreactors. Biotechnol Bioeng 95:169–175. doi: 10.1002/bit.21004. [DOI] [PubMed] [Google Scholar]
- 26.Hovanec TA, Taylor LT, Blakis A, DeLong EF. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol 64:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer HP. 2012. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol 7:1389–1399. doi: 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell HA, Rose LJ, Shams A, Bradley M, Arduino MJ, Rice EW. 2009. Variability of Burkholderia pseudomallei strain sensitivities to chlorine disinfection. Appl Environ Microbiol 75:5405–5409. doi: 10.1128/AEM.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard K, Inglis TJJ. 2005. Disinfection of Burkholderia pseudomallei in potable water. Water Res 39:1085–1092. doi: 10.1016/j.watres.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 30.Mogoa E, Bodet C, Morel F, Rodier M, Legube B, Hechard Y. 2011. Cellular response of the amoeba Acanthamoeba castellanii to chlorine, chlorine dioxide, and monochloramine treatments. Appl Environ Microbiol 77:4974–4980. doi: 10.1128/AEM.00234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howard K, Inglis TJJ. 2003. The effect of free chlorine on Burkholderia pseudomallei in potable water. Water Res 37:4425–4432. doi: 10.1016/S0043-1354(03)00440-8. [DOI] [PubMed] [Google Scholar]
- 32.Shaw JLA, Monis P, Fabris R, Ho L, Braun K, Drikas M, Cooper A. 2014. Assessing the impact of water treatment on bacterial biofilms in drinking water distribution systems using high-throughput DNA sequencing. Chemosphere 117:185–192. doi: 10.1016/j.chemosphere.2014.06.077. [DOI] [PubMed] [Google Scholar]
- 33.Lewis K. 2001. Riddle of biofilm resistance. Antimicrob Agents Chemother 45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun W, Lui W, Cui L, Zhang M, Wang B. 2013. Characterization and identification of a chlorine-resistant bacterium, Sphingomonas TS001, from a model drinking water distribution system. Sci Total Environ 458–460:169–175. doi: 10.1016/j.scitotenv.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 35.Inglis TJJ, Garrow SC, Adams C, Henderson M, Mayo M, Currie BJ. 1999. Acute melioidosis outbreak in Western Australia. Epidemiol Infect 123:437–443. doi: 10.1017/S0950268899002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Josephson J. 2001. Melioidosis: an emerging tropical health problem. Environ Sci Technol 35:454A–457A. doi: 10.1021/es0125265. [DOI] [PubMed] [Google Scholar]
- 37.Nachamkin I, Skirrow MB. 1998. Campylobacter, Arcobacter and Helicobacter, p 1237–1256. In Balows A, Duerden BI (ed), Topley & Wilson's microbiology and microbial infections: systematic bacteriology, 9th ed, vol 2 Arnold, London, United Kingdom. [Google Scholar]
- 38.Adams DG, Carr NG. 1981. The developmental biology of heterocyst and alkinete formation in cyanobacteria. Crit Rev Microbiol 9:45–100. doi: 10.3109/10408418109104486. [DOI] [PubMed] [Google Scholar]
- 39.Wnorowski AU. 1992. Tastes and odours in the aquatic environment: a review. Water SA 18:203–214. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.