Abstract
In the present study, microbial toluene degradation in controlled constructed wetland model systems, planted fixed-bed reactors (PFRs), was queried with DNA-based methods in combination with stable isotope fractionation analysis and characterization of toluene-degrading microbial isolates. Two PFR replicates were operated with toluene as the sole external carbon and electron source for 2 years. The bulk redox conditions in these systems were hypoxic to anoxic. The autochthonous bacterial communities, as analyzed by Illumina sequencing of 16S rRNA gene amplicons, were mainly comprised of the families Xanthomonadaceae, Comamonadaceae, and Burkholderiaceae, plus Rhodospirillaceae in one of the PFR replicates. DNA microarray analyses of the catabolic potentials for aromatic compound degradation suggested the presence of the ring monooxygenation pathway in both systems, as well as the anaerobic toluene pathway in the PFR replicate with a high abundance of Rhodospirillaceae. The presence of catabolic genes encoding the ring monooxygenation pathway was verified by quantitative PCR analysis, utilizing the obtained toluene-degrading isolates as references. Stable isotope fractionation analysis showed low-level of carbon fractionation and only minimal hydrogen fractionation in both PFRs, which matches the fractionation signatures of monooxygenation and dioxygenation. In combination with the results of the DNA-based analyses, this suggests that toluene degradation occurs predominantly via ring monooxygenation in the PFRs.
INTRODUCTION
Biology-based remediation technologies (bioremediation) for the treatment of groundwater and soils polluted with organic compounds have been receiving high interest due to their low cost, high efficiency, and relative operational simplicity (1). Rhizoremediation is one such effective bioremediation approach, where the transformation of contaminants to innocuous products may be enhanced by plant-microbe interactions occurring in the rhizosphere (2–4). The plants provide the rhizospheric microbial community with root exudates such as carbohydrates, amino acids/amines, and organic acids (5) and thus promote the microbes' establishment and proliferation. Rhizoremediation is particularly effective in constructed wetlands. These treatment systems have been applied for the removal of even high loads of contaminants present in inflow waters (6–8). In addition to the input of labile organic carbon, the helophytes used in these systems have the capacity of channeling significant amounts of oxygen from the atmosphere through a specific tissues, the aerenchyma, into their roots, and thereby foster aerobic microbial activities in the rhizosphere (9).
In order to enhance the performance of constructed wetlands through engineering optimizations of the systems, it is deemed necessary to understand the functionality of the relevant microbial community present in the rhizosphere, and some efforts have been pursued in this direction (7, 10). However, due to the presence of steep chemical gradients and variable environmental conditions, it has been difficult to dissect many of the microbial processes occurring in constructed wetlands. To overcome the difficulties in studying such complex systems, a laboratory-scale rhizosphere reactor was designed and tested (11). Within these planted fixed-bed reactors (PFRs) a macro-gradient-free flow is generated and an efficient turnover of organic carbon occurs. PFRs have been used successfully to study the effect of different redox cycles on the contaminant removal from waste- and groundwater (12, 13).
In our laboratory, two PFRs were operated for 2 years using toluene as the sole external carbon and electron source. Among the BTEX compounds (benzene, toluene, ethylbenzene, and the three isomers of xylene), the biodegradation of toluene has been most extensively studied and several bacterial strains able to degrade toluene aerobically or anaerobically are known (14–16) (see Fig. S1 in the supplemental material). In the three main aerobic pathways, toluene is activated through monooxygenation either of the methyl group (side-chain monooxygenases, pathway 1 in Fig. S1 in the supplemental material) or the phenyl ring (soluble diiron monooxygenases, pathways 2, 3, and 4) or by dioxygenation of the phenyl ring (Rieske non-heme iron oxygenases, pathway 5) (17). Most of the aerobic pathways, except for the toluene 4-monooxygenase pathway of Pseudomonas mendocina KR1 (pathway 4), converge in catecholic intermediates which are subsequently cleaved by extradiol dioxygenases (typically type I or vicinal oxygen chelate superfamily of extradiol dioxygenases enzymes) (15, 18). Anaerobically, the degradation of toluene is initiated by the addition of fumarate to the methyl group to form benzylsuccinate (pathway 6), which is further metabolized to benzoyl-coenzyme A (benzoyl-CoA) as the central intermediate in the anaerobic degradation of many aromatic compounds (16).
Our aim here was to identify the dominant toluene biodegradation pathway in the PFR model wetland systems. To this purpose, the genetic potential for microbial toluene transformation was assessed by deciphering the microbial diversity via next generation sequencing and the catabolic gene landscape using a recently developed microarray (19), substantiated by quantitative PCR (qPCR) analysis of selected genes. These results on catabolic potentialities were combined with 13C and 2H stable isotope fractionation analysis to characterize the system regarding the predominant toluene degradation pathway (20). The combination of molecular biology and stable isotope analysis allowed identifying for the first time the microbial toluene degradation mechanisms in these complex wetland-like systems.
MATERIALS AND METHODS
PFR setups.
The test systems have been previously described in detail (11), and a picture of a planted-reactor is shown in Fig. S2 in the supplemental material. Briefly, two PFRs (PFR-1 and -2) consisting of a cylindrical glass vessel (diameter, 30 cm; height, 30 cm) and containing a metal basket (diameter, 26 cm; height, 28 cm) filled with ∼20 kg of gravel were operated for a 2-year period. The reactors were planted with soft rush, Juncus effusus, and closed with a Teflon lid with five openings for the rush. The plants were replaced in spring of each year, but the gravel matrix was kept intact as much as possible to lessen the impact on the established microbial communities in the reactors. The handling, feeding regime, and running conditions were identical for both reactors. Before starting with toluene addition, an acclimation period of 1 month for the new plants was necessary. During this time, acetate (102.4 mg/liter) and/or benzoate (53.5 mg/liter) were used as additional carbon source to sustain the microbial community. Each reactor was constantly fed with distilled water supplemented with toluene (10 to 50 mg/liter) as an external carbon source and 1 ml/liter of a trace element solution (0.25% HCl, 151 mg/liter FeCl2·4H2O, 68.1 mg/liter ZnCl2, 98.9 mg/liter MnCl2·4H2O, 6.18 mg/liter H3BO3, 190.3 mg/liter CoCl2·6H2O, 1.7 mg/liter CuCl2·2H2O, 23.77 mg/liter NiCl2·6H2O, 36.3 mg/liter Na2MoO4·2H2O). The concentration of the feeding toluene solution was increased gradually every 2 to 3 weeks, when no toluene was detected in the circulation flow. Additional nutrients (NO3−, 135 mg/liter; NH4+, 25.71 mg/liter; PO43−, 15.29 mg/liter) were constantly injected into the outer inflow ring of the reactor using a syringe pump (model KDS 200; KD Scientific, Inc.). The reactors possess a pore volume of ∼10 liters, and the hydraulic retention time was 5 days. Together with the constant inflow feeding, an internal circulation flow was used for the mixing of the liquid inside the reactor, thus generating macrogradient-free conditions in the fixed bed. The level of liquid inside the reactors and therefore the outflow were controlled by a level control system that was comprised of a bottom pressure sensor, which controls the outlet valve in the internal circulation flow. Redox potential (Pt-metal redox potential electrode, 2ME-2G-PtK-1; Jumo), pH (pH-electrode, 2G E-2-G-U-1; Jumo), and dissolved oxygen concentration (membrane electrode, SIPAN 34, 7MA3100-8EF; Siemens) were constantly measured in the circulation flow and automatically recorded. The reactors were operated in a greenhouse under defined environmental conditions to simulate an average summer day in a moderate climate. The temperature set points were 22°C from 6 a.m. to 9 p.m. and 16°C at night. The light intensity was continuously monitored on the roof of the greenhouse by means of a phototransistor (Adolf Thies GmbH). Whenever the light intensity fell below 60 kLux, one lamp (Master SON-PIA 400 W; Phillips) per reactor was switched on during daytime as an additional light source.
Toluene measurements in liquid phase.
Inflow and circulation flow water samples were taken twice per week. Toluene concentration was measured using a gas chromatograph with a flame ionization detector (GC-FID) (Agilent HP6890) equipped with a 30 m by 0.45 mm by 2.55-μm film thickness Agilent DB-MTBE column. The temperature program was as follows: 70°C for 2 min, followed by heating by 30°C min−1 until reaching 260°C and then holding for 2 min. The FID detection temperature was 280°C, and helium was used as the carrier gas (5.8 ml/min). The injection was via an automated headspace autosampler (DANI HSS 86.50) after equilibration of the samples for 10 min at 50°C. Water samples of 5 ml were collected in 20-ml glass vials (Supelco) containing 5 ml of sodium azide solution (final concentration, 0.65 g/liter) to inhibit microbial activity. Vials were closed with a Teflon-coated butyl rubber septum. The samples were stored at −20°C until measurement.
DNA extraction.
Pore water samples were collected from five different points inside each PFR (180 ml in total) with a syringe. A 200-mm-long metal needle (2 mm in diameter; Glasgerätebau Ochs) was used to reach the rhizospheric area. In order to detach biofilms settled on the gravel or roots, water was flushed up and down the syringe several times before taking the respective sample. The five samples were filtered together to collect the biomass using polyethersulfone filters with a pore size of 0.2 μm (Pall Corp.). The filters were cut into small pieces and distributed to 2-ml bead solution tubes from an UltraClean soil DNA isolation kit (MO BIO Laboratories, Inc.). The alternative protocol for maximum yields recommended by the manufacturer was followed for DNA extraction.
Illumina sequencing and diversity analysis.
The bacterial community structure of the PFRs was analyzed by Illumina sequencing in June of 2011. The amplicon library preparation and the bioinformatics analysis was essentially performed as described before (21), with the differences that in the present study, the variable regions V5-V6 of the bacterial 16S rRNA gene were amplified using the primers 807F and 1050R (22), and both the forward and reverse sequence reads (85 bp each) were processed. The PCR products were purified and reamplified to add barcodes and adapters for Illumina sequencing (21). Totals of 141,084 and 36,201 reads were obtained for PFR-1 and PFR-2, respectively. To trim the 3′ ends of the reads that fell below a quality score of 15, a quality filter that runs a sliding window of 10% of the read length at a time and calculates the local average score based on the Phred quality score of the fastq file was used (http://wiki.bioinformatics.ucdavis.edu/index.php/Trim.pl). All reads with nucleotide ambiguity, mismatches with primers or barcodes, or homopolymer stretches of >10 nucleotides (nt) were discarded using mothur (23), leaving a total of 106,966 reads. These remaining reads were pairwise combined, leaving a gap in the middle, and clustered, allowing for two mismatches, using the mothur programs unique.seqs and pre.cluster. Phylogenetic analysis was carried out using the Ribosomal Data Project (http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp). All mitochondria sequences and sequences that did not appear to be of 16S rRNA gene origin were deleted. Since there is no program available to identify chimeras in short sequence reads with high quality, chimera analysis was performed manually. Phylotypes where the reverse read and the forward read were previously observed in different abundant phylotypes were considered putative chimers. Only low-abundance phylotypes were considered chimers, and a phylotype was discarded if its abundance was <2% of the apparent “parent” phylotypes of the two reads. The data set of 106,966 sequences was then filtered to consider only phylotypes that appeared in at least one of the bioreactors at >0.02% abundance (a total of 90,661 sequences: PFR-1, 71,306; PFR-2, 19,355). Normalization to obtain the same number of reads per sample (19,355) and rarefaction analyses were performed using R (24).
DNA microarray for catabolic transformation of aromatic compounds.
To investigate the catabolic potential of the microbial community in the planted reactors, a catabolome microarray designed and tested previously was used (19). Three technical replicates of each sample were analyzed using the microarray. Briefly, DNA amplification was performed with 100 ng of template DNA using a REPLI-g Ultrafast kit (Qiagen), and amplified DNA was purified, suspended in 100 μl of Milli-Q water, and heat fragmented at 95°C for up to 1.5 h. After precipitation, DNA was suspended in 45 μl of Milli-Q water and labeled using terminal transferase (Roche) and Cy5-dUTP (GE Healthcare). Labeled DNA fragments were precipitated and suspended in 20 μl of Milli-Q water, and 80 μl of hybridization buffer was added. For hybridization, slides (CodeLink activated slides; SurModics, Eden Praine, MN) were inserted into the hybridization chamber (Slide-Booster SB401/800; Advalytics/Beckman Coulter Biomedical GmbH, Munich, Germany) and covered by coverslips (Implen GmbH, Munich, Germany). Labeled DNA was hybridized at 55°C over 18 h, using hybridization buffer comprising 15% (vol/vol) dimethyl sulfoxide, 25% (vol/vol) formamide, 1.25× SSC (190 mM sodium chloride plus 20 mM trisodium citrate), 0.15% sodium dodecyl sulfate (SDS), 0.15% Tween20, 880 mM Betaine, 5× TE buffer (50 mM Tris-HCl, 5 mM EDTA), and 0.1 mg of bovine serum albumin (BSA)/ml in aqueous solution, corresponding to a final concentration of 1× SSC. The slides were subsequently washed in 1× SSC–0.3% SDS (5 min, 42°C), twice in 1× SSC (1 min each, 20°C), once in 0.5× SSC (1 min, 20°C) and 0.1× SSC–0.3% SDS (1 min, 42°C), and twice in 0.1× SSC (1 min, 20°C). Slides were dried by centrifugation (ArrayIt microarray high-speed centrifuge; Arrayit Corp., Sunnyvale, CA) and scanned using a high-resolution microarray scanner (Agilent Technologies, Life Sciences and Chemical Analysis Group, Santa Clara, CA). After scanning, spots corresponding to the internal controls 50-mer A to D were used to generate a standard curve, and spots with an intensity higher than 50-mer A were further analyzed (19).
PCR detection and sequencing of catabolic genes in toluene-degrading isolates and qPCR.
Nine aerobic toluene-degrading bacterial strains were isolated from PFR-1 and -2. Briefly, pore water samples taken in December 2011 were serially diluted, plated on M9 mineral medium agar, and placed in hermetically closed jars with toluene vapor as the sole carbon source. Subsequently, colonies with different phenotypes were transferred to toluene-amended mineral broth, and the purity of the toluene-degrading isolates was tested on several rich media (LB, R2A, and CM149). The 16S rRNA gene of each strain was amplified by PCR using universal primers (25), the purified PCR products were sequenced, and the sequences were phylogenetically assigned using the BLASTn tool (http://www.ncbi.nlm.nih.gov/BLAST/). The axenic cultures were examined for the presence of catabolic genes involved in toluene degradation using several previously described degenerate primers (26–29) and the newly designed primers PHE-F (5′-GAYCCBTTYCGYHTRACCATGGA) and PHE-R (5′-GGCARCATGTARTCCWKCATCAT) for amplifying a 700-bp gene fragment of the α-subunit of phenol-methylphenol monooxygenases of the soluble diiron monooxygenases (19, 30). The PHE-F and PHE-R primers were used at a final concentration of 0.5 μM according to the following PCR program: 95°C for 15 min; 35 cycles of 95°C for 45 s, 52°C for 45 s, and 72°C for 1 min; and a final elongation at 72°C for 8 min. Three isolates (AET-5-01, AET-5-VIII, and AET-6-14) representing three different aerobic toluene degradation pathways were chosen as reference strains, and the corresponding catabolic genes were sequenced (see Table S1 in the supplemental material). The closest homologs of the sequences were identified using the blastx program via the NCBI website.
New primer sets suitable for qPCR were designed using Beacon Designer program (Premier Biosoft) targeting the genes for xylene monooxygenase (xylM-AET01-F/-R), toluene dioxygenase (TOD-AET18-F/-R), catechol-2,3-dioxygenase (EXDO-AET18-F/-R and EXDO-AET14-F/-R), toluene monooxygenase (TMO-AET14-F/-R), and phenol hydroxylase (PHE-AET14-F/-R) (see Table S2 in the supplemental material). For quantification of the total number of bacteria, universal eubacterial primer sets previously described by Nadkarni et al. (25) were used. SYBR green was used as a fluorescent dye (SensiMix SYBR Low-ROX kit; Bioline); a PCR mix consisted of 1× of SensiMix SYBR Low-ROX kit, forward and reverse primer, 1% BSA, 300 nM ROX, 1 μl of template, and DNase-free water to complete a reaction volume of 12.5 μl. Thermal cycling conditions for the selected primers were as follows: an initial cycle of 95°C for 10 min; 40 cycles of 95°C for 15 s, an annealing step for 20 s (acquisition data step), and an elongation step at 72°C for 20 s; 1 cycle at 95°C for 15 s and 60°C for 1 min; and 80 cycles from 60°C for 15 s at +0.3°C cycle−1 (melting curve). A standard curve was prepared using purified and quantified PCR products of the corresponding catabolic genes of reference strains Pseudomonas putida mt-2 (xylM and 16S rRNA gene), Pseudomonas putida F1 (todC1), isolate AET-6-14 (tmoA), and Ralstonia eutropha JM134 (phe). The copy number per microliter of the purified PCR product was calculated according to the following equation: (6.022 × 1023 molecules/mol × PCR product concentration [g/μl])/(number of base pairs × 660 Da). A freshly prepared 10-fold dilution series of the PCR product, covering from 108 to 101 copies μl−1, was used as calibration curve for each run. Triplicates of each sample were loaded in each run, and the standard deviation was calculated. Six blanks containing only the master mix were also used per array. The formation of primer dimers and/or unspecific PCR products was monitored using a melting curve analysis.
13C and 2H isotope fractionation.
At the end of second year of operation (September 2012), a pulse experiment was performed to measure the 13C and 2H isotope fractionation in the PFRs. For the isotope fractionation experiment, 330 mg of toluene dissolved in 1 liter of water was injected directly into the gravel beds of the systems, in addition to the constant inflow feed of 40 mg/liter in order to reach an initial concentration in the reactor of ∼30 mg/liter. The subsequent decrease in toluene concentration in the PFRs was measured by taking duplicate pore water samples (5 ml of each) three times during each day and night from two different sampling points in the reactors. After 31 h, additional toluene (330 mg of in 1 liter of water) was injected into the reactors, and again a continuous decrease in the toluene concentration was observed. The samples for the isotope analysis (13 ml of pore water) were pipetted into 15-ml screw-cap vials to which subsequently 0.5 ml of HCl (37%) and 1 ml of cool pentane were added for toluene extraction. The vials were stored inverted at 4°C until measurement. The carbon and hydrogen isotope signatures of toluene were analyzed by gas chromatography-isotope ratio mass spectroscopy as described elsewhere (20). The samples were analyzed in triplicates. Carbon and hydrogen isotope signatures are given in δ-notation (‰) relative to the Vienna Pee Dee Belemnite standard (VPDB) and Vienna standard mean ocean water (VSMOW), respectively, according to the following equation: δ13Csample or δ2Hsample (‰) = (Rstandard − Rsample/Rstandard) × 1,000, where Rsample and Rstandard are the 13C/12C and 2H/1H ratios of each sample and an international standard, respectively. Calculations of isotopic enrichment factors (ε), which describe the relationship between changes in isotope composition and the concentrations during the course of the experiment, were determined from the natural logarithm of the simplified Rayleigh equation for closed systems: (Rt/R0) = (Ct/C0)ε/1,000. The errors of the enrichment factors are given as 95% confidence interval and determined using a linear regression analysis (31).
Toluene volatilization measurements during isotope fractionation experiment.
During the 13C and 2H isotope fractionation experiment, gaseous emissions of toluene from the PFRs were determined. A cylindrical flowthrough test chamber made of ethylene tetrafluoroethylene foil (internal diameter, 34 cm; height, 85 cm) was placed upon the plants in the reactors, leaving an air entrance at the bottom of ∼5 cm. Air with a flow rate of about 110 liters/min was pumped through the flow chamber to collect all gaseous emission from the plants. To determine background toluene concentrations, the ambient air was sampled next to the air entrance of the test chamber. At the upper funnel-shaped end of the chamber, a subsample of the airflow through the chamber was collected on Tenax TA test tubes using a gas sampling device with a flow rate of 60 ml/min. Samples were collected every 8 h during the 3-day experiment on Tenax TA (150 mg) filled into glass tubes (SKC, 110 by 8 mm; Eighty Four PA). Two tubes were installed in series to avoid potential pass-through of substances. Samples were analyzed using a Thermo-desorption gas chromatography-mass spectrometry system (EM 640/S; Bruker) equipped with a DB1 column (30 m by 0.25 mm [inner diameter] by 1-μm film). The desorption step was carried out at 250°C (180 s), and the following temperature program was applied for gas chromatography analysis: 40°C (3 min) and 15°C min−1 to 250°C (1 min). Injection was splitless (120 s) with an inlet temperature of 260°C and helium as the carrier gas. To calculate the losses due to volatilization from the PFRs, the total free mass of toluene in the pore water was calculated by integration of the concentration during the measuring period of 8 h by using equations 1 and 2. The peak-like toluene concentration after the first toluene injection in both reactors (t = 0) was used to estimate the rate constants, whereas the second injection peak was used to validate the constants:
| (1) |
| (2) |
The total mass of toluene present in the pore water sample during an 8-h period corresponded to the hundred percentage of toluene available for evaporation. The percentage of evaporated toluene can be calculated as follows: (mass of toluene evaporated in mg × 100)/(free mass of toluene in pore water in mg).
Nucleotide accession numbers.
The nucleotide sequences of the Illumina analysis were deposited in the European Nucleotide Archive under accession numbers LN852401 to LN852658. The 16S rRNA gene sequences and the corresponding catabolic gene sequences from the toluene-degrading isolates have been deposited in the GenBank database under accession numbers KR902601 to KR902609 and KT021789 to KT021794, respectively.
RESULTS
Toluene degradation in PFRs.
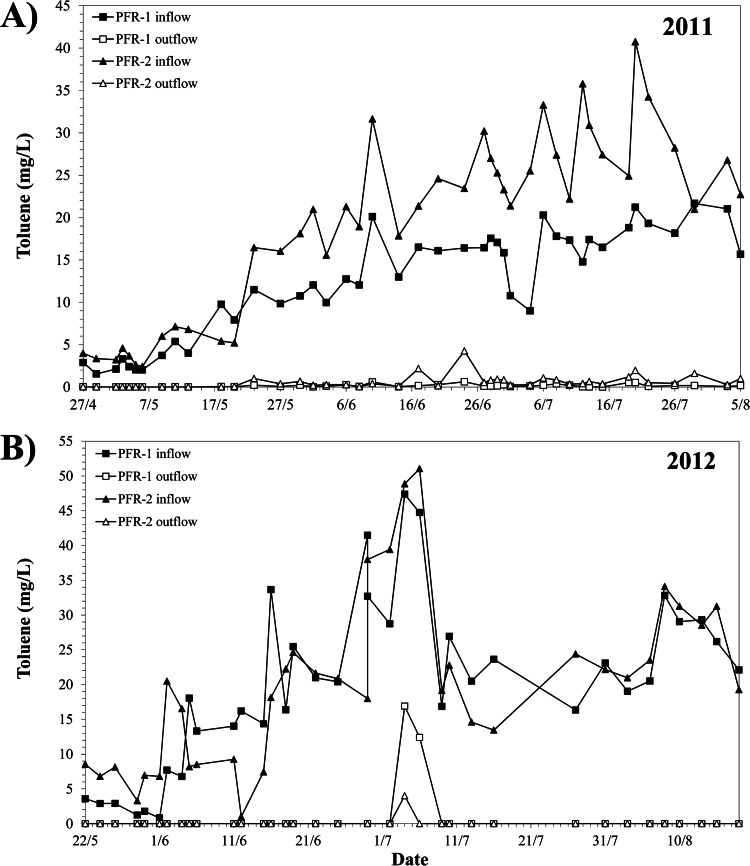
Over 2 years of operation (2011 and 2012) the toluene concentration of the inflow and circulation flow of PFR-1 and -2 was monitored weekly during the months of higher photosynthetic plant activity (approximately May to August). Toluene removal was equally efficient in both reactors, and independent of the toluene concentration in the inflow (Fig. 1). Only residual amounts of toluene were detected in the circulation flow of the PFRs after each increment of the inflow concentration. During the first year, these peaks of toluene reached at maximum 18% (4.3 mg/liter) and were typically below 3% of the inflow concentration (Fig. 1A). Likewise, in the subsequent year of operation toluene concentrations in the circulation flows were predominantly low and only reached ca. 8 and 36% of that of the inflow for a few days when the inflow concentration was increased up to 50 mg/liter (Fig. 1B).
FIG 1.
Toluene concentration measured in the inflow (solid symbols) and the outflow (open symbols) of PFR-1 and PFR-2 during the operation period in 2011 (A) and 2012 (B).
The respective bulk aqueous phase of the PFRs was largely hypoxic and even anoxic with measured molecular oxygen levels fluctuating between zero and about 1 mg/liter (see Fig. S3 in the supplemental material). Periods of anoxia were more pronounced during the first year (e.g., July 2011; see Fig. S3B in the supplemental material). During some time frames (e.g., 21 March 2012 to 18 June 2012 in Fig. S3C in the supplemental material), the O2 concentration oscillated concomitantly with the day/night rhythm, as described previously (9). The redox potential, Eh, reflected the measured oxygen condition (see Fig. S3 in the supplemental material). During the first year, the Eh ranged mostly around +300 mV, except during the period of prolonged anoxia, while in 2012 the redox potential was shifted to more positive values.
Biodiversity and potential transformation capabilities for aromatic compounds.
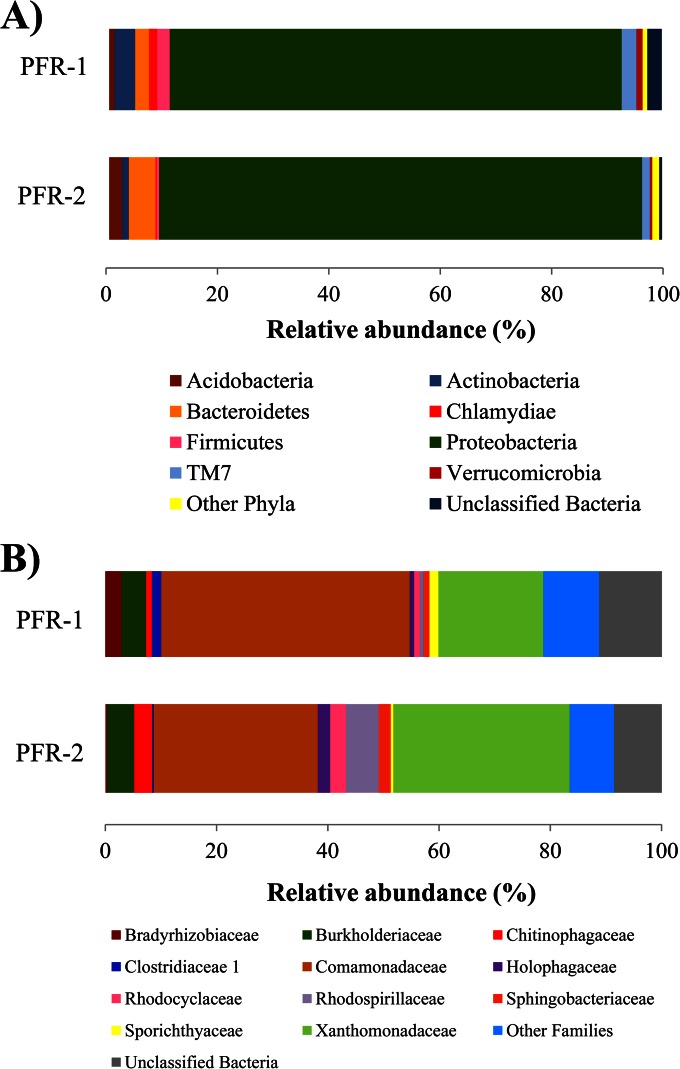
In June of the first year of operation (2011), DNA was isolated from pore water samples of both reactors, and biodiversity and catabolic profiles were recorded from these systems. Illumina sequencing of the V5-V6 16S rRNA gene regions resulted in a total of 258 phylotypes identified (the total numbers of filtered sequences in PFR-1 and PFR-2 were 71,306 and 19,355, respectively). The phylotype sequences and their abundances (as a percentage), together with their corresponding classifications, are shown in Table S3 in the supplemental material. Rarefaction curves of all high-quality sequences showed a plateau, which demonstrates that the sequencing depth captured sufficiently the bacterial communities in the PFRs (see Fig. S4 in the supplemental material). The richness (S) values were 213 and 183 for PFR-1 and -2, respectively. Shannon diversity (H′) and Pielou's evenness (J′) values were very similar for both reactors (H′, 3.1 and 3.4, respectively; J′, 0.59 and 0.65, respectively). A total of 239 phylotypes corresponding to 97.5 and 99.5% of the total reads could be assigned to the phylum level. Of the 16 phyla detected, Proteobacteria was the most abundant, with relative abundances of 81.8 and 87.3% for PFR-1 and PFR-2, respectively (Fig. 2A). Far less represented were Bacteroidetes (2.5 and 4.8%) and Actinobacteria (3.7 and 1.2%) and with <3% relative abundance the phyla Acidobacteria, Firmicutes, and TM7, with minor distribution differences in both systems (Fig. 2A). The majority of phylotypes (195 phylotypes accounting for 91.0 and 92.8% of reads, respectively) could be classified to the family level. Just two families accounted for more than 50% of the reads in both reactors, Comamonadaceae (Betaproteobacteria; 44.6 and 29.4%) and Xanthomonadaceae (Gammaproteobacteria; 18.8 and 31.7%) (Fig. 2B). Next to the variant proportions of these two families, the reactors differed in the abundance of Rhodospirillaceae (Alphaproteobacteria; 5.9% in PFR-2 versus 0.7% in PFR-1), whereby almost all of those sequences affiliated with Magnetospirillum magnetotacticum. The family Burkholderiaceae (Betaproteobacteria) was similarly distributed in both reactors, with relative abundances of 4.4 and 4.8% in PFR-1 and -2, respectively. The families Sphingobacteriaceae and Chitinophagaceae from the Sphingobacteria, Rhodocyclaceae (Betaproteobacteria), and Bradyrhizobiaceae (Alphaproteobacteria) were also part of the microbial community, but their relative abundances were ca. 3% or less, and their distributions varied strongly between both PFRs (Fig. 2B).
FIG 2.
Bacterial community composition of PFR-1 and PFR-2. Relative abundance of bacterial classes (A) and families (B) in DNA extracted from pore water samples of PFR-1 and -2. The variable region V5-V6 of the 16S rRNA gene was sequenced using Illumina. Only phyla and families with a representation of ≥1% in at least one of the rectors are represented here. The categories “other phyla” and “other families” represent taxa with a representation of <1% in both reactors.
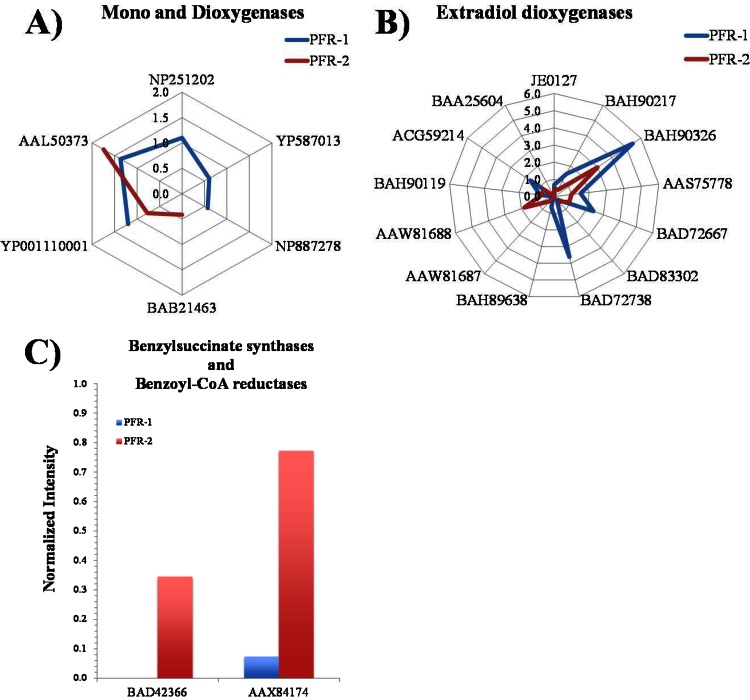
In agreement with the biodiversity profiles, the DNA microarray-based query of the genomic potential for aromatic compound transformation showed mostly hybridization signals with probes that represent genes from Proteobacteria (for a description of the enzymes associated to the accession numbers used to describe the results see Table S4 in the supplemental material). Of the 1,595 probes represented in the microarray, 37 showed hybridization, but only a few of these were directly related to toluene degradation. Although the aerobic toluene degradation pathways detected by the arrays differed slightly between the reactors, both reactors shared a set of signals that indicate the presence of proteobacterial phenol hydroxylase encoding genes (i.e., AAL50373 and YP_001110001) (Fig. 3A). Phenol hydroxylases belong to the superfamily of soluble diiron monooxygenases (30) and have a preference for phenol and methyl-substituted phenols as the substrates. However, some representatives of this group of enzymes may act also on toluene (32, 33), and the presence of a phenol hydroxylase could enable bacterial strains to mineralize toluene or benzene via two successive ring monooxygenations (34). In accordance with this pathway, probes representing genes encoding a subtype of catechol ring-cleavage extradiol dioxygenases (BAH90326 and AAS75778), typically encoded in an operon with phenol hydroxylase encoding genes, also showed hybridization (Fig. 3B). Hybridization with probe AAB70825 (see Table S4 in the supplemental material) indicates the presence of xylene monooxygenase and thus toluene degradation via hydroxylation of the methyl group in PFR-1 only (see Fig. S5 and pathway 1 in Fig. S1 in the supplemental material). BAD72667, BAD72738, and AAW81688 (Fig. 3B), targeting the subfamily I.2.A (18) of extradiol dioxygenase also showed hybridization. Even though this subfamily comprises genes encoded in the same gene cluster as xylene monooxygenases (35), respective genes were also observed in salicylate catabolic gene clusters (36) or phenol-degrading strains (37), and does not necessarily give an indication of the presence of the so called TOL pathway, but rather they confirm the presence of Gammaproteobacteria in PFR-1. Probes targeting benzene/toluene/isopropylbenzene/biphenyl Rieske non-heme iron oxygenases did not show hybridization, indicating that the dioxygenation pathway (pathway 5 in Fig. S1 in the supplemental material) is of minor importance, if any, in the PFRs. Three other probes targeting dioxygenases involved in the degradation of benzoate or substituted benzoates (NP_251202, YP_587013, and BAB21463) showed hybridization (Fig. 3A); however, the respective genes typically belong to the core genome of Proteobacteria, and therefore they are probably fortuitously enriched and not directly involved in toluene degradation.
FIG 3.
Abundance of catabolic genes involved in aromatic compounds degradation present in PFR-1 and PFR-2 expressed as the normalized intensity. (A) Rieske non-heme iron oxygenases (dioxygenases) and soluble diiron monooxygenases (YP_001110001, methane-phenol-toluene hydroxylase [Burkholderia vietnamiensis G4]; AAL50373, TomA3 [Burkholderia cepacia]; NP_251202, anthranilate dioxygenase large subunit [Pseudomonas aeruginosa PAO1]; BAB21463, chlorobenzoate 1,2-dioxygenase [Burkholderia sp. strain NK8]; YP_587013, benzoate 1,2-dioxygenase [Cupriavidus metallidurans CH34]; NP_887278, hydroxylating alpha subunit of a dioxygenase system [Bordetella bronchiseptica RB50]). (B) Extradiol dioxygenases of the vicinal chelate superfamily (catechol 2,3-dioxygenases; BAD72667, BAD83302, BAD72738, BAH89638, and BAH90326, catechol 2,3-dioxygenase [uncultured bacterium]; AAW81687, catechol 2,3-dioxygenase [Pseudomonas fluorescens PC36]; AAW81688, catechol 2,3-dioxygenase [Pseudomonas fluorescens PC37]; AAS75778, catechol 2,3-dioxygenase [Comamonas testosteroni CC F3]; BAH90217, 2,3-dihydroxybiphenyl-1,2-dioxygenase [uncultured bacterium]; JE0127, catechol 2,3-dioxygenase [EC 1.13.11.2] [Pseudomonas sp. strain Y2]; BAH90119, 2,3-dihydroxybiphenyl-1,2-dioxygenase [uncultured bacterium]; BAA25604, 2,3-dihydroxybiphenyl 1,2-dioxygenase [Rhodococcus erythropolis TA421]; ACG59214, 3-isopropylcatechol-2,3-dioxygenase-like protein [uncultured bacterium]). (C) Probes involved in anaerobic toluene degradation: benzylsuccinate synthase (BAD42366, putative benzylsuccinate synthase alpha subunit [Magnetospirillum sp. strain TS-6]); and benzoyl-CoA reductase (AAX84174, putative benzoyl-CoA reductase [Thauera aromatica]).
The potential for the anaerobic degradation of toluene was present only in PFR-2, where probes for benzylsuccinate synthase (BAD42366) and benzoyl-CoA reductase (AAX84174) similar to those of Magnetospirillum sp. strain TS6, a toluene degrader under nitrate-reducing conditions (38), showed hybridization (Fig. 3C). These results are in accordance with the microbial community structure data, where Magnetospirillum sequences were more abundant in PFR-2.
Some probes targeting intradiol dioxygenase also hybridized with the DNA of the planted reactors (data not shown). This is highly expected, as these belong to the core genome of various Proteobacteria, including Burkholderiales (39) and would only give indications of the presence of this phyla in the PFRs. Similarly, the significant number of probes representing the alkane monooxygenases indicated the enrichment of strains with the capacity of aliphatic hydrocarbons degradation (see Fig. S5 in the supplemental material), which are compounds normally found as part of plant-derived hydrocarbons (40).
Detection of catabolic genes from aerobic isolates in PFRs.
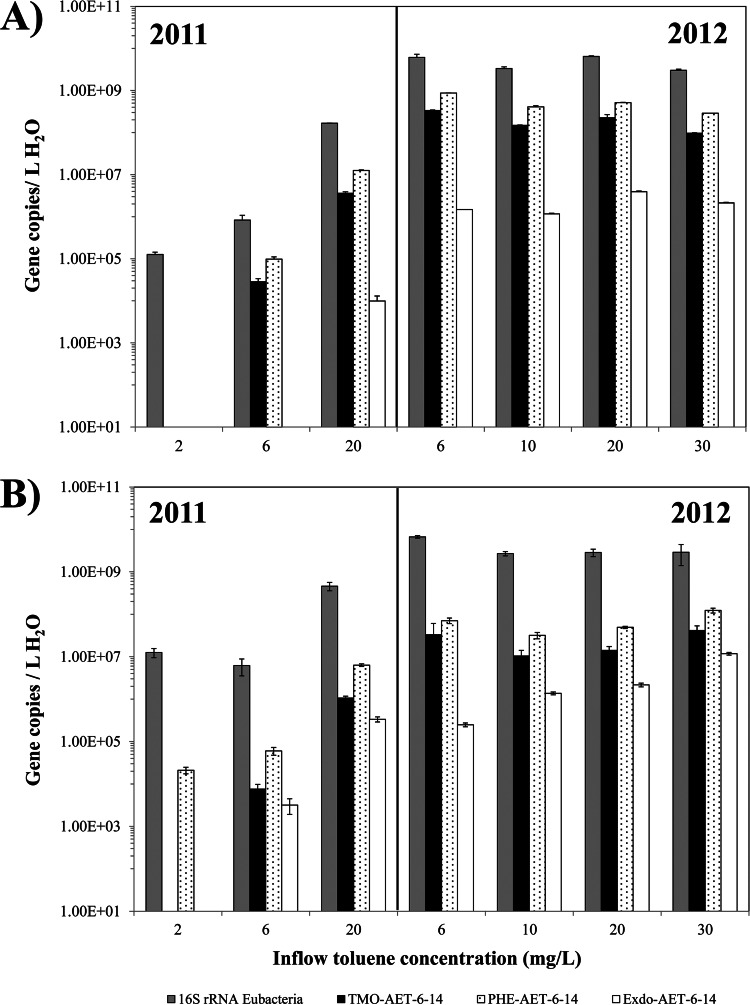
In order to quantify independently selected genes that were identified via microarray analysis, qPCR was used using primers targeting the catabolic genes detected in toluene-degrading bacteria isolated from the PFRs. A total of nine aerobic toluene-degrading strains were obtained from pore water samples of the planted reactors (see Table S1 in the supplemental material). From those isolates, Pseudomonas sp. strain AET-5-01, Pseudomonas sp. strain AET-5-VIII, and Ralstonia sp. strain AET-6-14 were selected as reference strains carrying the xylene monooxygenase (xylM), toluene dioxygenase (tod), and ring-hydroxylating (tmo and phe) genes, respectively (see Table S1 in the supplemental material). Primer sets suitable for qPCR analysis were designed to determine the abundance of the isolates' catabolic genes inside the PFRs over the 2-year period of operation at the different toluene feeding concentrations (Fig. 4). Total eubacterial 16S rRNA gene copy numbers were quantified to have an estimation of the whole bacterial population as reference. According to the qPCR results, only the genes belonging to the ring-hydroxylating pathway as in the isolate AET-6-14 (tmo, phe, and exdo-aet-14; pathway 3 in Fig. S1 in the supplemental material) were present in significant amounts in both reactors, which is in accordance with the low abundance of Pseudomonas and with the significant presence of Burkholderiaceae in both reactors. During the operation period of the first year (2011) a gradual increase in total bacteria numbers and catabolic genes was observed concomitant with the increasing toluene concentrations, while in the second year (2012) gene copy numbers did not fluctuate greatly. The tmo gene was first detected in both reactors a month after the feeding with toluene started (at a 6-mg/liter toluene feeding concentration), whereas the phe gene representing the phenol hydroxylase was detected already a couple of weeks after the feeding started (toluene feeding concentration of 2 mg/liter), at least in PFR-2 (Fig. 4B). The exdo-aet-14 gene was detected 2 and 1 month after the feeding with toluene started in PFR-1 and PFR-2, respectively. The presence of these genes in different abundances indicated that they are present in different organisms. The abundance of the detected catabolic genes represents a small fraction of the community compared to the total number of bacteria. For example, tmo genes reached a maximum of 5 and 1.4% of representation in PFR-1 and -2, respectively. These percentages fit well with the relative abundances of Burkholderiaceae in both PFRs (4.4 and 4.8%, respectively), suggesting that members of that family are probably involved in the first step of toluene degradation. The phe genes were slightly higher represented in both reactors with a maximum of 14 and 4% in PFR-1 and -2, respectively. Although several members of Burkholderiaceae have been described to carry phenol hydroxylase genes, the abundance of phe genes exceeds the representation of this family in PFR-1. Those phenol hydroxylases genes probably belong to members of the Comamonadaceae family, which are highly abundant in both reactors and where some members have been described to carry those genes in their genomes (39). The least represented gene was exdo-aet-14 with <1% in each reactor.
FIG 4.
Quantification of total number of bacteria (16S rRNA Eubacteria) and the catabolic genes belonging to the ring-hydroxylating pathway, as in the isolate AET-6-14 tmo (toluene monooxygenase), phe (phenol hydroxylase), and exdo-aet-14 (extradiol-dioxygenase) through qPCR, during 2 years of operation of PFR-1 (A) and PFR-2 (B). Each bar represents the average of three technical replicates of the qPCR. Samples were taken on 6 May (2 mg/liter), 20 May (6 mg/liter), and 22 June (20 mg/liter) in 2011 and on 1 June (6 mg/liter), 14 June (10 mg/liter), 25 June (20 mg/liter), and 13 August (30 mg/liter) in 2012.
Stable isotope fractionation analysis: pulse experiment.
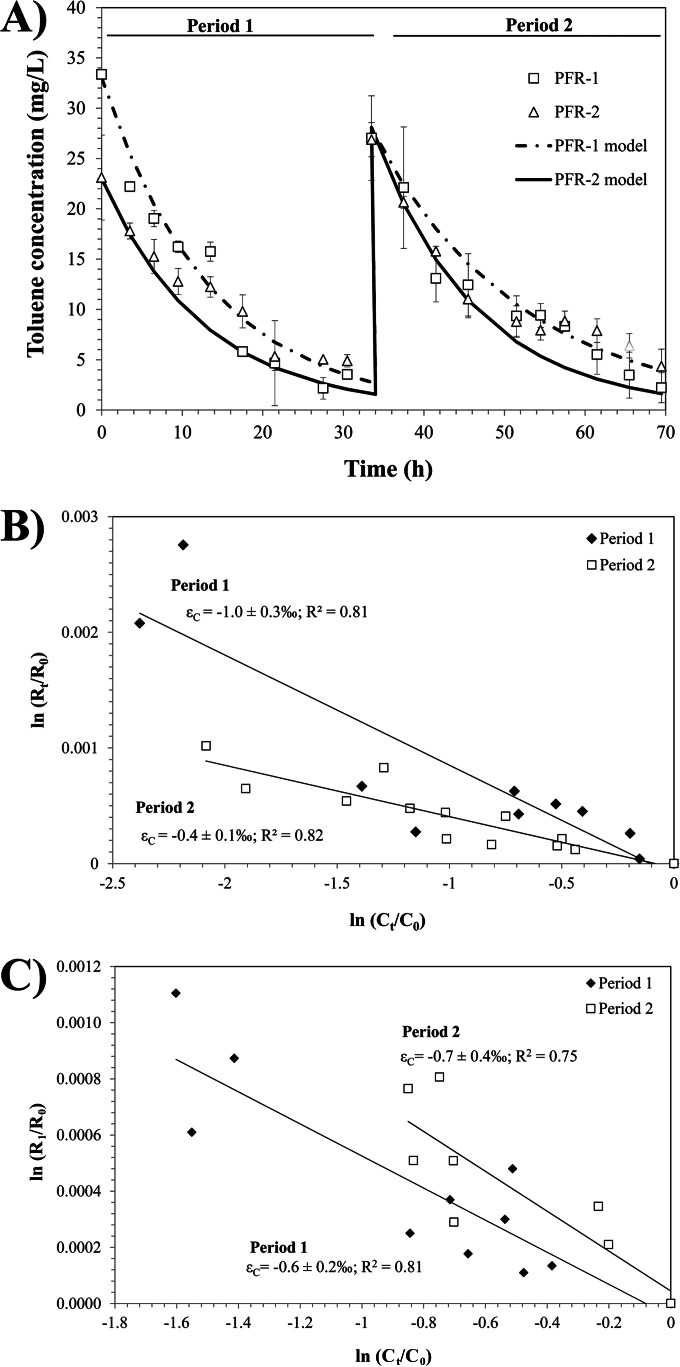
In order to monitor whether the genetic potential for ring monooxygenation was translated into the respective in situ biochemical activity, toluene stable isotope fractionation analysis was carried out. This method relies on the shift in the compounds' stable isotope composition (e.g., 13C/12C or 2H/1H) during some enzymatically catalyzed reactions, leading to an enrichment of heavier isotopes in the residual fraction of the initial compound. The reaction-dependent compound-specific isotope enrichment factor, ε, for different toluene degradation mechanisms under aerobic and anaerobic conditions has been determined with model strains and may be used for discriminating between different biodegradation pathways in the environment (20, 41). In our study, toluene was injected twice into the reactors, and samples were taken after each pulse, which are defined as periods 1 and 2 (Fig. 5A). After each injection of toluene, the concentration decreased rapidly and continuously following first-order kinetics in both reactors. The emission rate of toluene fluctuated between 0.1 and 0.6 mg/h, and no specific trend was observed during day and night periods. Since evaporation processes were minimal, we used the simplified Rayleigh equation to quantify the isotope fractionation described for closed systems. Low, but significant enrichment in 13C occurred during degradation of toluene during the first period in both systems, with εC of −1.0‰ ± 0.3‰ (R2 = 0.81) and −0.6‰ ± 0.2‰ (R2 = 0.81) for PFR-1 and -2, respectively (Fig. 5B and C). During the second degradation period, an εC corresponding to −0.4‰ ± 0.1‰ (R2 = 0.82) was observed in PFR-1 (Fig. 5B). In the case of PFR-2, during the period 2 of degradation a similar εC to period 1 was observed corresponding to −0.7‰ ± 0.4‰, but the correlation factor (R2) between toluene concentrations and isotope signatures was lower (R2 = 0.75). No significant 2H enrichment was measured except in PFR-1 during period 1, where an εH of −13.6‰ ± 3.7‰ was estimated (see Fig. S6 in the supplemental material). Comparing the obtained enrichment factors to those reported for toluene-degrading reference strains, it can be concluded that toluene degradation was not driven by anaerobic degradation or degradation via the TOL plasmid-encoded pathway initiating degradation by oxidation of the methyl substituent. These mechanisms would result in much higher εC and εH than that observed in PFR-1 and -2. Thus, the isotope fractionation data suggest that ring hydroxylation or dioxygenation, which cannot be differentiated solely based on isotope fractionation analysis, or a mixture of both pathways was driving the toluene degradation process.
FIG 5.
Isotope fractionation experiment carried out in PFR-1 and -2 in order to determine active degradation pathways in the systems. (A) Measured toluene concentration (open symbols) and first-order kinetics models describing the toluene degradation in PFR-1 (solid line) and PFR-2 (dashed line). The first sample to measure toluene concentration was taken at 6:30 p.m. (time = 0). (B and C) Double logarithmic plot of the carbon isotopic composition versus the residual concentration of toluene in PFR-1 and PFR-2, respectively. Closed symbols represent the measurements during the first pulse period (period 1) and open symbols the ones during the second pulse period (period 2). A linear regression was used in order to determine carbon enrichment factors (εC).
DISCUSSION
Efficient removal of organic contaminants was previously observed in pilot-scale constructed wetlands (6–8). Likewise, high removal of toluene during a 2-year period was accomplished in the PFRs used in the present study. One of the particular characteristics of constructed wetlands and PFRs is that the plants grown in these systems may efficiently provide molecular oxygen to the rhizosphere in dependence of the day/night rhythm (9). In the present study, diurnal changes in the oxygen concentration (hypoxic to anoxic) and redox potential were observed during some time periods as well.
The PFRs harbor a broad phylogenetic diversity that is numerically dominated by Proteobacteria, which represented more than 80% of the phyla in both reactors, followed by Bacteroidetes, Acidobacteria, and Actinobacteria, and thus is in accord with previously described constructed wetland systems (42–44). Specifically, in the PFR systems, the families Comamonadaceae and Xanthomonadaceae comprised more than 50% of the population, followed by the Burkholderiaceae family. Members of the Xanthomonadaceae family are obligate aerobes, and some of them have been directly or indirectly related to petroleum hydrocarbon degradation (45, 46). The family Comamonadaceae belonging to the order Burkholderiales harbor a remarkable phenotypic diversity, while members of the Burkholderiaceae family from the same order have been found in diverse ecological niches. A comprehensive review of aerobic aromatic compound degradation in the Burkholderiaceae and Comamonadaceae family, and in general the order Burkholderiales, is available (39). Toluene degraders such as Comamonas testosteroni R5, Methylibium petroleiphilum, Ralstonia pickettii PKO1, and Burkholderia vietnamensis G4 are prominent members of these families (47–50). Bacteria belonging to the less represented families (≤3%) Sphingobacteriaceae, Chitinophagaceae, Rhodocyclaceae, and Bradyrhizobiaceae have been detected in diverse environments, including soil, rhizosphere, and as endophytes (51–53), and some of them have been also associated with aromatic compound degradation (54, 55). Although the bacterial compositions of the two PFRs were globally similar, one remarkable difference was the higher representation (6%) of the family Rhodospirillaceae in PFR-2. The reads belonging to this family were mostly associated with the genus Magnetospirillum. Microbes from this genus can be frequently found at the oxic-anoxic conditions in freshwater sediments (56), and isolates such as Magnetospirillum sp. strain TS-6 are able to degrade toluene under nitrate-reducing conditions but apparently not under aerobic conditions (38).
The catabolic gene landscape present in the PFRs was investigated using a recently developed microarray which contains probes for key aromatic degradation families and key alkane degradation genes (19). In agreement with the amplicon sequencing results, the majority of the hybridized probes represented proteobacterial catabolic genes. Within the soluble diiron monooxygenase superfamily, two probes representing phenol hydroxylases found in the Burkholderiaceae family showed hybridization with DNA derived from both PFRs. The phenol hydroxylase group is comprised of multicomponent enzymes able to hydroxylate phenol and methyl-substituted derivatives (17) but also by the toluene 2-monooxygenase of Burkholderia cepacea G4, which is able to hydroxylate toluene to 2-methylphenol and further to 3-methylcatechol (57). This type of toluene degradation mechanism, two-successive monooxygenation of toluene, has also been proposed for the recently sequenced Alycycliphilus denitrificans (34). The presence of a complete monooxygenation pathway for the degradation of toluene is also supported by the presence of genes encoding extradiol dioxygenases (catechol 2,3-dioxygenases; (BAH90326 and AAS75778), which commonly cluster together with phenol hydroxylase encoding genes in toluene-degrading strains (27). Two other possible degradation pathways for toluene degradation were suggested by the microarray results, although each of them was found only in one of the reactors. In PFR-1, the microarray analysis provided evidence for the presence of genes related to those found in the TOL pathway from Pseudomonas species such as P. putida mt-2 (58–60). However, the presence of these genes, which are normally associated with the genus Pseudomonas, does not fit with the absence of the Pseudomonadaceae family in PFR-1. Meanwhile, probes representing benzylsuccinate synthase and benzoyl-CoA reductase, the enzymes for anaerobic toluene degradation similar to those of Magnetospirillum sp. strain TS-6, showed high hybridization signals only in PFR-2, a finding in agreement with the 6% relative abundance of the family Rhodospirillaceae (genus Magnetospirillum) shown by the Illumina data. Further investigations are ongoing in order to isolate this strain, to characterize it in detail, and to determine the role of Magnetospirillum in this system.
The quantification of catabolic genes from aerobic toluene-degrading isolates in DNA samples retrieved from the PFRs showed that genes from the monooxygenation pathway present in the isolate Ralstonia sp. strain AET-6-14 were the only ones detected in both PFRs (pathway 3 in Fig. S1 in the supplemental material). This isolate harbors catabolic genes closely related to those encoding toluene monooxygenase (tmo) from Ralstonia pickettii PKO1 and the phenol monooxygenase (phe) and catechol 2,3-dioxygenase (exdo-aet-14) from Ralstonia pickettii strain 12D. Initially, toluene monooxygenase of R. pickettii PKO1 was reported to hydroxylate toluene at the meta position, producing primarily 3-methylphenol (61). However, later studies have shown that this monooxygenase hydroxylates toluene predominantly at the para position producing 4-methylphenol (62). The different temporal and absolute occurrences of the tmo, phe, and exdo-aet-14 genes indicate the presence of microbes with different gene inventories than AET-6-14. In addition, comparison of the relative abundance of these genes with the 16S rRNA gene sequencing results indicates that bacteria carrying phe genes are probably members of families Burkholderiaceae and Comamonadaceae. The concomitant increase in tmo, phe, and exdo-aet-14 gene copy numbers over time with increasing toluene concentrations in the feeding of the reactors emphasizes how microbial communities present in constructed wetlands systems can adapt to and can specialize on degradation of organic pollutants, which makes these systems an effective remediation technique as previously shown (6, 8, 63). Several strains carrying xylene monooxygenase-, toluene dioxygenase-, and toluene monooxygenase-encoding genes were isolated from the PFRs. However, most isolates especially from PFR-1 were members of the genus Pseudomonas, which were only in low relative abundance according to the Illumina data. Cultivation-dependent methods are known to poorly reflect microbial community diversity (64). The isolates harboring the xylene monooxygenase and toluene dioxygenase pathway are evidently not dominant members of the toluene-degrading community.
In order to test whether the degradation pathways suggested by the microarray and qPCR analyses are indeed the active mechanism predominant in the PFRs, two-dimensional stable isotope analysis was used. This method has been successfully used to assess biodegradation of BTEX contaminants in groundwater (65), as well as in constructed wetlands (6, 63). The carbon and hydrogen isotopic profiles observed in PFR-1 during period 1 were very similar to those observed during toluene degradation by R. pickettii PKO1, which uses a monooxygenation mechanism as a first step of the toluene degradation pathway (20, 41). During period 2, a low observed εC would be associated with an aromatic ring-dioxygenation mechanism, such as the one used by P. putida F1, according to previous studies using toluene-degrading model bacteria (41). In addition, the absence of hydrogen fractionation during this period in PFR-1 correlates with the small εC observed, which could be the result of dioxygenation as the degradation mechanism. In both types of mechanisms (toluene monooxygenation and dioxygenation), the first step of degradation involves the hydroxylation and dihydroxylation of the aromatic nucleus to form a cresol isomer and toluene cis-dihydrodiol, respectively (15). These reactions are not linked to the cleavage of a C-H bond, and thus a small or no carbon and hydrogen isotopic fractionation would be expected (20). The low carbon enrichment factors observed during isotope fractionation analysis suggests that ring hydroxylation, dioxygenation, or a mixture of these two pathways were present. However, genes related to the dioxygenation pathway were not detected in the PFRs.
The combined analyses of the genomic potential for aromatic compound degradation, biodiversity, and isotope fractionation allowed for the identification of the predominant active toluene degradation pathway in the PFRs, i.e., members of the betaproteobacterial community carry out sequential monooxygenation of the aromatic ring, followed by ring cleavage through an extradiol dioxygenase as previously described for R. pickettii PKO1. The monooxygenation pathway has been also proposed as the predominant mechanism for the degradation of benzene-contaminated groundwater in a bench-scale constructed wetland (63). The dominant role of this pathway in the PFRs may be due to the high oxygen affinity of enzymes such as toluene monooxygenases compared to methyl-monooxygenases or toluene dioxygenases, which would allow the microorganisms harboring this type of enzymes to be adapted to grow on some aromatic compounds in this and other low oxygen environments (66, 67). Likewise, the catechol 2,3-dioxygenases encoded in the catabolic operons, together with this type of monooxygenase, have also been reported to perform efficiently under low-oxygen conditions (68). The appearance of genes encoding multicomponent monooxygenase and extradiol dioxygenases from the subfamily I.2.C type (18) has been detected in other hypoxic environments, indicating that these types of genes could have a relevant function in these type of systems, which has not been well studied or exploited yet (69, 70).
The use of PFRs and the combination of molecular methods with stable isotope fractionation analysis have allowed us to describe the adapted microbial community and its catabolic gene potential and to obtain evidence on the removal pathway(s) of toluene degradation occurring in complex planted systems used for bioremediation. Our results are useful to take into account when engineering constructed wetland system for the remediation of aromatic pollutants.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carsten Vogt for his help with isotope fractionation data analysis, Ursula Günther and Ines Mäusezahl for their technical support, and Henrike Beck and Iris Adam for their support with software usage. We also thank the anonymous reviewers for critically reading the manuscript and suggesting many improvements.
This study was supported by a collaborative project (BACSIN, contract 211684) of the European Commission within its Seventh Framework Programme. R.V.-V. was supported as a postdoctoral fellow from the Belgian Science Policy Office (BELSPO).
This article is dedicated to Peter Kuschk, who passed away in October 2014.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01822-15.
REFERENCES
- 1.Andreoni V, Gianfreda L. 2007. Bioremediation and monitoring of aromatic-polluted habitats. Appl Microbiol Biotechnol 76:287–308. doi: 10.1007/s00253-007-1018-5. [DOI] [PubMed] [Google Scholar]
- 2.Khan S, Afzal M, Iqbal S, Khan QM. 2013. Plant–bacteria partnerships for the remediation of hydrocarbon contaminated soils. Chemosphere 90:1317–1332. doi: 10.1016/j.chemosphere.2012.09.045. [DOI] [PubMed] [Google Scholar]
- 3.Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJJ. 2004. Rhizoremediation: a beneficial plant-microbe interaction. Mol Plant Microbe Interact 17:6–15. doi: 10.1094/MPMI.2004.17.1.6. [DOI] [PubMed] [Google Scholar]
- 4.Segura A, Ramos JL. 2013. Plant–bacteria interactions in the removal of pollutants. Curr Opin Biotechnol 24:467–473. doi: 10.1016/j.copbio.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Hütsch BW, Augustin J, Merbach W. 2002. Plant rhizodeposition: an important source for carbon turnover in soils. J Plant Nutr Soil Sci 165:397–407. doi:. [DOI] [Google Scholar]
- 6.Braeckevelt M, Rokadia H, Imfeld G, Stelzer N, Paschke H, Kuschk P, Kästner M, Richnow HH, Weber S. 2007. Assessment of in situ biodegradation of monochlorobenzene in contaminated groundwater treated in a constructed wetland. Environ Pollut 148:428–437. doi: 10.1016/j.envpol.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Imfeld G, Braeckevelt M, Kuschk P, Richnow HH. 2009. Monitoring and assessing processes of organic chemicals removal in constructed wetlands. Chemosphere 74:349–362. doi: 10.1016/j.chemosphere.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 8.Seeger EM, Kuschk P, Fazekas H, Grathwohl P, Kästner M. 2011. Bioremediation of benzene-, MTBE-, and ammonia-contaminated groundwater with pilot-scale constructed wetlands. Environ Pollut 159:3769–3776. doi: 10.1016/j.envpol.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 9.Wiessner A, Kappelmeyer U, Kuschk P, Kästner M. 2005. Influence of the redox condition dynamics on the removal efficiency of a laboratory-scale constructed wetland. Water Res 39:248–256. doi: 10.1016/j.watres.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 10.Imfeld G, Aragonés CE, Fetzer I, Mészáros E, Zeiger S, Nijenhuis I, Nikolausz M, Delerce S, Richnow HH. 2010. Characterization of microbial communities in the aqueous phase of a constructed model wetland treating 1,2-dichloroethene-contaminated groundwater. FEMS Microbiol Ecol 72:74–88. doi: 10.1111/j.1574-6941.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- 11.Kappelmeyer U, Wiessner A, Kuschk P, Kästner M. 2002. Operation of a universal test unit for planted soil filters–planted fixed bed reactor. Eng Life Sci 2:311–315. doi:. [DOI] [Google Scholar]
- 12.Wiessner A, Kappelmeyer U, Kästner M, Schultze-Nobre L, Kuschk P. 2013. Response of ammonium removal to growth and transpiration of Juncus effusus during the treatment of artificial sewage in laboratory-scale wetlands. Water Res 47:4265–4273. doi: 10.1016/j.watres.2013.04.045. [DOI] [PubMed] [Google Scholar]
- 13.Wu SB, Kuschk P, Wiessner A, Kästner M, Pang C, Dong RJ. 2013. Response of removal rates to various organic carbon and ammonium loads in laboratory-scale constructed wetlands treating artificial wastewater. Water Environ Res 85:44–53. doi: 10.2175/106143012X13415215907293. [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty R, Coates JD. 2004. Anaerobic degradation of monoaromatic hydrocarbons. Appl Microbiol Biotechnol 64:437–446. doi: 10.1007/s00253-003-1526-x. [DOI] [PubMed] [Google Scholar]
- 15.Parales RE, Parales JV, Pelletier DA, Ditt JL. 2008. Diversity of microbial toluene degradation pathways. Adv Appl Microbiol 64:1–73. doi: 10.1016/S0065-2164(08)00401-2. [DOI] [PubMed] [Google Scholar]
- 16.Spormann AM, Widdel F. 2000. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 11:85–105. doi: 10.1023/A:1011122631799. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Pantoja D, González B, Pieper DH. 2010. Aerobic degradation of aromatic hydrocarbons, p 799–837. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, New York, NY. [Google Scholar]
- 18.Eltis L, Bolin JF. 1996. Evolutionary relationships among extradiol dioxygenases. J Bacteriol 178:5930–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilchez-Vargas R, Geffers R, Suarez-Diez M, Conte I, Waliczek A, Kaser V, Kralova M, Junca H, Pieper DH. 2013. Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ Microbiol 15:1016–1039. doi: 10.1111/j.1462-2920.2012.02752.x. [DOI] [PubMed] [Google Scholar]
- 20.Vogt C, Cyrus E, Herklotz I, Schlosser D, Bahr A, Herrmann S, Richnow HH, Fischer A. 2008. Evaluation of toluene degradation pathways by two-dimensional stable isotope fractionation. Environ Sci Technol 42:7793–7800. doi: 10.1021/es8003415. [DOI] [PubMed] [Google Scholar]
- 21.Camarinha-Silva A, Jauregui R, Chaves-Moreno D, Oxley APA, Schaumburg F, Becker K, Wos-Oxley ML, Pieper DH. 2014. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol 16:2939–2952. doi: 10.1111/1462-2920.12362. [DOI] [PubMed] [Google Scholar]
- 22.Bohorquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, Kolter R, Junca H, Baena S, Zambrano MM. 2012. In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol 63:103–115. doi: 10.1007/s00248-011-9943-3. [DOI] [PubMed] [Google Scholar]
- 23.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Peter Solymos M, Stevens HH, Wagner H. 2013. vegan: community ecology package. R package version 2.0-10. http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 25.Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266. [DOI] [PubMed] [Google Scholar]
- 26.Baldwin BR, Nakatsu CH, Nies L. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl Environ Microbiol 69:3350–3358. doi: 10.1128/AEM.69.6.3350-3358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennerova MV, Josefiova J, Brenner V, Pieper DH, Junca H. 2009. Metagenomics reveals diversity and abundance of meta-cleavage pathways in microbial communities from soil highly contaminated with jet fuel under air-sparging bioremediation. Environ Microbiol 11:2216–2227. doi: 10.1111/j.1462-2920.2009.01943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrickx B, Dejonghe W, Faber F, Boënne W, Bastiaens L, Verstraete W, Top EM, Springael D. 2006. PCR-DGGE method to assess the diversity of BTEX mono-oxygenase genes at contaminated sites. FEMS Microbiol Ecol 55:262–273. doi: 10.1111/j.1574-6941.2005.00018.x. [DOI] [PubMed] [Google Scholar]
- 29.Hendrickx B, Junca H, Vosahlova J, Lindner A, Rüegg I, Bucheli-Witschel M, Faber F, Egli T, Mau M, Schlömann M, Brennerova MV, Brenner V, Pieper DH, Top EM, Dejonghe W, Bastiaens L, Springael D. 2006. Alternative primer sets for PCR detection of genotypes involved in bacterial aerobic BTEX degradation: distribution of the genes in BTEX-degrading isolates and in subsurface soils of a BTEX contaminated industrial site. J Microbiol Methods 64:250–265. doi: 10.1016/j.mimet.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Leahy JG, Batchelor PJ, Morcomb SM. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev 27:449–479. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 31.Elsner M, McKelvie J, Lacrampe-Couloume G, Sherwood Lollar B. 2007. Insight into methyl tert-butyl ether (MTBE) stable isotope fractionation from abiotic reference experiments. Environ Sci Technol 41:5693–5700. doi: 10.1021/es070531o. [DOI] [PubMed] [Google Scholar]
- 32.Cafaro V, Izzo V, Scognamiglio R, Notomista E, Capasso P, Casbarra A, Pucci P, Di Donato A. 2004. Phenol hydroxylase and toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1: interplay between two enzymes. Appl Environ Microbiol 70:2211–2219. doi: 10.1128/AEM.70.4.2211-2219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman L, Wackett LP. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066–14076. [DOI] [PubMed] [Google Scholar]
- 34.Oosterkamp MJ, Veuskens T, Talarico Saia F, Weelink SA, Goodwin LA, Daligault HE, Bruce DC, Detter JC, Tapia R, Han CS, Land ML, Hauser LJ, Langenhoff AA, Gerritse J, van Berkel WJ, Pieper DH, Junca H, Smidt H, Schraa G, Davids M, Schaap PJ, Plugge CM, Stams AJ. 2013. Genome analysis and physiological comparison of Alicycliphilus denitrificans strains BC and K601(T.). PLoS One 8:e66971. doi: 10.1371/journal.pone.0066971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franklin FC, Bagdasarian M, Bagdasarian MM, Timmis KN. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A 78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch R, García-Valdés E, Moore ER. 2000. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 245:65–74. doi: 10.1016/S0378-1119(00)00038-X. [DOI] [PubMed] [Google Scholar]
- 37.Merimaa M, Heinaru E, Liivak M, Vedler E, Heinaru A. 2006. Grouping of phenol hydroxylase and catechol 2,3-dioxygenase genes among phenol- and p-cresol-degrading Pseudomonas species and biotypes. Arch Microbiol 186:287–296. doi: 10.1007/s00203-006-0143-3. [DOI] [PubMed] [Google Scholar]
- 38.Shinoda Y, Akagi J, Uchihashi Y, Hiraishi A, Yukawa H, Yurimoto H, Sakai Y, Kato N. 2005. Anaerobic degradation of aromatic compounds by magnetospirillum strains: isolation and degradation genes. Biosci Biotechnol Biochem 69:1483–1491. doi: 10.1271/bbb.69.1483. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Pantoja D, Donoso R, Agulló L, Córdova M, Seeger M, Pieper DH, González B. 2012. Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ Microbiol 14:1091–1117. doi: 10.1111/j.1462-2920.2011.02613.x. [DOI] [PubMed] [Google Scholar]
- 40.Giebler J, Wick LY, Schloter M, Harms H, Chatzinotas A. 2013. Evaluating the assignment of alkB terminal restriction fragments and sequence types to distinct bacterial taxa. Appl Environ Microbiol 79:3129–3132. doi: 10.1128/AEM.04028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morasch B, Richnow HH, Schink B, Vieth A, Meckenstock RU. 2002. Carbon and hydrogen stable isotope fractionation during aerobic bacterial degradation of aromatic hydrocarbons. Appl Environ Microbiol 68:5191–5194. doi: 10.1128/AEM.68.10.5191-5194.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arroyo P, Ansola G, Sáenz de Miera LE. 2013. Effects of substrate, vegetation and flow on arsenic and zinc removal efficiency and microbial diversity in constructed wetlands. Ecol Eng 51:95–103. doi: 10.1016/j.ecoleng.2012.12.013. [DOI] [Google Scholar]
- 43.Elsayed OF, Maillarda E, Vuilleumier S, Imfeld G. 2014. Bacterial communities in batch and continuous-flow wetlands treating the herbicide S-metolachlor. Sci Total Environ 499:327–335. doi: 10.1016/j.scitotenv.2014.08.048. [DOI] [PubMed] [Google Scholar]
- 44.Ligi T, Oopkaup K, Truu M, Preem J-K, Nõlvak H, Mitsch WJ, Mander Ü Truu J. 2014. Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput16SrRNA amplicon sequencing. Ecol Eng 72:56–66. doi: 10.1016/j.ecoleng.2013.09.007. [DOI] [Google Scholar]
- 45.Cerqueira VS, Hollenbach EB, Maboni F, Vainstein MH, Camargo FA, do Carmo R, Peralba M, Bento FM. 2011. Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol 102:11003–11010. doi: 10.1016/j.biortech.2011.09.074. [DOI] [PubMed] [Google Scholar]
- 46.Kim JM, Le NT, Chung BS, Park JH, Bae J-W, Madsen EL, Jeon CO. 2008. Influence of soil components on the biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Pseudoxanthomonas spadix BD-59. Appl Environ Microbiol 74:7313–7320. doi: 10.1128/AEM.01695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kukor JJ, Olsen RH. 1990. Molecular cloning, characterization, and regulation of a Pseudomonas pickettii PKO1 gene encoding phenol hydroxylase and expression of the gene in Pseudomonas aeruginosa PAO1c. J Bacteriol 172:4624–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakatsu CH, Hristova K, Hanada S, Meng XY, Hanson JR, Scow KM, Kamagata Y. 2006. Methylibium petroleiphilum gen. nov., sp. nov, a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int J Syst Evol Microbiol 56:983–989. doi: 10.1099/ijs.0.63524-0. [DOI] [PubMed] [Google Scholar]
- 49.Nelson MJK, Montgomery SO, Mahaffey WR, Pritchard PH. 1987. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl Environ Microbiol 53:949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teramoto M, Futamata H, Harayama S, Watanabe K. 1999. Characterization of a high-affinity phenol hydroxylase from Comamonas testosteroni R5 by gene cloning, and expression in Pseudomonas aeruginosa PAO1c. Mol Gen Genet 262:552–558. doi: 10.1007/s004380051117. [DOI] [PubMed] [Google Scholar]
- 51.Chen K, Tang SK, Wang GL, Nie GX, Li QF, Zhang JD, Li WJ, Li SP. 2013. Olivibacter jilunii sp. nov., isolated from DDT-contaminated soil. Int J Syst Evol Microbiol 63:1083–1088. doi: 10.1099/ijs.0.042416-0. [DOI] [PubMed] [Google Scholar]
- 52.Hurek T, Handley LL, Reinhold-Hurek B, Piché Y. 2002. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol Plant-Microbe Interact 15:233–242. doi: 10.1094/MPMI.2002.15.3.233. [DOI] [PubMed] [Google Scholar]
- 53.Madhaiyan M, Poonguzhali S, Senthilkumar M, Pragatheswari D, Lee JS, Lee KC. 2015. Arachidicoccus rhizosphaerae gen. nov., sp. nov., a plant-growth-promoting bacterium in the family Chitinophagaceae isolated from rhizosphere soil. Int J Syst Evol Microbiol 65:578–586. doi: 10.1099/ijs.0.069377-0. [DOI] [PubMed] [Google Scholar]
- 54.Achong GR, Rodriguez AM, Spormann AM. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol 183:6763–6770. doi: 10.1128/JB.183.23.6763-6770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tschech A, Fuchs F. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol 148:213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- 56.Geelhoed JS, Sorokin DY, Epping E, Tourova TP, Banciu HL, Muyzer G, Stams AJ, van Loosdrecht MC. 2009. Microbial sulfide oxidation in the oxic-anoxic transition zone of freshwater sediment: involvement of lithoautotrophic Magnetospirillum strain J10. FEMS Microbiol Ecol 70:54–65. doi: 10.1111/j.1574-6941.2009.00739.x. [DOI] [PubMed] [Google Scholar]
- 57.Newman LM, Wackett LP. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 58.Nakai C, Kagamiyama H, Nozaki M, Nakazawa T, Inouye S, Ebina Y, Nakazawa A. 1983. Complete nucleotide sequence of the metapyrocatechase gene on the TOL plasmid of Pseudomonas putida mt-2. J Biol Chem 258:2923–2928. [PubMed] [Google Scholar]
- 59.Nakazawa T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ Microbiol 4:782–786. doi: 10.1046/j.1462-2920.2002.00310.x. [DOI] [PubMed] [Google Scholar]
- 60.Ramos JL, Marqués S, Timmis KN. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol 51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 61.Olsen RH, Kukor JJ, Kaphammer B. 1994. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol 176:3749–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fishman A, Tao Y, Wood TK. 2004. Toluene 3-monooxygenase of Ralstonia pickettii PKO1 is a para-hydroxylating enzyme. J Bacteriol 186:3117–3123. doi: 10.1128/JB.186.10.3117-3123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rakoczy J, Remy B, Vogt C, Richnow HH. 2011. A bench-scale constructed wetland as a model to characterize benzene biodegradation processes in freshwater wetlands. Environ Sci Technol 45:10036–10044. doi: 10.1021/es2026196. [DOI] [PubMed] [Google Scholar]
- 64.Garcia-Armisen T, Anzil A, Cornelis P, Chevreuil M, Servais P. 2013. Identification of antimicrobial resistant bacteria in rivers: insights into the cultivation bias. Water Res 47:4938–4947. doi: 10.1016/j.watres.2013.05.036. [DOI] [PubMed] [Google Scholar]
- 65.Meckenstock RU, Morasch B, Griebler C, Richnow HH. 2004. Stable isotope fractionation analysis as a tool to monitor biodegradation in contaminated aquifers. J Contam Hydrol 75:215–255. doi: 10.1016/j.jconhyd.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Duetz WA, de Jong C, Williams PA, van Andel JG. 1994. Competition in chemostat culture between Pseudomonas strains that use different pathways for the degradation of toluene. Appl Environ Microbiol 60:2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olsen RH, Mikesell MD, Kukor JJ, Byrne AM. 1995. Physiological attributes of microbial BTEX degradation in oxygen-limited environments. Environ Health Perspect 103:49–51. doi: 10.2307/3432345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kukor JJ, Olsen RH. 1996. Catechol-2,3-dioxygenases functional in oxygen-limited (hypoxic) environments. Appl Environ Microbiol 62:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larentis M, Hoermann K, Lueders T. 2013. Fine-scale degrader community profiling over an aerobic/anaerobic redox gradient in a toluene-contaminated aquifer. Environ Microbiol Rep 5:225–234. doi: 10.1111/1758-2229.12004. [DOI] [PubMed] [Google Scholar]
- 70.Táncsics A, Szoboszlay S, Szabó I, Farkas M, Kovács B, Kukolya J, Mayer Z, Kriszt B. 2012. Quantification of subfamily I.2.C catechol 2,3-dioxygenase mRNA transcripts in groundwater samples of an oxygen-limited BTEX-contaminated site. Environ Sci Technol 46:232–240. doi: 10.1021/es201842h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.