Abstract
Detection of free radicals in biological systems is challenging due to their short half-lives. We have applied electron spin resonance (ESR) spectroscopy combined with spin traps using the probes PBN (N-tert-butyl-α-phenylnitrone) and DMPO (5,5-dimethyl-1-pyrroline N-oxide) to assess free radical formation in the human pathogen Staphylococcus aureus treated with a bactericidal antibiotic, vancomycin or ciprofloxacin. While we were unable to detect ESR signals in bacterial cells, hydroxyl radicals were observed in the supernatant of bacterial cell cultures. Surprisingly, the strongest signal was detected in broth medium without bacterial cells present and it was mitigated by iron chelation or by addition of catalase, which catalyzes the decomposition of hydrogen peroxide to water and oxygen. This suggests that the signal originates from hydroxyl radicals formed by the Fenton reaction, in which iron is oxidized by hydrogen peroxide. Previously, hydroxyl radicals have been proposed to be generated within bacterial cells in response to bactericidal antibiotics. We found that when S. aureus was exposed to vancomycin or ciprofloxacin, hydroxyl radical formation in the broth was indeed increased compared to the level seen with untreated bacterial cells. However, S. aureus cells express catalase, and the antibiotic-mediated increase in hydroxyl radical formation was correlated with reduced katA expression and catalase activity in the presence of either antibiotic. Therefore, our results show that in S. aureus, bactericidal antibiotics modulate catalase expression, which in turn influences the formation of free radicals in the surrounding broth medium. If similar regulation is found in other bacterial species, it might explain why bactericidal antibiotics are perceived as inducing formation of free radicals.
INTRODUCTION
The mechanism by which bactericidal antibiotics kill bacterial cells is highly debated (1–5). Interest was spurred by findings suggesting that bacterial cells exposed to lethal concentrations of bactericidal antibiotics are killed in part by reactive oxygen species (ROS), such as hydroxyl radicals generated independently of the primary targets of the antibiotics (6). The initial observation was followed by a number of studies implicating free radicals in killing of bacterial cells by antimicrobial compounds (7–14). Also reported was the contribution of intrinsic factors providing protection against antibiotics such as guanosine pentaphosphate (ppGpp), namely, extracellular hydrogen sulfide and endogenous nitric oxide in biofilms and planktonic cells, respectively (15–17). Although the hypothesis of a common pathway for killing by bactericidal antibiotics was later disputed by several research groups (3, 4, 18–22), bacterial metabolism and metabolites undoubtedly influence antibiotic susceptibility, and in some situations ROS may still promote lethality of antimicrobial compounds or be formed as part of the killing process (5).
The main challenges in monitoring free radicals are their low steady-state concentrations and short half-lives (23). Therefore, their detection relies on the activation of probes with which they react in order to yield relatively stable derivatives that can be measured (24). However, indirect detection via probe activation can be challenging, as was demonstrated for the hydroxyphenyl fluorescein (HPF) probe, which, when applied in combination with flow cytometry for hydroxyl radical detection in bacteria treated with antibiotics, gave a false-positive result due to increased autofluorescence in filamenting cells after exposure to fluoroquinolone or beta-lactam antibiotics (25, 26). Another approach to detect free radicals is electron spin resonance (ESR) spectroscopy (27). ESR is often applied in combination with spin traps, which react with the short-lived free radicals to yield relatively long-lived and detectable radical spin adducts (28). In ESR, the unpaired electrons in radicals are exposed to an external magnetic field and their absorption of definite electromagnetic radiation can be used to obtain structural and quantitative characteristics of individual radical species present in the testing system (28).
In this study, we examined if ESR/spin trapping is a suitable method for the detection of free radicals in the Gram-positive bacterium Staphylococcus aureus when exposed to hydrogen peroxide and vancomycin or ciprofloxacin antibiotics. S. aureus is a facultative anaerobic bacterium that causes a variety of serious infections and is increasingly difficult to treat due to antibiotic resistance (29). It protects itself against reactive oxygen species by expressing scavenging enzymes such as catalase, encoded by katA (30, 31). Yet hydroxyl radicals have been implicated in antibiotic-mediated killing of S. aureus in a process mitigated by the presence of the iron-chelating compound 2,2′-bipyridyl, which reduces free radical formation via the Fenton reaction and by the radical scavenger thiourea (6). Here, we have monitored intracellular and extracellular free radical formation in S. aureus cells employing the lipophilic spin trap probes PBN (N-tert-butyl-α-phenylnitrone), which mainly reacts with radicals buried in cell membrane, and DMPO (5,5-dimethyl-1-pyrroline N-oxide), which is relatively hydrophilic and applicable for detecting radicals in the water phase (32). For the PBN probe, sensitivity is significantly enhanced by ethanol, which reacts with hydroxyl and alkoxyl radicals to form relatively stable 1-hydroxyethyl radicals, which give highly stable spin adducts (33–35). Our results suggest that bactericidal antibiotics enhance extracellular free radical formation through repression of catalase expression, and they highlight the role of hydrogen peroxide-scavenging enzymes in the monitoring of free radical formation in bacterial cells.
MATERIALS AND METHODS
Strain, chemicals, and cultivation conditions.
Staphylococcus aureus Newman (NTCT 8178) was cultivated in 25 ml of tryptic soy broth (TSB) or brain heart infusion broth (BHI) (Oxoid, Denmark) in a 250-ml narrow-neck Erlenmeyer flask at 37°C and 200 rpm. Ethanol (Kemetyl; 2% [vol/vol]) and PBN (40 mM) were present in all cultivations for ESR experiments. The spin traps PBN and DMPO (no further purification required) were obtained from Sigma-Aldrich. The iron chelator 2,2′-bipyridyl (Sigma-Aldrich) and Chelex 100 (Bio-Rad) were used to remove the free iron from the cell broth media. Catalase from bovine liver (Sigma-Aldrich) (2,000 to 5,000 U/mg powder) and Triton X-100 (Sigma-Aldrich) were used for catalase quantification assay. Plasma pure nitric acid (SCP Science, Denmark) was used for sample preparation of inductively coupled plasma optical emission spectrometry (ICP-OES).
Growth was followed in TSB, and, when the early exponential phase (optical density at 600 nm [OD600] = 0.1, 108 CFU/ml) was reached, hydrogen peroxide (Sigma-Aldrich) (6 mM), vancomycin (Sigma-Aldrich) (2.5 μg/ml), or ciprofloxacin (Bayer Schering Pharma) (0.4 μg/ml) was added to the bacterial cultures. The samples for determination of CFU were taken immediately before the addition of chemicals and every 2 h afterward. CFU levels were assessed after 24 h of incubation at 37°C on tryptic soy agar (TSA) or brain heart infusion agar (BHIA) (Oxoid, Denmark) plates.
ESR spectroscopy measurement.
Four different protocols were used for the electron spin resonance (ESR) spectroscopy measurements. In the first protocol, bacterial culture (500 μl) was collected as described for CFU determination except with sampling every hour. All samples were adjusted to an OD600 of 0.1 for ESR measurement. The rest of the culture was centrifuged at 1.2 × 104 × g at room temperature for 30 s, and then the supernatant and the cell pellet resuspended with fresh medium were measured by ESR as well. The second protocol was modified from a previous study (36) with the following changes. Cells were collected when the OD600 reached 0.1 and were resuspended in 50 mM sodium sulfate buffer to a final concentration of 2.5 × 109 CFU/ml. DMPO (66 mM) and hydrogen peroxide (6 mM) or antibiotics (vancomycin at 2.5 μg/ml or ciprofloxacin at 0.4 μg/ml) were added to the suspension at the same time to reach a total volume of 150 μl for ESR measurement, and the reaction was initiated with a sweep time of 300 s and was continued overall for 50 min, at which time water was added to the bacterial cell suspension as a blank control. While the preceding reactions were all recorded in the cavity of the ESR equipment at room temperature throughout the measurement, the third protocol entailed 2.5 × 109 cells that were vigorously mixed with DMPO (66 mM) or PBN (40 mM) for 5 min and incubated at 37°C for 30 min before ESR measurement. The fourth protocol was utilized to test the effect of catalase on the radical formation in broth medium. Catalase (1 U/ml) was added into BHI medium, followed by incubation at 37°C with ethanol (2% [vol/vol]) and PBN (40 mM). Samples (50 μl) were collected every 30 min for ESR analysis. The ESR spectra were obtained with a MiniScope MS200 spectrometer (MagnetTech, Berlin, Germany) and the settings were as follows: magnetic field, 3,370 G; field sweep, 97 G; sweep time, 30 s; modulation amplitude, 2 G. The ESR spectra were analyzed with MiniScope Control 6.51 and Analysis 2.02 software.
Removal of iron from bacterial broth media.
BHI broth medium, iron-depleted BHI medium treated with Chelex 100, and BHI medium with iron chelator 2,2′-bipyridyl (5 mM or 50 mM) were shaken at 37°C and 200 rpm for 6 h in total with 2% (vol/vol) ethanol and 40 mM PBN (37). A 50-μl volume was taken from each sample every hour for ESR measurement. The same setup was tested with TSB medium.
Quantification of intracellular iron by ICP-OES.
The bacteria were cultivated in BHI medium under the same conditions as those described above, and samples were taken immediately before the addition of chemicals and 6 h after. A 1-ml volume of each sample was digested at 230°C with 2.5 ml 70% nitric acid and 1 ml 15% hydrogen peroxide in a pressurized microwave oven (Ultrawave; Milestone Inc., Italy). The digest was analyzed by ICP-OES (Optima 5300 DV; PerkinElmer, Waltham, MA, USA), and the data were processed with PerkinElmer Winlab32 3.1.0.0107.
Catalase assay.
The quantification of cellular catalase activity was tested as reported previously (38). After treatment with hydrogen peroxide or antibiotics for 3 h in TSB medium, 2 × 109 cells were collected from each sample, resuspended with FKP buffer (8.5 g/liter sodium chloride, 1 g/liter peptone), and thoroughly mixed with 100 μl Triton X-100 and 100 μl hydrogen peroxide (undiluted, 30 weight percent) in a Pyrex tube (Assistent, Germany) (16-mm diameter by 160-mm height). The mixture was incubated at room temperature until the reaction completed after approximately 10 min, and the height of foam was measured with a ruler. To define the catalase activity, 100 μl of catalase solution with predefined concentrations replaced the cell suspension in the reaction mixture.
Quantification of catalase expression by real-time qPCR.
After 3 h of treatment with hydrogen peroxide or antibiotics, RNA was extracted from cells grown in TSB using an RNeasy minikit (Qiagen). mRNA was reverse transcribed (RT) to cDNA by the use of a RevertAid H Minus First Strand cDNA synthesis kit (Thermo Scientific). For real-time quantitative PCR (qPCR), FastStart Essential DNA Green Master (Roche) was used with the primers listed in Table S1 in the supplemental material. The PCR products were detected and analyzed with a LightCycler 96 real-time PCR system (Roche). The relative expression levels were normalized to the expression of the ileS reference gene, and the ratios between samples were normalized to the one without hydrogen peroxide or antibiotics. Means of biological triplicates with standard deviations are shown, each measured with technical triplicates.
RESULTS
ESR detection of free radicals in S. aureus broth culture.
We examined growth in two bacterial broth media, BHI and TSB, in the presence of the spin trap PBN and ethanol (see Fig. S1 in the supplemental material) and selected concentrations of bactericidal agents to obtain a reduction in CFU comparable to what has previously been described (see Fig. S2 in the supplemental material) (6). From the hydrogen peroxide-treated cells, we obtained the characteristic spectra of 1-hydroxyethyl radical spin adducts, which confirms the existence of hydroxyl radicals (Fig. 1) (34). Monitored over time, the greatest and most consistent level of PBN spin adducts accumulated in the absence of bacterial cells for both types of broth medium (Fig. 2). In contrast, the presence of bacterial cells essentially eliminated spin adduct formation, whereas when the cells were treated with either of the bactericidal antibiotics vancomycin and ciprofloxacin, spin adduct formation was increased compared to the levels seen with untreated cells (Fig. 2). After addition of hydrogen peroxide, the formation of PBN spin adducts peaked immediately, reflecting the fact that hydrogen peroxide reacts with iron in the broth medium, yielding hydroxyl radicals via the Fenton reaction (27). Collectively, these results indicate that the hydroxyl radicals monitored by the PBN probe and ESR spectroscopy may be formed in the growth medium rather than inside the bacterial cells and that bactericidal antibiotics affect their formation.
FIG 1.
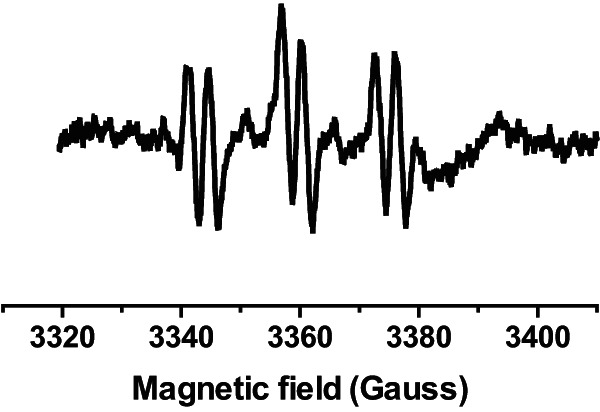
ESR spectra of spin adduct of PBN and 1-hydroxyethyl radicals (aN ≈ 16.1 G, aH ≈ 3.4 G) generated by hydroxyl radicals in a suspension of S. aureus cells treated with 6 mM hydrogen peroxide for 1 h and containing 2% (vol/vol) ethanol and 40 mM PBN.
FIG 2.
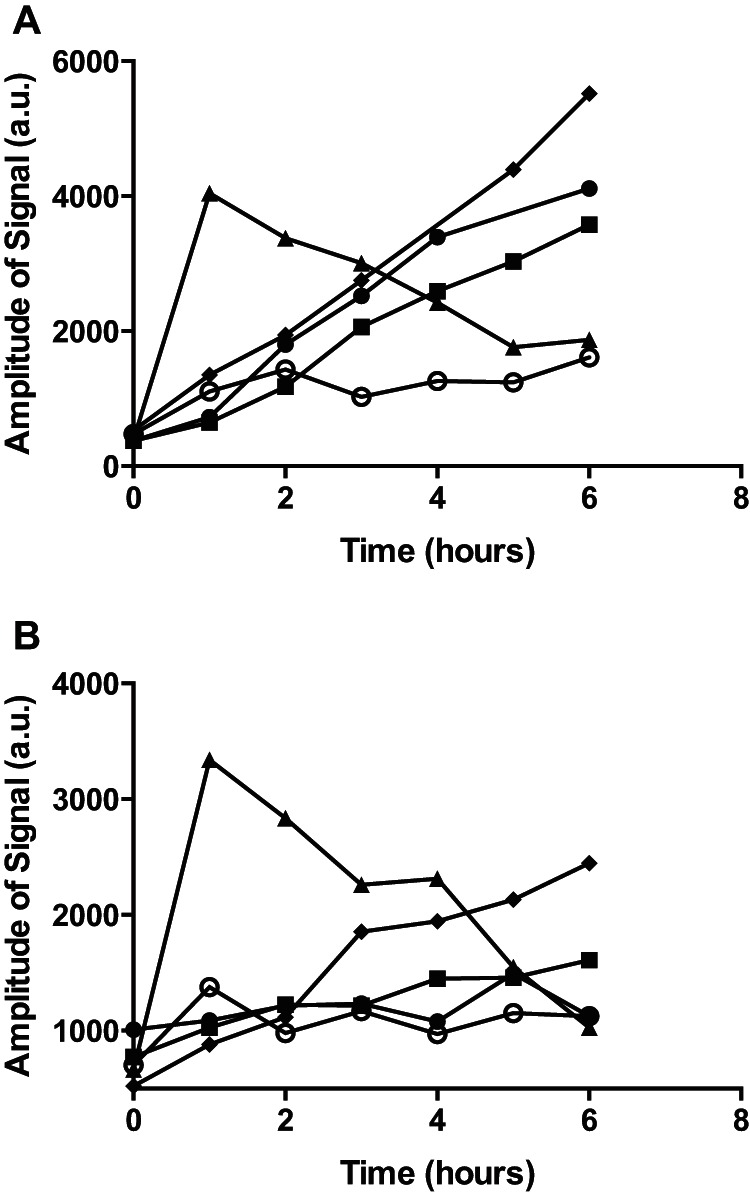
ESR determination of hydroxyl radicals formed in cultures of S. aureus treated with hydrogen peroxide or antibiotics. Hydroxyl-radical signals were monitored in BHI (A) or TSB (B) medium (filled diamonds) or in cultures of S. aureus grown in the corresponding medium left untreated (open circles) or treated with 6 mM hydrogen peroxide (filled triangles), 2.5 μg/ml vancomycin (filled circles), or 0.4 μg/ml ciprofloxacin (filled squares). a.u., arbitrary units.
To confirm that hydroxyl radicals are formed in the cell-free broth medium as a result of the Fenton reaction, we reduced the free iron concentration by use of the chelator, 2,2′-bipyridyl, or of the ion exchange resin, Chelex 100 (39). In both cases, hydroxyl radical formation was essentially abolished compared to untreated BHI medium results (Fig. 3A). Also, addition of purified catalase at a concentration of 1 U/ml immediately halted PBN spin adduct formation (Fig. 3B). These results show that the ESR signals were formed by hydrogen peroxide and iron in the broth medium entering the Fenton reaction, and they indicate that catalase activity of S. aureus was reducing the hydroxyl radical formation.
FIG 3.
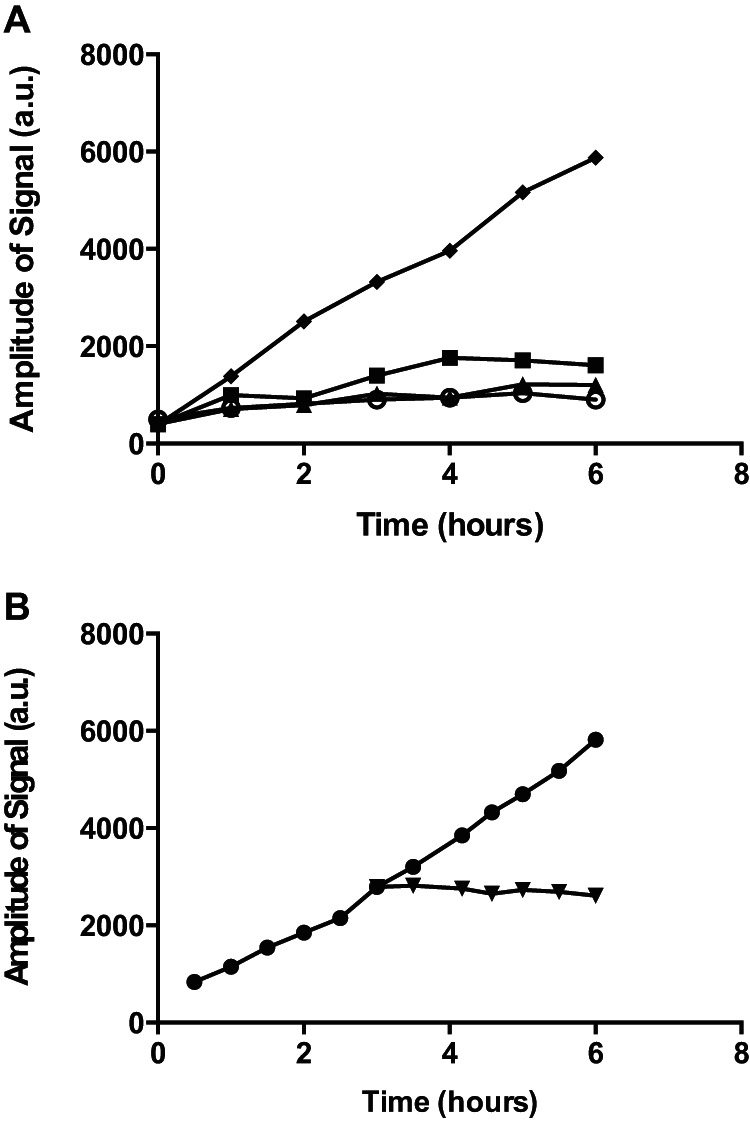
Iron sequestration and catalase abolish hydroxyl-radical formation. Hydroxyl radicals formed in BHI broth medium were detected by ESR (40 mM PBN and 2% [vol/vol] ethanol) in the presence of iron chelator 2,2′-bipyridyl (5 mM [filled triangles] or 50 mM [open circles]) or Chelex 100 (filled squares) or under untreated conditions (filled diamonds) (A) or after addition of catalase at 3 h (1 U/ml) (filled upside-down triangles) compared to the untreated control (filled circles) (B).
S. aureus cells do not produce detectable levels of hydroxyl radicals.
Since the ESR signal recorded by the PBN probe in bacterial cultures appeared to be highly influenced by the radicals formed in the broth medium itself, we attempted to detect cellular free radicals using the cell-permeative spin trap probe, DMPO, applied to bacterial cells suspended in sodium sulfate buffer (36). Compared with PBN, the DMPO probe is relatively hydrophilic, with a partitioning coefficient of 0.15 in octanal/water (32). However, with this probe, there was no significant difference in spin adduct signal between untreated cells and those exposed to hydrogen peroxide, vancomycin, or ciprofloxacin (Fig. 4). This result confirms that the spin adducts observed for antibiotic-treated broth cultures are not produced by the cells but are formed in the broth medium.
FIG 4.
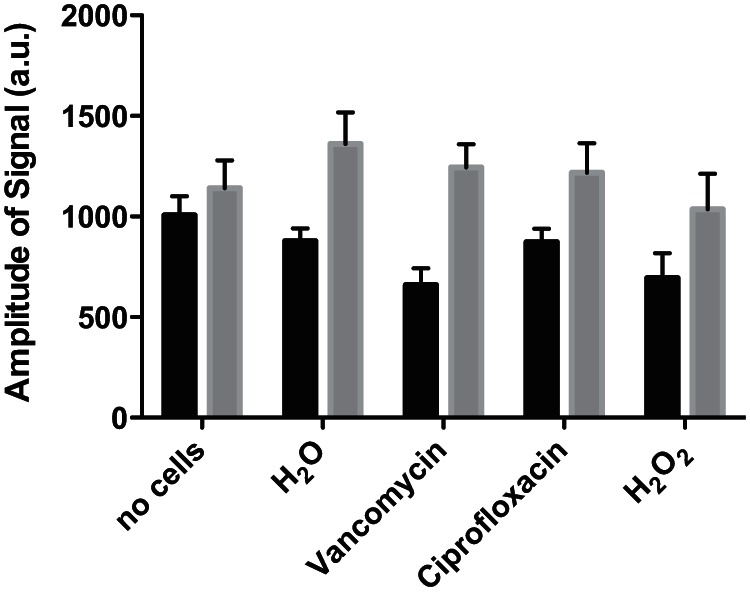
ESR determination of hydroxyl radicals formed in S. aureus. Hydroxyl-radical signals were detected in S. aureus cells resuspended in 50 mM sodium sulfate buffer using the DMPO probe. Cells were treated with 6 mM hydrogen peroxide, 2.5 μg/ml vancomycin, or 0.4 μg/ml ciprofloxacin. Measurements at 15 min (black columns) and 35 min (gray columns) of treatment are shown.
S. aureus catalase activity and katA expression are influenced by antibiotics.
To address how bactericidal antibiotics affect the ability of S. aureus to mitigate free radical formation in the broth medium, we monitored trace metal content in spent supernatants by inductively coupled plasma optical emission spectrometry (ICP-OES) (40). However, we found that Cu and Fe concentrations were approximately identical in bacterial spent supernatant from untreated cells and in supernatant from those treated with antibiotics, which precludes the possibility that antibiotic treatment influences the ability of S. aureus to bind or consume metal ions (Fig. 5).
FIG 5.
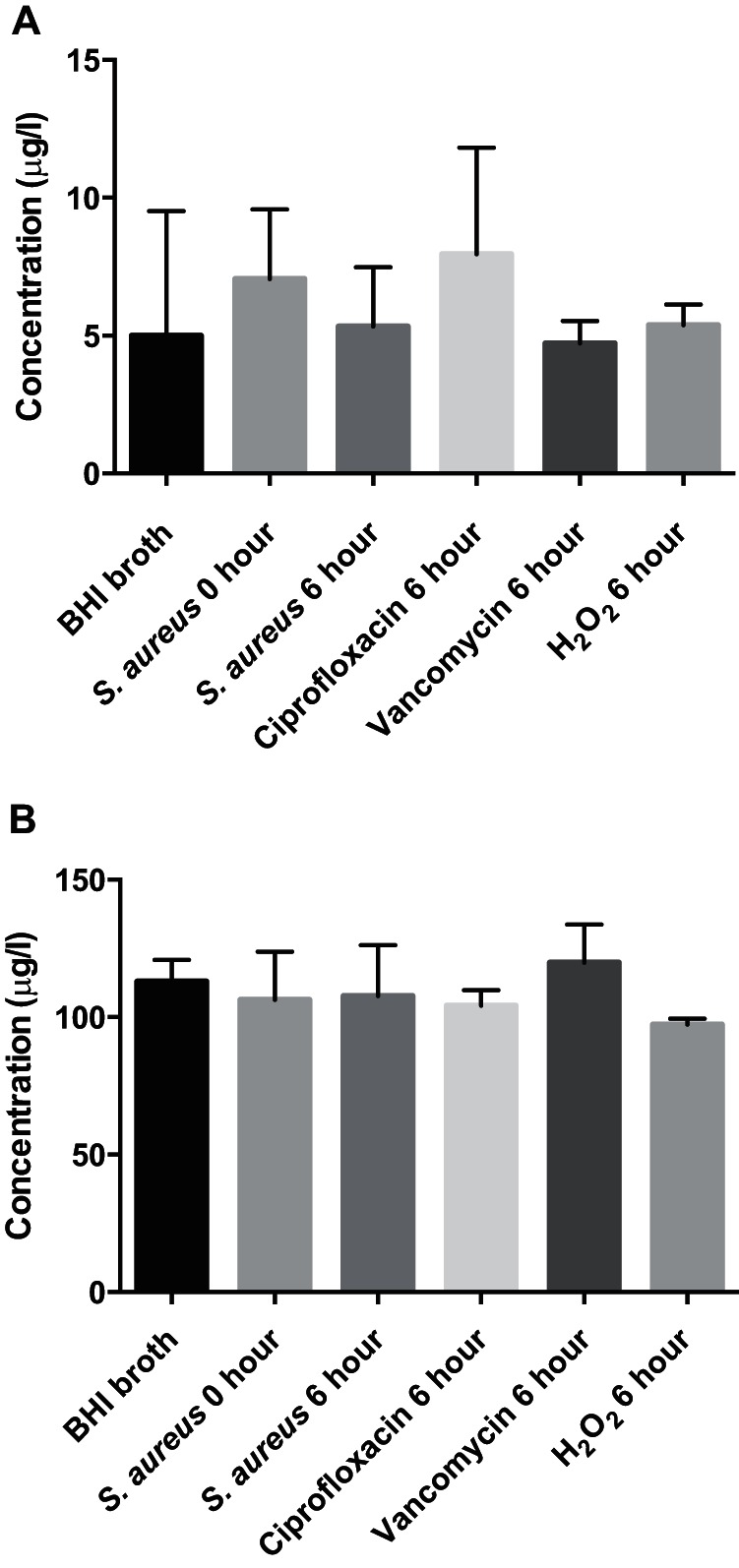
Trace metal concentrations in the supernatants of S. aureus culture. The concentrations of Cu (A) and Fe (B) remaining in the supernatant of bacterial cultures at 0 and 6 h without any treatment or with treatment with 6 mM hydrogen peroxide, 2.5 μg/ml vancomycin, or 0.4 μg/ml ciprofloxacin were measured by ICP-OES.
Alternatively, S. aureus is known to produce catalase; therefore, the effect of bactericidal antibiotics on free radical formation could be mediated through altered catalase production (30). To examine this, we monitored catalase activity as well as transcription of the katA gene and found that both vancomycin and ciprofloxacin reduced the activity (Fig. 6A) as well as expression (Fig. 6B) of catalase compared to the results seen with untreated cells. This reduction correlates with the increased ESR signal observed in the presence of either of the antibiotics compared to untreated cells (Fig. 2). Thus, our results show that catalase activity in S. aureus is affected by antibiotics and explain the effect that antibiotics exert on the ability of S. aureus to reduce broth-mediated ESR signals.
FIG 6.
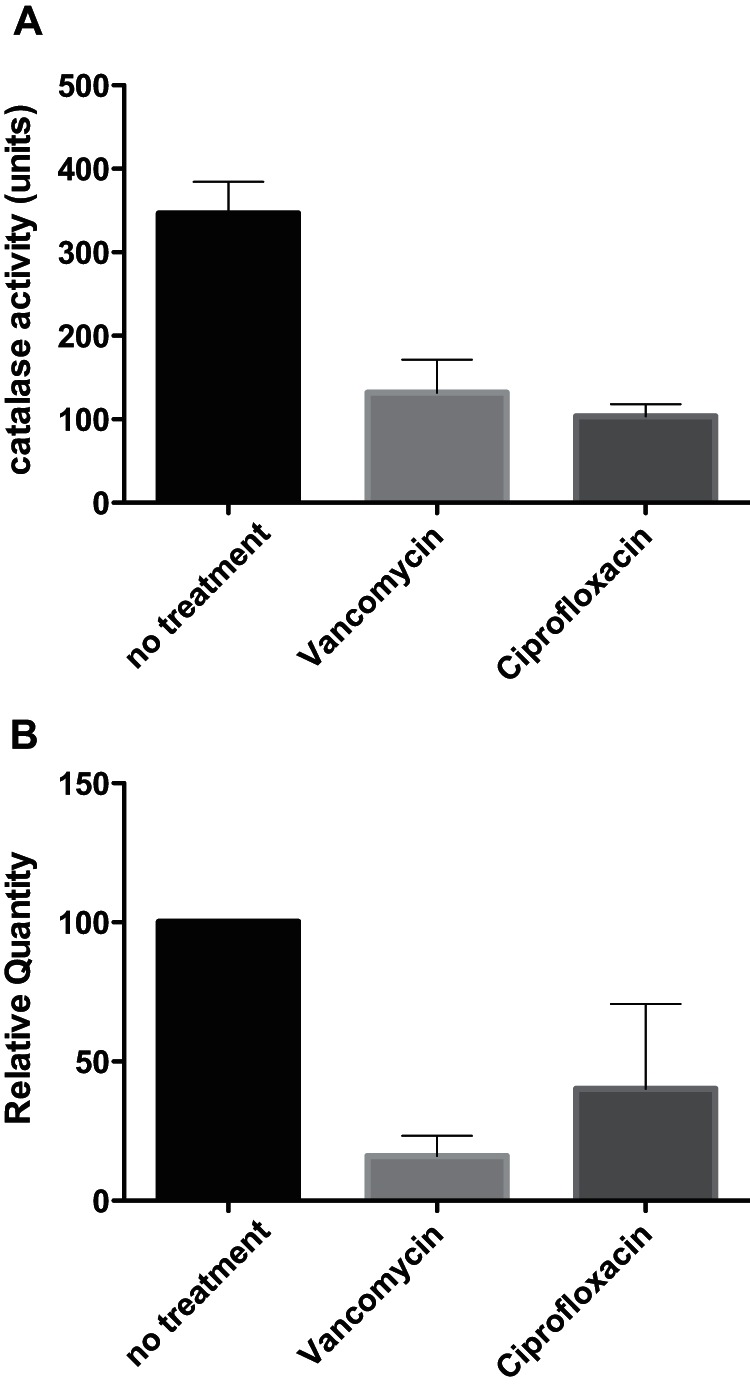
Catalase activity and expression in the presence of antibiotics. The activity (A) or expression (B) of catalase was monitored by foam formation or RT-qPCR, respectively, in untreated cells or in cells treated with 2.5 μg/ml vancomycin or 0.4 μg/ml ciprofloxacin after 3 h of exposure.
DISCUSSION
Free radicals play important roles in biological processes, but detection is complicated by their low concentrations and intracellular locations (23). For this purpose, radical-sensitive probes are often employed. Yet free radical detection is still challenging, as was recently demonstrated for the HPF probe, which in Escherichia coli cells displays autofluorescence and masks the probe oxidation tentatively taking place in response to, for example, bactericidal antibiotics (25, 26).
In the current study, we employed ESR spectroscopy and spin trapping to monitor hydroxyl radical formation in S. aureus using PBN and DMPO spin traps. In comparison to other radical detection methods, an advantage of the ESR/spin-trapping detection is that it relies on absorption of microwave irradiation, which interferes less with biological materials than UV or visible light, whereas, for example, the fluorescent HPF probe requires irradiation with intense excitation light (27). Using ESR/spin trapping, we were unable to detect free radicals formed inside S. aureus cells, but the uninoculated broth medium provided high levels of spin-adduct signals with the PBN spin trap, demonstrating the spontaneous formation of free radicals in the BHI and TSB media during aerobic incubation at 37°C. The level of free radicals was greatly reduced in the presence of S. aureus cells. This reduction was not due to bacterial consumption or binding of prooxidative transition metal ions such as iron ions, which are present in culture media in order to support growth, but was mediated via hydrogen peroxide-scavenging enzymes produced by S. aureus such as catalase, as addition of catalase to the broth medium immediately stopped the formation of new spin adducts. Catalase is commonly produced by aerobically growing bacteria, and it has been reported to be found extracellularly (41, 42). Importantly, catalase production by S. aureus was reduced by treatment with the bactericidal antibiotics vancomycin and ciprofloxacin and this reduction correlated with the increased PBN spin-adduct signal that was observed compared to the level seen for untreated cells. ESR and spin trapping have previously been applied to analyze radical formation in cultures of Lactobacillus acidophilus, Listeria innocua, Escherichia coli, S. aureus, and Streptococcus pneumoniae (34, 36, 43). All of those studies focused on the detection of hydroxyl radicals, which mostly have been claimed to be present intracellularly in the bacteria. In the case of L. acidophilus, production of hydroxyl radicals was shown to take place extracellularly (34). In an early study, the presence of intracellular radicals in bacterial cells was inferred from the ability of the DMPO probe to reduce killing by peracetic acid but direct detection of hydroxyl radicals was complicated by the presence of differing degrees of membrane permeability and large background signals (36). Our data reveal additional challenges in detection of free radicals in bacterial cells, namely, the formation of free radicals in the broth medium and the hydrogen peroxide-scavenging enzymes produced by some bacteria. Also complicating free radical detection is the recent finding that the oxidative stress levels in broth media differ between suppliers, as was observed for E. coli and Salmonella spp., where growth of oxidative stress response mutants depended on the source of the LB broth (44). Thus, careful consideration should be made prior to the interpretation of data from the monitoring of free radicals in bacterial cells.
Within the eukaryotic host, reactive oxygen species, such as hydrogen peroxide, are produced as part of the innate immune response (45). The primary host defense against S. aureus is represented by the neutrophils, which rely on ROS for antimicrobial activity (46). Since our results show that antibiotics influence catalase expression, this modulation may affect the efficacy of antimicrobial therapy. For the investigated antibiotics, the reduced catalase expression could help eliminate S. aureus infections by reducing the organism's ability to cope with oxidative stress (47, 48).
In summary, we show that the free radicals detected by ESR and spin trapping are produced extracellularly as a consequence of the presence of hydrogen peroxide and iron in the broth medium driving the Fenton reaction and radical formation. Given our inability to detect free radicals within bacterial cells, we were unable to assess the degree to which bactericidal antibiotics induce such radical production. For S. aureus, however, the antibiotic-mediated reduction in hydrogen peroxide-scavenging enzymes resulted in an increase in free radicals in the broth medium. Our findings may explain previous reports of free radicals in biological systems, and they highlight the need for reliable methods for detection of intracellular free radicals in bacterial cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Henriette Rifbjerg Erichsen and Thomas Hesselhøj Hansen for technical support. We also thank Oana Ciofu (Department of Clinical Microbiology, University of Copenhagen) and Peter Østrup Jensen (Department of International Health, University of Copenhagen) for providing an introduction to HPF detection and James Imlay for helpful comments on the manuscript.
H.I. is supported by grants from the Danish Research Council of Independent Research (274-08-0531). W.P. is supported by grants from the Danish Research Council of Independent Research (09-069656). A.B.H. is supported by the Ministry of Food, Agriculture and Fisheries of Denmark. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01199-15.
REFERENCES
- 1.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 5.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 7.Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. 2012. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother 56:5642–5649. doi: 10.1128/AAC.00756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Liu X, Qu Y, Wang X, Li L, Zhao X. 2012. Inhibitors of reactive oxygen species accumulation delay and/or reduce the lethality of several antistaphylococcal agents. Antimicrob Agents Chemother 56:6048–6050. doi: 10.1128/AAC.00754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhao X. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother 53:1395–1402. doi: 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Zhao X, Malik M, Drlica K. 2010. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J Antimicrob Chemother 65:520–524. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belenky P, Camacho D, Collins JJ. 2013. Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep 3:350–358. doi: 10.1016/j.celrep.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ. 2013. Silver enhances antibiotic activity against gram-negative bacteria. Sci Transl Med 5:190ra81. doi: 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu IF, Annamalai T, Sutherland JH, Tse-Dinh YC. 2009. Hydroxyl radicals are involved in cell killing by the bacterial topoisomerase I cleavage complex. J Bacteriol 191:5315–5319. doi: 10.1128/JB.00559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalghatgi S, Spina CS, Costello JC, Liesa M, Morones-Ramirez JR, Slomovic S, Molina A, Shirihai OS, Collins JJ. 2013. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci Transl Med 5:192ra85. doi: 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 17.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 19.Mahoney TF, Silhavy TJ. 2013. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol 195:1869–1874. doi: 10.1128/JB.02197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricci V, Loman N, Pallen M, Ivens A, Fookes M, Langridge GC, Wain J, Piddock LJ. 2012. The TCA cycle is not required for selection or survival of multidrug-resistant Salmonella. J Antimicrob Chemother 67:589–599. doi: 10.1093/jac/dkr515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosato RR, Fernandez R, Paz LI, Singh CR, Rosato AE. 2014. TCA cycle-mediated generation of ROS is a key mediator for HeR-MRSA survival under beta-lactam antibiotic exposure. PLoS One 9:e99605. doi: 10.1371/journal.pone.0099605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imlay JA. 2015. Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin Microbiol 24:124–131. doi: 10.1016/j.mib.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy MP, Holmgren A, Larsson NG, Halliwell B, Chang CJ, Kalyanaraman B, Rhee SG, Thornalley PJ, Partridge L, Gems D, Nystrom T, Belousov V, Schumacker PT, Winterbourn CC. 2011. Unraveling the biological roles of reactive oxygen species. Cell Metab 13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halliwell B, Whiteman M. 2004. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol 142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renggli S, Keck W, Jenal U, Ritz D. 2013. Role of autofluorescence in flow cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J Bacteriol 195:4067–4073. doi: 10.1128/JB.00393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulander W, Wang Y, Folkesson A, Charbon G, Lobner-Olesen A, Ingmer H. 2014. Bactericidal antibiotics increase hydroxyphenyl fluorescein signal by altering cell morphology. PLoS One 9:e92231. doi: 10.1371/journal.pone.0092231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halliwell B, Gutteridge JMC. 2007. Free radicals in biology and medicine. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 28.Brustolon M, Giamello E. 2009. Electron paramagnetic resonance: a practitioner's toolkit. John Wiley & Sons Inc, Hoboken, NJ. [Google Scholar]
- 29.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 30.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das D, Bishayi B. 2009. Staphylococcal catalase protects intracellularly survived bacteria by destroying H2O2 produced by the murine peritoneal macrophages. Microb Pathog 47:57–67. doi: 10.1016/j.micpath.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Berliner LJ, Khramtsov V, Fujii H, Clanton TL. 2001. Unique in vivo applications of spin traps. Free Radic Biol Med 30:489–499. doi: 10.1016/S0891-5849(00)00491-3. [DOI] [PubMed] [Google Scholar]
- 33.Pou S, Ramos CL, Gladwell T, Renks E, Centra M, Young D, Cohen MS, Rosen GM. 1994. A kinetic approach to the selection of a sensitive spin trapping system for the detection of hydroxyl radical. Anal Biochem 217:76–83. doi: 10.1006/abio.1994.1085. [DOI] [PubMed] [Google Scholar]
- 34.Hougaard AB, Arneborg N, Andersen ML, Skibsted LH. 2013. ESR spin trapping for characterization of radical formation in Lactobacillus acidophilus NCFM and Listeria innocua. J Microbiol Methods 94:205–212. doi: 10.1016/j.mimet.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Ramos CL, Pou S, Britigan BE, Cohen MS, Rosen GM. 1992. Spin trapping evidence for myeloperoxidase-dependent hydroxyl radical formation by human neutrophils and monocytes. J Biol Chem 267:8307–8312. [PubMed] [Google Scholar]
- 36.Clapp PA, Davies MJ, French MS, Gilbert BC. 1994. The bactericidal action of peroxides; an E.P.R. spin-trapping study. Free Radic Res 21:147–167. doi: 10.3109/10715769409056566. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez JE, Padilla R, Avila J, Carrascosa JL. 1990. Removal of the carboxy terminus of beta-tubulin subunit produces lateral annealing of microtubules with different orientations. Int J Biochem 22:1419–1425. doi: 10.1016/0020-711X(90)90232-R. [DOI] [PubMed] [Google Scholar]
- 38.Iwase T, Tajima A, Sugimoto S, Okuda K, Hironaka I, Kamata Y, Takada K, Mizunoe Y. 2013. A simple assay for measuring catalase activity: a visual approach. Sci Rep 3:3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadurugamuwa JL, Anwar H, Brown MR, Shand GH, Ward KH. 1987. Media for study of growth kinetics and envelope properties of iron-deprived bacteria. J Clin Microbiol 25:849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burkhardt EM, Bischoff S, Akob DM, Buchel G, Kusel K. 2011. Heavy metal tolerance of Fe(III)-reducing microbial communities in contaminated creek bank soils. Appl Environ Microbiol 77:3132–3136. doi: 10.1128/AEM.02085-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naclerio G, Baccigalupi L, Caruso C, De Felice M, Ricca E. 1995. Bacillus subtilis vegetative catalase is an extracellular enzyme. Appl Environ Microbiol 61:4471–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol 185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ezraty B, Henry C, Herisse M, Denamur E, Barras F. 2014. Commercial lysogeny broth culture media and oxidative stress: a cautious tale. Free Radic Biol Med 74:245–251. doi: 10.1016/j.freeradbiomed.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 45.Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 46.Gaupp R, Ledala N, Somerville GA. 2012. Staphylococcal response to oxidative stress. Front Cell Infect Microbiol 2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Chen S, Zhang J, Rothenbacher FP, Jiang T, Kan B, Zhong Z, Zhu J. 2012. Catalases promote resistance of oxidative stress in Vibrio cholerae. PLoS One 7:e53383. doi: 10.1371/journal.pone.0053383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


