Abstract
The control of multihost pathogens, such as Coxiella burnetii, should rely on accurate information about the roles played by the main hosts. We aimed to determine the involvement of the red deer (Cervus elaphus) in the ecology of C. burnetii. We predicted that red deer populations from broad geographic areas within a European context would be exposed to C. burnetii, and therefore, we hypothesized that a series of factors would modulate the exposure of red deer to C. burnetii. To test this hypothesis, we designed a retrospective survey of 47 Iberian red deer populations from which 1,751 serum samples and 489 spleen samples were collected. Sera were analyzed by enzyme-linked immunosorbent assays (ELISA) in order to estimate exposure to C. burnetii, and spleen samples were analyzed by PCR in order to estimate the prevalence of systemic infections. Thereafter, we gathered 23 variables—within environmental, host, and management factors—potentially modulating the risk of exposure of deer to C. burnetii, and we performed multivariate statistical analyses to identify the main risk factors. Twenty-three populations were seropositive (48.9%), and C. burnetii DNA in the spleen was detected in 50% of the populations analyzed. The statistical analyses reflect the complexity of C. burnetii ecology and suggest that although red deer may maintain the circulation of C. burnetii without third species, the most frequent scenario probably includes other wild and domestic host species. These findings, taken together with previous evidence of C. burnetii shedding by naturally infected red deer, point at this wild ungulate as a true reservoir for C. burnetii and an important node in the life cycle of C. burnetii, at least in the Iberian Peninsula.
INTRODUCTION
Coxiella burnetii is a Gram-negative intracellular bacterium that causes Q fever, a disease that affects both humans and animals. Whereas the epidemiological status of C. burnetii in European domestic ruminants is well known (1), information for wildlife is mostly local and scattered (2, 3). Although the majority of human Q fever outbreaks are linked to the transmission of C. burnetii from domestic ruminants (4, 5), the ability of C. burnetii to infect wild hosts (3, 6) and its high environmental resistance (1) make wildlife species potential reservoirs of C. burnetii. Based on this hypothesis, wildlife could maintain C. burnetii and transmit it to wildlife (7), domestic animals (8), or humans (9). It is therefore of paramount relevance (i) to identify those potential wild reservoir species that could, through direct and indirect interactions, transmit C. burnetii to target species (domestic animals and humans) and (ii) to determine which environmental factors are the main drivers of C. burnetii within the most relevant wild reservoirs. Efficient prevention of C. burnetii transmission at the wildlife–domestic-animal–human interface can be approached only once the main reservoirs have been identified and the driving risk factors are known (10).
Several wild ruminant species are present and well distributed in Europe; on the premise that they are susceptible to infection by C. burnetii, these could constitute important wild reservoirs of C. burnetii. However, among European wild ruminants, the red deer (Cervus elaphus) could perhaps constitute a potential wild reservoir for C. burnetii due to its geographic distribution, demographic status, importance as game, and behavior. The red deer displays broad global (11, 12) and European (13) geographic distribution, with trends to increasing distribution and density (14, 15). It is currently one of the most important game species among European large mammals (16). Many red deer populations in Europe are subjected to management for hunting (17), and red deer farming has expanded in recent decades as a consequence of the demand for venison and live individuals for population-restocking programs (18). Additionally, the gregarious behavior of the red deer (19, 20) promotes the aggregation of individuals. In domestic animals, host density and aggregation are important drivers of C. burnetii transmission (21, 22), and some Iberian red deer populations reach densities higher than 70 deer/km2 (14). Increasing red deer densities, deer management (including artificial feeding), and gregarious behavior constitute the main factors favoring the transmission of circulating pathogens in red deer populations (23, 24).
Taken together, distribution, demography, management, and behavior point at red deer as one of the most concerning reservoirs of shared pathogens among European wild ruminants; e.g., 44% of red deer in Italy were found to be infected by piroplasms (25), and >60% of Slovakian red deer carried Anaplasma spp. (26). Therefore, we predicted that C. burnetii would be circulating in red deer populations in Iberia, and we hypothesized that particular environmental, management, and host factors would contribute to the exposure of red deer to C. burnetii. To test these hypotheses, we designed a retrospective epidemiological survey targeting Iberian (Spanish and Portuguese) red deer populations within their geographic distribution range.
MATERIALS AND METHODS
Survey design.
Sera from 47 red deer populations were collected from 2000 to 2012 in mainland Spain and Portugal (Fig. 1). Study populations were selected on the basis of (i) management systems, including unmanaged, naturally free-ranging populations (in game reserves and natural and national parks), managed free-ranging populations, and farms, (ii) geographic location (location within the different bioregions established for wildlife disease surveillance schemes in mainland Spain [27] and from different regions in mainland Portugal), and (iii) the range of geographic distribution of red deer (Fig. 1), in order to obtain spatial representativeness.
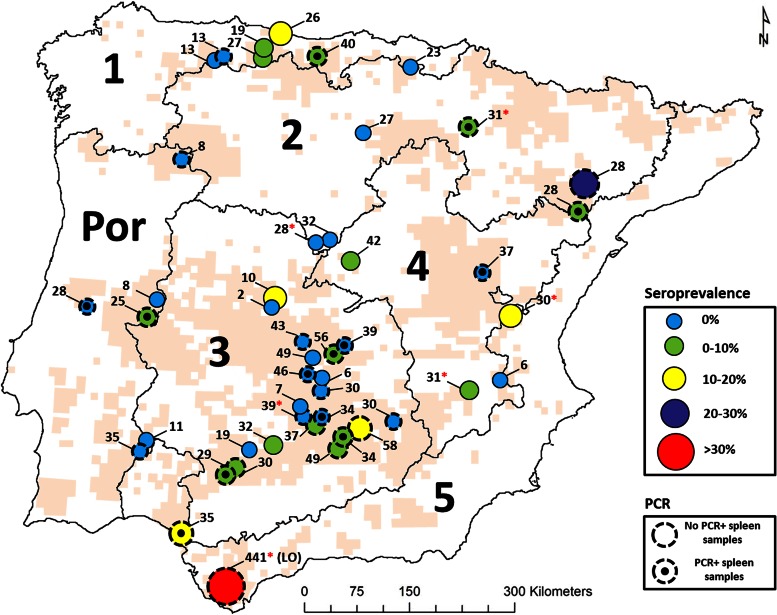
FIG 1.
Spatial distribution of Coxiella burnetii seroprevalence in Iberian red deer and presence of C. burnetii DNA in spleen samples. Each dot represents a surveyed red deer population. The current geographic distribution of the red deer in the Iberian Peninsula is shown in pale orange (54, 55). The number of sera analyzed per population is shown. A red asterisk next to the sampling size indicates red deer farms. The map of Spain has been divided into the bioregions established in the current Spanish wildlife disease surveillance program (27). Por, Portugal.
Serological analyses.
The presence of specific antibodies against C. burnetii phase I and II antigens in deer sera was analyzed with a commercial indirect enzyme-linked immunosorbent assay (ELISA) (LSIVet Ruminant Q Fever Serum/Milk ELISA kit; Life Technologies, USA) with an in-house modification in the secondary antibody (protein G-horseradish peroxidase; Sigma-Aldrich, USA) (28) that was previously validated for wild and domestic ungulates (29). Briefly, for validation, we employed positive (n = 8) and negative (n = 6) red and roe deer sera analyzed by indirect immunofluorescence assay (IFA), as well as ELISA-positive, PCR-positive and ELISA-negative, PCR-negative cattle (n, 14 and 12, respectively) and sheep (n, 16 and 17, respectively) sera. For each sample, the sample-to-positive-control (SP) ratio was calculated as (ODs − ODnc)/(ODpc − ODnc) × 100, where ODs is the optical density of the sample as measured using a dual-wavelength spectrophotometer (first at 450 nm and then at 620 nm), ODnc is the optical density of the negative control, and ODpc is the optical density of the positive control. All SP values of ≤40% were considered negative, whereas SP values of >40% were considered positive.
PCR analyses.
Spleen samples were collected from a subset of the populations studied during necropsies performed on hunter-harvested or euthanized farmed deer. Spleen samples from seropositive and seronegative deer were selected for PCR analyses. Total DNA from spleen samples was purified with the DNeasy blood and tissue kit (Qiagen, Germany) according to the manufacturer's protocol (http://mvz.berkeley.edu/egl/inserts/DNeasy_Blood_&_Tissue_Handbook.pdf). The DNA concentration in aliquots was quantified (NanoDrop 2000c/2000 spectrophotometer; Thermo Scientific, USA), and aliquots were frozen at −20°C until the PCR was performed. Sample cross-contamination during DNA extraction was excluded by including negative controls (nuclease-free water; Promega, USA) that were also tested by PCR.
DNA samples were analyzed by a quantitative real-time PCR (qPCR) targeting a transposon-like repetitive region of C. burnetii as described previously (Table 1) (30). SsoAdvanced universal probes supermix (Bio-Rad, USA) was used in qPCR according to the specifications of the manufacturers. DNA extraction and PCR were performed in separate laboratories under biosafety level II conditions (Bio II A cabinet; Telstar, Spain) to avoid cross-contamination. As a positive control in this real-time PCR, we used a DNA extract of Coxiella burnetii from the Coxevac vaccine (CEVA Santé Animale, France). We considered a sample to be positive at a threshold cycle (CT) value below 40 (30).
TABLE 1.
Primers and probe used in the qPCR
| Primer or probe | Sequence (5′–3′)a | Locationb | Amplicon size (bp) |
|---|---|---|---|
| QKF3 | GTGGTGCCAAGCGATTTTAT | 7216–7235 | 78 |
| QKR3 | GTTTCATCCGCGGTGTTAAT | 7293–7274 | |
| QKP3 | FAM-TTTAGCGAGCGAAGCGGTGG-TAMRA | 7253–7272 |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Positions in the whole-genome sequence of C. burnetii RSA493 (GenBank accession number AE016828) that encode the transposase gene of the C. burnetii-specific IS1111a insertion element.
Risk predictor variables.
In order to identify factors modulating the risk of exposure of individual red deer to C. burnetii infection, a set of abiotic and biotic variables within three main factors—environment, management, and host (Table 2)—were gathered on the basis of their potential impact on C. burnetii ecology.
TABLE 2.
Set of variables gathered for risk factor modeling of exposure of individual deer to Coxiella burnetii
| Factor | Variable codea | Description of variable (unit of measurement) |
|---|---|---|
| Environment | X* | Longitude (m) |
| Y* | Latitude (m) | |
| Se* | Season (Sp, spring; Su, summer; Au, autumn; Wi, winter) | |
| AvSpT* | Avg spring temp (°C) | |
| Management | EsT*b | Estate type (Um, unmanaged free-ranging; Mg, managed free-ranging; Fd, farmed) |
| Host | CFd* | Density of cattle farms in the municipality (farms/km2) |
| SFd | Density of sheep farms in the municipality (farms/km2) | |
| GFd | Density of goat farms in the municipality (farms/km2) | |
| SrFd* | Density of small-ruminant farms in the municipality (farms/km2) | |
| RuFd | Density of ruminant farms in the municipality (farms/km2) | |
| Cd | Density of cattle in the municipality (animals/km2) | |
| Sd | Density of sheep in the municipality (animals/km2) | |
| Gd | Density of goats in the municipality (animals/km2) | |
| Srd | Density of small ruminants in the municipality (animals/km2) | |
| Rud* | Density of ruminants in the municipality (animals/km2) | |
| RdFi* | Environmental favorability for red deer | |
| RoFi | Environmental favorability for roe deer | |
| WbFi | Environmental favorability for wild boar | |
| HUd* | Density of humans in the municipality (people/km2) | |
| HsDi | Distance to the nearest human settlement (km) | |
| Sx* | Sex (M, male; F, female) | |
| Ag* | Age class (Cf, calf; Yr, yearling; Sa, subadult; Ad, adult) | |
| Unclassified | Sy* | Sampling yr |
Variables included in the statistical modeling process are marked with asterisks.
This variable was included only in the overall (unmanaged plus managed plus farmed) deer data set.
(i) Environmental factors.
Both spatial and meteorology-related variables were considered for risk factor modeling. Longitude (X) and latitude (Y) were considered as spatial factors to control for any potential spatial autocorrelation of data. Coordinates were recorded at the sampling-site level with portable global-positioning-system (GPS) devices (Garmin Ltd., Cayman Islands), so all deer from a sampling site were assigned the same X and Y values. The average spring temperature (AvSpT) and the season (Se) in which deer were surveyed (4 categorical classes: spring [Sp], April to June; summer [Su], July to September; autumn [Au], October to December; and winter [Wi], January to March) were considered as meteorology-related variables. AvSpT was considered as a potential proxy for C. burnetii environmental survival and as a potential driver of airborne transmission of C. burnetii—probably dependent on air moisture, which is highly correlated with temperature (31)—in the expected shedding season. The prevalence of C. burnetii shedding is expected to be higher in the spring, when calving takes place, as a recent study suggests (32). The season was considered as a proxy of potential year-round variability in infection risk because of the expected predominance of C. burnetii shedding in the spring.
(ii) Management factors.
Human interference in deer ecology and behavior was considered on the basis of deer population management systems: (i) unmanaged free-ranging deer populations (Um), (ii) free-ranging deer populations managed for hunting purposes (Mg) (high-wire fencing restriction and year-round supplementary feeding), and (iii) farmed deer populations (Fd) (extensively produced red deer in 6- to 10-ha enclosures).
(iii) Host factors.
Different host population and individual host variables were considered, because C. burnetii is a multihost pathogen (33):
(a) Densities of domestic ruminants and domestic ruminant farms in the municipality to which individual deer belong.
Domestic ruminant density (densities of cattle [Cd], sheep [Sd], and goats [Gd], and of combinations of these ruminants [Rud]) and farm density (CFd, SFd, GFd, small-ruminant farm density [SrFd], and RuFd) values at the municipality level were calculated on the basis of livestock census data gathered by the Spanish and Portuguese National Statistics Institutes (http://www.ine.es and http://www.ine.pt, respectively) in 2009.
(b) Environmental favorability index.
The environmental favorability index (Fi) ranged from 0 (minimum favorability) to 1 (maximum favorability) for red deer (RdFi), roe deer (RoFi), and wild boar (WbFi) at Universal Transverse Mercator (UTM) 10- by 10-km resolution squares, calculated for peninsular Spain (34). This index is a measure of the suitability of a land surface for a particular species and is well correlated with the real abundance of the species (35). Environmental favorability indices of wild ungulates have not been estimated for Portugal. Therefore, Portuguese populations that were close to the Spanish border (n = 6) (Fig. 1) were linked to the favorability indices of the closest Spanish UTM 10- by 10-km square. The only population surveyed in central Portugal could not be associated with any wild ungulate favorability index and was not considered for risk factor analyses. Red deer, roe deer, and wild boar have been found to be infected by C. burnetii previously (36, 37, 38). No favorability indices for any other potential wild host of C. burnetii are available for the study area.
(c) Density of humans in the municipality (HUd).
Updated human demographic data were obtained from the Spanish and Portuguese National Statistics Institutes in 2011 and 2010, respectively.
(d) Straight-line Euclidean distance to the nearest human settlement (HsDi).
The distance to the nearest human settlement (town or city) was measured with Geographic Information Systems (Quantum GIS; http://www.qgis.org/es/site/). Human and their pets may be hosts for C. burnetii and may potentially modulate the risk of exposure of deer (33). For this reason, HUd and HsDi were considered for modeling analyses.
(e) Host sex (Sx; male [M] versus female [F]).
Among farmed deer, the number of stags reared was significantly lower than the number of females reared, and therefore, there was a sex bias in the sample.
(f) Host age (Ag).
For free-ranging deer, tooth eruption patterns (39) were used to estimate the ages of animals <2 years old, whereas for animals >2 years old, age was determined by the number of cementum annuli of the incisor 1 root (40). Farm keepers provided the year of birth for farmed deer. For analytical purposes, four age classes were established: calf (Cf; 0 to 1 years old), yearling (Yr; 1 to 2 years old), subadult (Sa; 2 to 3 years old), and adult (Ad; >3 years old). In free-ranging populations, a conscious negative bias against calves existed according to reported age-related C. burnetii seroprevalence patterns (32, 41).
The year in which each individual deer was sampled (Sy) was additionally considered as a survey-associated factor modulating the risk of exposure of deer to C. burnetii (22).
Statistical analyses.
Four different data sets were employed to test for the main hypothesis of this study—the modulating effect of risk factors on the exposure of deer to C. burnetii—in order to seek major driving factors, including or not including the management system (an expected major epidemiological driver according to existing literature on wild ungulate pathogen dynamics). Data sets included (i) overall deer populations studied, (ii) unmanaged free-roaming deer populations, (iii) managed free-roaming deer populations, and (iv) deer farms. The deer management system was included in modeling of the data set that included all deer in order to test for the effect of management on the risk of exposure of deer to C. burnetii. Within each data set and with the aim of reducing the interference of multicollinearity among predictor variables in modeling output, a correlation matrix (Spearman's rank tests) of continuous variables was built. Therefore, only uncorrelated variables (Spearman's rho, <0.4) were selected for statistical modeling (Table 2).
For risk factor modeling, multivariate logistic regression models—generalized linear mixed models (42) fitted with a binomial distribution and a logit link function—were built (lme4 package for R) to test the influence of different potential risk factors (Table 2) on the risk of exposure of individual deer to C. burnetii. The individual status of anti-Coxiella burnetii antibodies was entered as a response variable (coded as 0 for an animal testing negative and as 1 for an animal testing positive) in the model. The location of origin of deer was entered as a random variable in the modeling process. Models were built by following a forward stepwise procedure with the aim of identifying the main modulating factors of the exposure of deer to C. burnetii. The Akaike information criterion (AIC) and the AIC increment (ΔAIC) were considered in order to select the best-fitted model (i.e., with the lowest AIC value and with a ΔAIC of >2 [43]). The statistical uncertainty associated with the estimation of individual prevalence values was assessed by calculating the associated Clopper-Pearson exact 95% confidence interval (95% CI).
RESULTS
A total of 1,751 serum samples were analyzed: 822 (46.9%) from unmanaged populations (n = 27), 329 (18.8%) from managed populations (n = 14), and 600 (34.3%) from farmed populations (n = 6). Of the 1,629 samples for which sex could be recorded, 1,147 (70.4%) were from females and 482 (29.6%) were from males. For 1,593 samples, age could be recorded; 100 samples (6.3%) belonged to calves, 240 (15.1%) belonged to yearlings, 251 (15.7%) belonged to subadults, and 1,002 (62.9%) belonged to adults. Age and sex could be recorded for 1,560 individuals at the time. Average individual seroprevalences by bioregion and deer management system are shown in Table 3.
TABLE 3.
Average individual seroprevalence, number of positive samples over sampling size, and associated 95% confidence interval throughout each sampling bioregiona and deer management system
| Country or bioregion | Seroprevalence (%) (Pos/n)b (95% CI) |
|||
|---|---|---|---|---|
| All deer populations | Unmanaged deer | Managed deer | Farmed deer | |
| Spain | ||||
| Bioregion 1 | 4.3 (7/161) (1.8–8.8) | 4.3 (7/161) (1.8–8.8) | NAc | NA |
| Bioregion 2 | 5.7 (10/174) (2.8–10.3) | 14.3 (8/56) (6.4–26.2) | 0.0 (0/59) (0.0–6.1) | 3.4 (2/59) (0.4–11.7) |
| Bioregion 3 | 2.7 (18/675) (1.6–4.2) | 3.8 (14/372) (2.1–6.2) | 1.5 (4/264) (0.4–3.8) | 0.0 (0/39) (0.0–9.0) |
| Bioregion 4 | 1.7 (2/116) (0.2–6.1) | 1.3 (1/79) (0.0–6.7) | 0.0 (0/6) (0.0–45.9) | 3.2 (1/31) (0.1–16.7) |
| Bioregion 5 | 34.6 (175/506) (30.4–38.9) | 14.3 (5/35) (4.8–30.2) | NA | 36.1 (170/471) (31.7–40.6) |
| Portugal | 1.7 (2/119) (0.2–5.9) | 1.7 (2/119) (0.2–5.9) | NA | NA |
| Total | 12.2 (214/1,751) (10.7–13.9) | 4.5 (37/822) (3.2–6.2) | 1.2 (4/329) (0.00–0.03) | 28.8 (173/600) (25.2–32.6) |
See reference 27.
Pos, number of positive samples; n, total number of samples.
NA, not applicable.
All IFA-positive red and roe deer sera presented SP values of >100, whereas IFA-negative sera had SP values of <25 (SP cutoff for positivity, >40). ELISA-positive, PCR-positive cattle and sheep sera displayed SP values of >70 and >100, respectively, whereas ELISA-negative, PCR-negative cattle and sheep sera had SP values of <30 (29). Therefore, with the controls employed in our validation approach, the ELISA reached 100% sensitivity and specificity for a positive cutoff SP of >40.
Twenty-three of the 47 deer populations surveyed (48.9%) had at least one seropositive sample; Four out of six deer farms (66.7%) and 55.6% of unmanaged free-ranging populations (15/27) had seropositive animals, in contrast to 21.4% of managed free-ranging deer populations (3/14). Seven of the 47 red deer populations (14.9%) had average individual seroprevalences of >10%. Average seroprevalence values by sex and age are shown in Table 4.
TABLE 4.
Average seroprevalence values, number of positive samples over sampling size, and associated exact 95% confidence interval by deer sex and age
| Sex | Age class | Seroprevalence (%) (Pos/n)a (95% CI) |
|---|---|---|
| Male | Calf | 2.7 (1/37) (0.1–14.2) |
| Yearling | 1.6 (1/61) (0.0–8.8) | |
| Subadult | 3.1 (1/32) (0.1–16.2) | |
| Adult | 3.9 (13/336) (2.1–6.5) | |
| Subtotal, male | 3.5 (17/482) (2.1–5.6) | |
| Female | Calf | 2.6 (1/38) (0.1–13.8) |
| Yearling | 9.0 (16/177) (5.3–14.3) | |
| Subadult | 33.0 (72/218) (26.8–39.7) | |
| Adult | 15.4 (102/661) (12.8–18.4) | |
| Subtotal, female | 16.9 (194/1,147) (14.8–19.2) |
Pos, number of positive samples; n, total number of samples.
A total of 489 spleen samples were analyzed by qPCR (Fig. 1); 305, 155, and 29 spleen samples came from unmanaged, managed, and farmed deer populations, respectively. Among all spleen samples, 5.7% (28/489) (95% CI, 3.8 to 8.2%) were qPCR positive (cycle threshold range for positive samples, 32.1 to 39.9). The prevalences of C. burnetii DNA in spleen were 6.2% (19/305) (95% CI, 3.8 to 9.6%), 5.2% (8/155) (95% CI, 2.3 to 9.9%), and 3.5% (1/29) (95% CI, 0.1 to 17.8%) in unmanaged, managed, and farmed deer populations, respectively. Ten of 140 males (7.1%; 95% CI, 3.5 to 12.6%) and 18 of 234 females (7.7%; 95% CI, 4.6 to 11.9%) were qPCR positive. One of 10 calves analyzed (10.0%; 95% CI, 0.3 to 44.5%), 5 of 41 juveniles (12.2%; 95% CI, 4.1 to 26.2%), 2 of 19 subadults (10.5%; 95% CI, 1.3 to 33.1%), and 20 of 302 adults (6.6%; 95% CI, 4.1 to 10.1%) were positive for C. burnetii DNA in the spleen by qPCR.
Twenty-six deer populations were studied for the prevalence of C. burnetii DNA in spleen samples. Of these, 12 were seronegative and 14 had at least one seropositive individual. Thirteen of those 26 deer populations (50.0%) had at least one positive spleen sample; 8 were seropositive and 5 were seronegative.
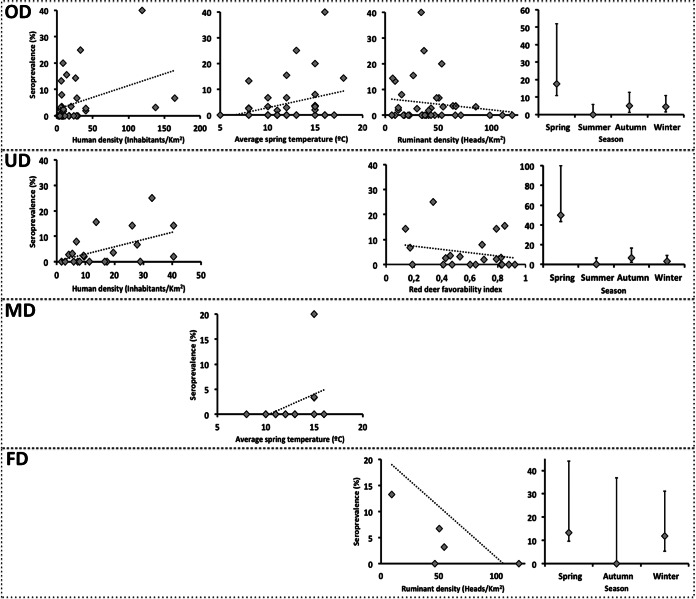
The best-fitted general model for risk factors for the exposure of deer to C. burnetii (Table 5) retained variables within the host and environment factors. Human density and the average spring temperature were positively related to increasing risks of exposure to C. burnetii (Fig. 2), and, in contrast to domestic-ruminant density, which showed a negative relationship, these relationships were statistically significant. The statistically significant negative effect of season was linked to the higher risk of exposure to C. burnetii in the spring (Fig. 2). According to outputs from partial models (for unmanaged, managed, and farmed deer data sets) (Table 5), host and environmental factors were also evidenced as relevant drivers of exposure to C. burnetii. However, the main drivers for each particular management system differed. Whereas human density, season, and the red deer environmental favorability index were retained by the best-fitted risk factor model for unmanaged deer, average spring temperature was retained by the best-fitted model for managed deer, and season and domestic-ruminant density appeared to be the main drivers of the risk of exposure to C. burnetii in red deer farms (Table 5; Fig. 2).
TABLE 5.
Best-fit model output throughout the deer data seta
| Data set | Variableb | Z | β | SE | Pc | AIC | ΔAIC | ED (%) |
|---|---|---|---|---|---|---|---|---|
| Unmanaged + managed + farmed | Intercept | −2.561 | −3.745 | 1.463 | * | 364.105 | 19.206 | 7.172 |
| HUd | 2.707 | 0.019 | 0.007 | ** | ||||
| Se | −2.476 | −0.690 | 0.279 | * | ||||
| AvSpT | 2.008 | 0.204 | 0.101 | * | ||||
| Rud | −1.573 | −0.017 | 0.011 | NS | ||||
| Unmanaged | Intercept | −1.331 | −1932 | 1.473 | NS | 220.417 | 2.525 | 3.723 |
| HUd | 1.788 | 0.053 | 0.030 | NS | ||||
| Se | −1.587 | −0.546 | 0.344 | NS | ||||
| RdFi | −0.862 | −1.193 | 1.382 | NS | ||||
| Managed | Intercept | −2.429 | −20.233 | 8.331 | * | 39.971 | 4.741 | 16.557 |
| AvSpT | 2.042 | 1.134 | 0556 | * | ||||
| Farmed | Intercept | −2.926 | −2.119 | 0.724 | ** | 91.102 | 16.224 | 19.572 |
| Se | 3.705 | 1.983 | 0.535 | *** | ||||
| Rud | −3.499 | −0.186 | 0.053 | *** |
The statistic (Z), the coefficient (β), its associated standard error (SE), the significance value (P), the model Akaike information criterion (AIC), the AIC increment (ΔAIC), and the explained deviance (ED) are shown.
Abbreviations of variables are explained in Table 2.
NS, P > 0.05; *, P <0.05; **, P < 0.01; ***, P < 0.001.
FIG 2.
Relationships between the seroprevalence of C. burnetii in the population and explanatory factors identified through risk factor modeling for each of the modeled data sets (overall deer populations [OD], unmanaged populations [UD], managed populations [MD], and farmed populations [FD]).
DISCUSSION
This study constitutes a transnational-scale survey of C. burnetii in European wild ruminants and a first approach to identifying the factors that drive the ecology of C. burnetii in red deer. We found that C. burnetii is present in approximately 50% of the free-ranging and farmed Iberian red deer populations and that systemic infections occur in 50% of them. These facts support the involvement of the red deer in the ecology of C. burnetii. Indeed, to support the notion that a particular host species is acting as a true reservoir for a specific pathogen, provided that the species is well distributed and abundant (44), one must determine that (i) the pathogen is widely distributed in populations of that host within a relatively large territory, (ii) the pathogen is able to cause systemic infections (33, 45), and (iii) the host is able to shed the pathogen. We confirm the first two requisites here; the third requisite was confirmed previously (7). Thus, the red deer may be confirmed as a true C. burnetii reservoir.
Methodological considerations.
The true seroprevalence of C. burnetii in free-ranging deer populations may have been underestimated because most deer sera (1,017 of 1,151) were collected from early autumn to midwinter, the main big-game-hunting season in Iberia. Recent data from LO farm (Fig. 1) suggest that annual individual seroprevalence fluctuates according to the red deer calving season, with the lowest values in winter and maximum values in late spring (32). Seroprevalence levels are higher in late spring to early summer, coinciding with the time of calving and, supposedly, with the main Coxiella burnetii excretion season.
Geographic distribution of C. burnetii in red deer populations.
The wide geographical distribution of C. burnetii in Iberian red deer populations is noteworthy. To date, to the best of our knowledge, no exhaustive national study of C. burnetii in wild ruminants has been performed in Europe. C. burnetii DNA was found in the tissues of 23% of the roe deer analyzed from 9 of the 12 Dutch provinces during the massive human Q fever epidemic affecting The Netherlands from 2007 to 2010 (36); however, the number of samples analyzed was low (n = 79). Comparisons with results from previous regional studies of Spanish red deer are difficult because of differing geographic scales and techniques for diagnosing pathogen exposure: an indirect immunofluorescence test by which 9.5% of wild red deer and 40.0% of farmed red deer were found to be seropositive (46) and molecular analyses (35) where none of the 22 red deer analyzed tested positive. In general terms, on the premise of possible underestimation of real seroprevalence values, we may conclude that C. burnetii circulates widely in Iberian red deer populations.
Factors modulating the exposure of red deer to C. burnetii.
The modeling output of the overall deer data set partly confirmed our second hypothesis; environmental and host factors were found to be significant drivers of C. burnetii transmission in red deer. However, no effect of the management system on the risk of exposure to C. burnetii was observed, although the main drivers in partial data sets differed slightly (Table 5). The three management categories of red deer considered in this study are related to deer abundance and aggregation (14, 47). Therefore, according to the observed effect of cattle density on the risk of exposure to C. burnetii (21, 22), we expected a clear effect of management. One would expect that in deer farms, and even in some intensively managed free-ranging deer populations, horizontal C. burnetii transmission would be enhanced due to the high animal-to-animal contact rate. This seems not to occur in general terms, perhaps due to the complexity of C. burnetii ecology and the existence of multiple reservoir hosts (3, 45). The low percentage of variance explained by the best-fitted model for unmanaged deer populations (Table 5) may reflect the existence of a complex scenario in environments with higher biological diversity. This would suggest that endemic cycles of C. burnetii implicating different wild (and domestic) host species might be established in Iberia.
Climatic conditions during the C. burnetii shedding season modulate the risk of exposure of deer to this pathogen. Nonetheless, partial models revealed that the average spring temperature is relevant only in free-ranging managed populations; this variable itself explained 16.5% of model variance. Whether this observation is related to a direct effect of temperature on C. burnetii survival or transmission, or to indirect, nonconsidered effects—e.g., an effect on transmission—cannot be determined with our findings and with the existing literature. Therefore, this finding should be the basis for further studies aiming to deepen in C. burnetii ecology.
Although a general effect of the season was evidenced, the risk of exposure to C. burnetii was higher in the spring for unmanaged deer and similar in the spring and winter for farmed deer. This observation for farmed deer may be caused by a seasonal bias in farmed deer sampling in this study; only deer from the LO farm, which had a high proportion of seropositive deer, were surveyed in the winter. The observation for unmanaged deer agrees with the higher level of shedding of C. burnetii expected at the time of deer calving in midspring, as mentioned above.
Finally, host effects were revealed by the general model and by models for unmanaged and farmed deer populations. The influence of human density in the general model may be slightly modulated by an apparently exceptional result (Fig. 2) from a red deer population with high seroprevalence. However, this factor also modulated the exposure of unmanaged deer to C. burnetii, thus showing the influence of human activities on the risk of exposure of deer to this pathogen. The density of coexisting domestic ruminants seems to dilute the risk of exposure of deer to C. burnetii. This result is shocking for a pathogen that is endemic in domestic ruminants in Iberia and whose transmission has been proven to be linked to host density (21, 22). This contrasting finding again suggests that the ecology of C. burnetii in wildlife is complex and that different wild and domestic species are involved in its maintenance, independently of the ability of the red deer to act as a true reservoir host. Indeed, the modeling output suggests that although an independent cycle of C. burnetii in red deer is possible without the intervention of a third species (susceptibility, systemic infection, and shedding demonstrated), other hosts may be implicated in the circulation of C. burnetii in wild foci.
Implications for animal and human health.
Haydon et al. (48) redefined the reservoir concept for multihost pathogens and stated that a specific pathogen of relevance for a target host of interest may be maintained through a high number of combinations of host populations or environments that keep the pathogen circulating. Therefore, defining the risk of transmission of C. burnetii from red deer to target hosts (livestock and humans) is difficult, preventing us from concluding whether the red deer plays a major role in C. burnetii maintenance in Iberia or not. We believe that red deer populations constitute a highly relevant node in the life cycle of C. burnetii, but particular scenarios of interaction with third species need to be further investigated. Wild lagomorphs and small mammals infected by C. burnetii, among others, may excrete infectious bacteria (45, 49, 50) and therefore constitute relevant pieces of the C. burnetii maintenance and transmission puzzle.
The risk of C. burnetii transmission from red deer to humans could be comparable to that from livestock if deer-human and livestock-human indirect interaction rates were similar. This is supported by the fact that both individual and population seroprevalences in red deer are similar to those found in domestic ruminants (21, 40, 51). Most effective livestock-human C. burnetii transmission events occur indirectly, through aerosols (33). We may expect that most deer-livestock and deer-human transmission events would occur indirectly (52). Therefore, the risk of transmission from deer to livestock and humans depends on the exposure rate of deer, suggesting that extensively produced domestic ruminants and humans involved in hunting and wild ungulate management and conservation face a higher risk (9, 53).
Our results point to free-ranging deer, perhaps in connection with other wild and domestic hosts, and deer farms as the main hot spots of circulation of C. burnetii among red deer in Iberia, and perhaps elsewhere in Europe. Further clarification of particular red deer-livestock or red deer-human interaction rates at different geographic scales should improve the chances of preventing C. burnetii transmission events.
ACKNOWLEDGMENTS
We are grateful to game estate owners, gamekeepers, natural and national park managers, and farm managers for their collaboration in sample collection. Special thanks go to José Antonio Ortiz from the LO farm and to Christian Gortázar for substantial support. We also acknowledge the great efforts of colleagues from the SaBio group at IREC in sample collection (Joaquín Vicente, Pelayo Acevedo, Isabel G. Fernández-de-Mera, Ursula Höfle, Paqui Talavera, Óscar Rodríguez, Álvaro Oleaga, Diego Villanúa, Vanesa Alzaga, Elisa Pérez, Raquel Jaroso, Raquel Sobrino, Encarnación Delgado, Jesús Carrasco, Ricardo Carrasco, Rafael Reyes García, Pablo Rodríguez, Mauricio Durán Martínez, Valeria Gutiérrez, and many others).
This work was funded by EU FP7 grant ANTIGONE (278976) and CDTI (Centro para el Desarrollo Tecnológico Industrial, Spanish Ministry for the Economy and Competitiveness [MINECO]). J. A. Barasona holds an FPU predoctoral scholarship from MECD. J. P. V. Santos and J. Queirós were supported by Ph.D. grants (SFRH/BD/65880/2009 and SFRH/BD/73732/2010, respectively) from the Portuguese Science and Technology Foundation (FCT). F. Ruiz-Fons is supported by a “Ramón y Cajal” contract from MINECO.
REFERENCES
- 1.Angelakis E, Raoult D. 2010. Q fever. Vet Microbiol 140:297–309. doi: 10.1016/j.vetmic.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 2.European Food Safety Authority (EFSA). 2010. Panel on Animal Health and Welfare (AHAW). Scientific opinion on Q fever. EFSA J 8:1595. doi: 10.2903/j.efsa.2010.1595. [DOI] [Google Scholar]
- 3.Ruiz-Fons F. 2012. Coxiella burnetii infection, p 409–412. In Gavier-Wid́en D, Duff JP, Meredith A (ed), Infectious diseases of wild mammals and birds in Europe, 1st ed Wiley-Blackwell, Chichester, United Kingdom. [Google Scholar]
- 4.Roest HIJ, Tilburg JJHC, van der Hoek W, Vellema P, van Zijderveld FG, Klaassen CHW, Raoult D. 2011. The Q fever epidemic in The Netherlands: history, onset, response and reflection. Epidemiol Infect 139:1–12. doi: 10.1017/S0950268810002268. [DOI] [PubMed] [Google Scholar]
- 5.Georgiev M, Afonso A, Neubauer H, Needham H, Thiéry R, Rodolakis A, Roest HJ, Stärk KD, Stegeman JA, Vellema P, van der Hoek W, More SJ. 2013. Q fever in humans and farm animals in four European countries, 1982 to 2010. Euro Surveill 18(8):pi=20407 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20407. [PubMed] [Google Scholar]
- 6.Babudieri B. 1959. Q fever: a zoonosis. Adv Vet Sci 5:81. [Google Scholar]
- 7.González-Barrio D, Almería S, Caro MR, Salinas J, Ortíz JA, Gortázar C, Ruiz-Fons J. 15 October 2013. Coxiella burnetii shedding by farmed red deer (Cervus elaphus). Transbound Emerg Dis doi: 10.1111/tbed.12179. [DOI] [PubMed] [Google Scholar]
- 8.Jado I, Carranza-Rodríguez C, Barandika JF, Toledo A, García-Amil C, Serrano B, Bolaños M, Gil H, Escudero R, García-Pérez AL, Olmeda AS, Astobiza I, Lobo B, Rodríguez-Vargas M, Pérez-Arellano JL, López-Gatius F, Pascual-Velasco F, Cilla G, Rodríguez NF, Anda P. 2012. Molecular method for the characterization of Coxiella burnetii from clinical and environmental samples: variability of genotypes in Spain. BMC Microbiol 12:91. doi: 10.1186/1471-2180-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tozer SJ, Lambert SB, Strong CL, Field HE, Sloots TP, Nissen MD. 2014. Potential animal and environmental sources of Q fever infection for humans in Queensland. Zoonoses Public Health 61:105–112. doi: 10.1111/zph.12051. [DOI] [PubMed] [Google Scholar]
- 10.Viana M, Mancy R, Biek R, Cleaveland S, Cross PC, Lloyd-Smith JO, Haydon DT. 2014. Assembling evidence for identifying reservoirs of infection. Trends Ecol Evol 29:270–279. doi: 10.1016/j.tree.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flueck WT, Smith-Flueck JM, Naumann CM. 2003. The current distribution of red deer (Cervus elaphus) in southern Latin America. Eur J Wildl Res 49:112–119. doi: 10.1007/BF02190451. [DOI] [Google Scholar]
- 12.Ludt CJ, Schroeder W, Rottmann O, Kuehn R. 2004. Mitochondrial DNA phylogeography of red deer (Cervus elaphus). Mol Phylogenet Evol 31:1064–1083. doi: 10.1016/j.ympev.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Zachos FE, Hartl GB. 2011. Phylogeography, population genetics and conservation of the European red deer Cervus elaphus. Mammal Rev 41:138–150. doi: 10.1111/j.1365-2907.2010.00177.x. [DOI] [Google Scholar]
- 14.Acevedo P, Ruiz-Fons F, Vicente J, Reyes-García AR, Alzaga V, Gortázar C. 2008. Estimating red deer abundance in a wide range of management situations in Mediterranean habitats. J Zool 276:37–47. doi: 10.1111/j.1469-7998.2008.00464.x. [DOI] [Google Scholar]
- 15.Apollonio M, Andersen R, Putman R. 2010. European ungulates and their management in the 21st century. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 16.Milner JM, Nilsen EB, Andreassen HP. 2007. Demographic side effects of selective hunting in ungulates and carnivores. Cons Biol 21:36–47. doi: 10.1111/j.1523-1739.2006.00591.x. [DOI] [PubMed] [Google Scholar]
- 17.Vicente J, Höfle U, Garrido JM, Fernández-De-Mera IG, Juste R, Barral M, Gortazar C. 2006. Wild boar and red deer display high prevalences of tuberculosis-like lesions in Spain. Vet Res 37:107–119. doi: 10.1051/vetres:2005044. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman LC, Wiklund E. 2006. Game and venison—meat for the modern consumer. Meat Sci 74:197–208. doi: 10.1016/j.meatsci.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Clutton-Brock TH, Guinness FE, Albon SP. 1982. Red deer: behaviour and ecology of two sexes. University of Chicago Press, Chicago, IL. [Google Scholar]
- 20.Vander Wal E, Paquet PC, Messier F, McLoughlin PD. 2013. Effects of phenology and sex on social proximity in a gregarious ungulate. Can J Zool 91:601–609. doi: 10.1139/cjz-2012-0237. [DOI] [Google Scholar]
- 21.Álvarez J, Pérez A, Mardones FO, Pérez-Sancho M, García-Seco T, Pagés E, Mirat F, Díaz R, Carpintero J, Domínguez L. 2012. Epidemiological factors associated with the exposure of cattle to Coxiella burnetii in the Madrid region of Spain. Vet J 194:102–107. doi: 10.1016/j.tvjl.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Piñero A, Ruiz-Fons F, Hurtado A, Barandika JF, Atxaerandio R, García-Pérez AL. 2014. Changes in the dynamics of Coxiella burnetii infection in dairy cattle: an approach to match field data with the epidemiological cycle of C. burnetii in endemic herds. J Dairy Sci 97:2718–2730. doi: 10.3168/jds.2013-7229. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz-Fons F, Reyes-García AR, Alcaide V, Gortázar C. 2008. Spatial and temporal evolution of Bluetongue virus in wild ruminants, Spain. Emerg Infect Dis 14:951–953. doi: 10.3201/eid1406.071586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boadella M, Carta T, Oleaga A, Pajares G, Muñoz M, Gortázar C. 2010. Serosurvey for selected pathogens in Iberian roe deer. BMC Vet Res 6:51. doi: 10.1186/1746-6148-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanet S, Trisciuoglio A, Bottero E, Fernandez De Mera IG, Gortázar C, Carpignano MG, Ferroglio E. 2014. Piroplasmosis in wildlife: Babesia and Theileria affecting free-ranging ungulates and carnivores in the Italian Alps. Parasite Vector 7:70. doi: 10.1186/1756-3305-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Víchová B, Majláthová V, Nováková M, Stanko M, Hviščová I, Pangrácová L, Chrudimský T, Čurlík J, Pet'ko B. 2014. Anaplasma infections in ticks and reservoir host from Slovakia. Infect Genet Evol 22:265–272. doi: 10.1016/j.meegid.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz PM, Boadella M, Arnal M, de Miguel MJ, Revilla M, Martínez D, Vicente J, Acevedo P, Oleaga T, Ruiz-Fons F, Marín CM, Prieto JM, de la Fuente J, Barral M, Barberán M, de Luco DF, Blasco JM, Gortázar C. 2010. Spatial distribution and risk factors of brucellosis in Iberian wild ungulates. BMC Infect Dis 10:46. doi: 10.1186/1471-2334-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stöbel K, Schönberg A, Staak C. 2002. A new non-species dependent ELISA for detection of antibodies to Borrelia burgdorferi s.l. in zoo animals. Int J Med Microbiol 291:88–89. doi: 10.1016/S1438-4221(02)80018-2. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Fons F, Astobiza I, Barral M, Barandika JF, García-Pérez AL. 2010. Modification of a commercial ELISA to detect antibodies against Coxiella burnetii in wild ungulates: application to population surveillance, abstr 6, p 16 Abstr 9th Biennial Conference, European Wildlife Disease Association https://docs.google.com/viewer?a=v&pid=sites&srcid=ZGVmYXVsdGRvbWFpbnxld2Rhd2Vic2l0ZXxneDo2ZDIyZmFjNmFjZjgxOWY1. [Google Scholar]
- 30.Tilburg JJHC, Melchers WJ, Petterson AM, Rossen JM, Hermans MH, van Hannen EJ, Nabuurs-Franssen MH, de Vries MC, Horrevorts AM, Klaassen CHW. 2010. Interlaboratory evaluation of different extraction and real-time PCR methods for detection of Coxiella burnetii DNA in serum. J Clin Microbiol 48:3923–3927. doi: 10.1128/JCM.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Fons F, Gilbert L. 2010. The role of deer as vehicles to move ticks, Ixodes ricinus, between contrasting habitats. Int J Parasitol 40:1013–1020. doi: 10.1016/j.ijpara.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 32.González-Barrio D, Queirós J, Fernández-de-Mera IG, Ruiz-Fons F. 2014. Dynamics of individual exposure to Coxiella burnetii infection in a Q fever endemic red deer (Cervus elaphus) farm, abstr 28, p 71 Abstr Joint 8th Ticks and Tick-Borne Pathogens and 12th Biennial Society for Tropical and Veterinary Medicine Conference http://www.savetcon.co.za/TTP8/files/TTP%20STVM%20Poster%20abstracts.pdf. [Google Scholar]
- 33.Maurin M, Raoult D. 1999. Q fever. Clin Microbiol Rev 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acevedo P, Ruiz-Fons F, Estrada R, Márquez AL, Miranda MA, Gortázar C, Lucientes J. 2010. A broad assessment of factors determining Culicoides imicola abundance: modelling the present and forecasting its future in climate change scenarios. PLoS One 5:e14236. doi: 10.1371/journal.pone.0014236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Real R, Barbosa AM, Rodríguez R, García FJ, Vargas JM, Palomo LJ, Delibes M. 2009. Conservation biogeography of ecologically interacting species: the case of the Iberian lynx and the European rabbit. Divers Distrib 15:390–400. doi: 10.1111/j.1472-4642.2008.00546.x. [DOI] [Google Scholar]
- 36.Astobiza I, Barral M, Ruiz-Fons F, Barandika JF, Gerrikagoitia X, Hurtado A, García-Pérez AL. 2011. Molecular investigation of the occurrence of Coxiella burnetii in wildlife and ticks in an endemic area. Vet Microbiol 147:190–194. doi: 10.1016/j.vetmic.2010.05.046. [DOI] [PubMed] [Google Scholar]
- 37.Rijks JM, Roest HIJ, van Tulden PW, Kik MJL, Ijzer J, Gröne A. 2011. Coxiella burnetii infection in roe deer during Q fever epidemic, The Netherlands. Emerg Infect Dis 17:2369–2371. doi: 10.3201/eid1712.110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ejercito CL, Cai L, Htwe KK, Taki M, Inoshima Y, Kondo T, Kano C, Abe S, Shirota K, Sugimoto T, Yamaguchi T, Fukushi H, Minamoto N, Kinjo T, Isogai E, Hirai K. 1993. Serological evidence of Coxiella burnetii infection in wild animals in Japan. J Wildl Dis 29:481–484. doi: 10.7589/0090-3558-29.3.481. [DOI] [PubMed] [Google Scholar]
- 39.Sáenz de Buruaga M, Lucio AJ, Purroy J. 1991. Reconocimiento de Sexo y Edad en Especies Cinegéticas. Diputación foral de Álava, Vitoria, Spain. [Google Scholar]
- 40.Hamlin KL, Pac DF, Sime CA, DeSimone RM, Dusek GL. 2000. Evaluating the accuracy of ages obtained by two methods for Montana ungulates. J Wildl Manage 64:441–449. doi: 10.2307/3803242. [DOI] [Google Scholar]
- 41.Ruiz-Fons F, Astobiza I, Barandika J, Hurtado A, Atxaerandio R, Juste R, García-Pérez AL. 2010. Seroepidemiological study of Q fever in domestic ruminants in semi-extensive grazing systems. BMC Vet Res 6:3. doi: 10.1186/1746-6148-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCulloch CE, Searle SR, Neuhaus JM. 2008. Generalized, linear, and mixed models, 2nd ed Wiley, Hoboken, NJ. [Google Scholar]
- 43.Burnham KP, Anderson DR. 2002. Model selection and multi-model inference. Springer, New York, NY. [Google Scholar]
- 44.Wobeser GA. 1994. Investigation and management of disease in wild animals. Plenum, New York, NY. [Google Scholar]
- 45.González-Barrio D, Maio E, Vieira-Pinto M, Ruiz-Fons F. 2015. European rabbits as reservoir for Coxiella burnetii. Emerg Infect Dis 21:1055–1058. doi: 10.3201/eid2106.141537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Fons F, Rodríguez O, Torina A, Naranjo V, Gortázar C, de la Fuente J. 2008. Prevalence of Coxiella burnetii infection in wild and farmed ungulates. Vet Microbiol 126:282–286. doi: 10.1016/j.vetmic.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 47.Gortázar C, Acevedo A, Ruiz-Fons F, Vicente J. 2006. Disease risk and overabundance of game species. Eur J Wildl Res 52:81–87. doi: 10.1007/s10344-005-0022-2. [DOI] [Google Scholar]
- 48.Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. 2002. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg Infect Dis 8:1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barandika J, Hurtado A, García-Esteban C, Gil H, Escudero R, Barral M, Jado I, Juste R, Anda P, García-Perez AL. 2007. Tick-borne zoonotic bacteria in wild and domestic small mammals in Northern Spain. Appl Environ Microbiol 73:6166–6171. doi: 10.1128/AEM.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson M, Mykytczuk N, Gooderham K, Schulte-Hostedde A. 2012. Prevalence of the bacterium Coxiella burnetii in wild rodents from a Canadian natural environment park. Zoonoses Public Health 59:553–560. doi: 10.1111/j.1863-2378.2012.01493.x. [DOI] [PubMed] [Google Scholar]
- 51.Astobiza I, Ruiz-Fons F, Piñero A, Barandika JF, Hurtado A, García-Pérez AL. 2012. Estimation of Coxiella burnetii prevalence in dairy cattle in intensive systems by serological and molecular analyses of bulk-tank milk samples. J Dairy Sci 95:1632–1638. doi: 10.3168/jds.2011-4721. [DOI] [PubMed] [Google Scholar]
- 52.Kukielka E, Barasona JA, Cowie CE, Drewe JA, Gortazar C, Cotarelo I, Vicente J. 2013. Spatial and temporal interactions between livestock and wildlife in South Central Spain assessed by camera traps. Prev Vet Med 112:213–221. doi: 10.1016/j.prevetmed.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 53.Whitney EAS, Massung RF, Candee AJ, Ailes EC, Myers LM, Patterson NE, Berkelman RL. 2009. Seroepidemiologic and occupational risk survey for Coxiella burnetii antibodies among US veterinarians. Clin Infect Dis 48:550–557. doi: 10.1086/596705. [DOI] [PubMed] [Google Scholar]
- 54.Salazar DC. 2009. Distribuição e estatuto do veado e corço em Portugal. Master's thesis. University of Aveiro, Aveiro, Portugal. [Google Scholar]
- 55.Palomo LJ, Gisbert J, Blanco JC. 2007. Atlas y libro rojo de los mamíferos terrestres de España. Dirección General para la Biodiversidad–SECEM–SECEMU, Madrid, Spain. [Google Scholar]