Abstract
Pathogenic enteric viruses are responsible for a wide range of infections in humans, with diverse symptoms. Raw and partially treated wastewaters are major sources of environmental contamination with enteric viruses. We monitored a municipal secondary wastewater treatment plant (New Orleans, LA) on a monthly basis for norovirus (NoV) GI and GII and enterovirus serotypes using multiplex reverse transcription-quantitative PCR (RT-qPCR) and microbial indicators of fecal contamination using standard plating methods. Densities of indicator bacteria (enterococci, fecal coliforms, and Escherichia coli) did not show monthly or seasonal patterns. Norovirus GII was more abundant than GI and, along with enterovirus serotypes, increased in influent during fall and spring. The highest NoV GI density in influent was in the fall, reaching an average of 4.0 log10 genomic copies/100 ml. Norovirus GI removal (0.95 log10) was lower than that for GII, enterovirus serotypes, and male-specific coliphages (1.48 log10) or for indicator bacteria (4.36 log10), suggesting higher resistance of viruses to treatment. Male-specific coliphages correlated with NoV GII densities in influent and effluent (r = 0.48 and 0.76, respectively) and monthly removal, indicating that male-specific coliphages can be more reliable than indicator bacteria to monitor norovirus GII load and microbial removal. Dominant norovirus genotypes were classified into three GI genotypes (GI.1, GI.3, and GI.4) and four GII genotypes (GII.3, GII.4, GII.13, and GII.21), dominated by GI.1 and GII.4 strains. Some of the seasonal and temporal patterns we observed in the pathogenic enteric viruses were different from those of epidemiological observations.
INTRODUCTION
Enteric viruses are responsible for a wide range of infections in humans with diverse symptoms. Infected individuals shed millions of virus particles in their feces or body fluids, which eventually enter sewage systems. Enteric viruses may naturally occur in aquatic environments as well, but human activities, in particular, sewage discharge, is the primary source of environmental contaminants (1–3). Among the pathogenic enteric viruses, norovirus (NoV), enterovirus (EV), adenovirus, astrovirus, and rotavirus have been found frequently in municipal wastewaters worldwide (1, 4–6). Several gastroenteritis outbreaks have been linked directly or indirectly to human exposure of raw or partially treated sewage-contaminated water or foods (2).
Municipal wastewaters usually undergo a secondary treatment before being discharged into the environment. The process involves a mechanical treatment for removing solids followed by biological and chemical treatments, nutrient removal, and discharge (7). Primary-treated (physically processed) wastewater or the effluent water that does not undergo a disinfection process may still harbor infectious enteric viruses, similar to the raw sewage (4, 8). To protect water quality and public safety, fecal coliforms and Escherichia coli have been used to monitor fecal pollution in wastewater discharge or environmental waters. In general, viruses are more resistant than bacteria to UV treatment or chlorination, two common disinfection strategies in secondary wastewater treatment (7, 9). The aggregation of viruses in water or other wastewater solids reduces the efficacy of disinfectants and helps viruses maintain their infectivity when discharged into environmental waters (7, 10). As a result, the bacterial indicators may not be able to reflect the occurrence of enteric viruses in water efficiently. In this regard, monitoring bacteriophages such as male-specific coliphages (MSC), or even direct measurement of enteric viruses such as enteroviruses and adenovirus, have been suggested as more reliable criteria to assess sewage pollutions of human origins in water (1, 11).
Norovirus (NoV; genus Norovirus, family Caliciviridae) has caused the most cases of human gastroenteritis worldwide. NoV is a positive-sense single-stranded RNA (+ssRNA) virus comprised of six genogroups (GI to GVI) and more than 32 genotypes. Genogroups I, II, and IV are responsible for disease in humans, whereas the rest have been found in animals (12–14). Despite the extensive genetically diverse nature, the GII.4 strains have been the predominant cause of the NoV outbreaks in humans globally during the past decade at least (13). Enteroviruses (genus Enterovirus, family Picornaviridae) are composed of poliovirus, coxsackievirus, echovirus, and the numbered enteroviruses (1). The genomic size and genomic and capsid structures of enteroviruses are similar to those of noroviruses. Enterovirus serotypes usually are transmitted through the fecal-oral routes and are responsible for a wide range of infections in humans with diverse clinical syndromes, such as gastroenteritis and meningitis (1, 15).
In this study, we monitored the loads of pathogenic enteric viruses and microbial indicators in a municipal secondary wastewater treatment plant (WWTP) in the United States (New Orleans, LA). The molecular techniques used to analyze viral genomes did not permit the absolute quantification of infectious viruses. This is due primarily to the negative effect of inhibitory compounds on the optimum amplification of viral RNA and the inability of PCR to distinguish intact from damaged virus particles. According to the available literature, this is the first report on the year-round monitoring of pathogenic enteric viruses, microbial indicators, and NoV diversity in a municipal WWTP influent and effluent in the United States. The identification of the dominant NoV genotypes provides important information for the development of vaccines that are more effective against circulating genetically diverse NoV strains (13).
MATERIALS AND METHODS
Wastewater treatment plant and sampling.
A municipal WWTP in New Orleans, LA, with an annual average flow of 98 million gallons per day (MGD) for 2013, was studied. The wastewater is generated from New Orleans on the east bank of the Mississippi River, with an estimated population of 378,715 (16). The sanitary sewage system collects wastewater using a gravity collection system and transfers it to the treatment plant through a series of pumping stations. A 24-h-a-day secondary treatment process employs a high-purity oxygen modification of the activated sludge system, including clarification through sedimentation, chlorination (0.5 mg/liter) for disinfection, and finally discharge of effluent directly into the Mississippi River (Louisiana). Solids are either returned to the process or wasted and disposed of through incineration and then landfill. Monthly samples from influent and effluent waters were obtained from July 2013 to June 2014. Each month, 2 liters of 24-h composite influent and effluent samples were collected (4°C). Even though the samples were collected on the same day, the influent and effluent samples were temporally separate due to the retention time for the influent water to go through the treatment. Effluent samples were collected prior to release into the Mississippi River and were dechlorinated using 0.5 ml of 0.63 M sodium thiosulfate per liter of water. The samples were stored on ice, transported to the Food Microbiology Laboratory at the Louisiana State University Agricultural Center, and analyzed within 24 h. Biochemical oxygen demand (BOD), total suspended solid (TSS), and volatile suspended solid (VSS) data were provided by the WWTP laboratory and were measured according to the Standard Methods for the Examination of Water and Wastewater protocols, no. 5210, 2540D, and 2540E, respectively (17).
Microbial indicators.
Microbial indicators of fecal contamination (fecal coliforms, thermotolerant E. coli, enterococci, and male-specific bacteriophages) were quantified in influent and effluent waters using U.S. Environmental Protection Agency (U.S. EPA) standard techniques. Fecal coliforms were enumerated on m-FC medium (Difco, Sparks, MD) after incubation at 45°C for 24 h (18). Thermotolerant E. coli cells were counted on modified membrane-thermotolerant E. coli agar (m-TEC; Difco) in which the plates were initially incubated at 35°C for 2 h to resuscitate injured or stressed bacteria and then incubated at 45°C for 24 h (19). Enterococci (Enterococcus faecalis, E. faecium, E. avium, and variants) were quantified on enterococcus membrane-indoxyl-β-d-glucoside agar (mEI; Difco) following incubation for 24 h at 41°C (20). Quantities of bacteria were reported as log10 CFU per 100 ml of wastewater sample. Single-agar-layer (SAL) plaque assay on 2× tryptic soy agar (Difco) containing 3.0 mg/ml of each ampicillin sodium salt and streptomycin sulfate salt (Sigma-Aldrich, Steinheim, Germany) was used for the male-specific coliphages plaque assay, using E. coli HS(pFamp)R (ATCC 700891) as the host strain. The plaque-forming units (PFU) were counted after incubating the plates for 16 to 24 h at 37°C and are reported as log10 PFU per 100 ml of wastewater sample. E. coli bacteriophage MS2 (ATCC 15597-B1) was used as a positive control along with the samples (21).
Enteric viruses. (i) Extraction and concentration of enteric viruses.
An ultracentrifuge method was used for the extraction and concentration of enteric viruses from 60 ml of the wastewater samples as developed by the U.S. FDA Gulf Coast Seafood Laboratory at Dauphin Island, AL (22). The RNA was extracted using an RNeasy mini kit (Qiagen, Germantown, MD) by following the manufacturer's instruction, with minor modifications (22) in which a 15-min hold time was given for each washing step. The extracted RNA was eluted in 40 μl THE RNA storage solution (1 mM sodium citrate, pH 6.4; Ambion) and immediately analyzed or stored frozen at −80°C until required.
(ii) Determination of pathogenic enteric viruses.
The detection and quantification method followed a multiplex real-time quantitative reverse transcription-PCR (RT-qPCR) for simultaneous detection of NoV GI, GII, and EV serotypes, along with a heterogeneous internal amplification control (IAC) optimized by Burkhardt et al. (23) and Nordstrom et al. (24). The sequence of the primers and probes for NoV GI and GII target sensitive and broadly reactive ORF1-ORF2 junctions, as designed by Kageyama et al. (25). The primers for EV amplified the 5′ untranslated region (UTR) of the enteroviral genome with a panenterovirus primer set (26). The reaction mixture used a Qiagen OneStep RT-PCR kit (Valencia, CA) for a total volume of 25 μl per reaction mixture and 3.0 μl of RNA template. A Cepheid SmartCycler II system (Sunnyvale, CA) was used for all RT-qPCR analyses. The templates were reverse transcribed at 50°C for 50 min, and then the DNA polymerase was activated at 95°C for 15 min, followed by thermal cycling for 10 s at 95°C, 25 s at 53°C, and 70 s at 62°C for a total of 50 cycles, and then a final extension at 72°C for 10 min (threshold, 10). Reactions were considered positive when the emission intensities exceeded the threshold during the first 46 cycles. All reactions were carried out in duplicate. NoV GI and GII RNA standards (109 genomic copies [GC]/μl) were kindly provided by Christine Moe's laboratory at Emory University (Atlanta, GA), and human poliovirus 3 stock (attenuated Sabin strain) was kindly provided by William Burkhardt's laboratory (U.S. FDA Gulf Coast Seafood Laboratory, Dauphin Island, AL) and were utilized as positive controls and for RNA quantification. The RNA extracted from the poliovirus stock was quantified by analyzing serial decimal dilutions of extracted RNA and assigning the value of one RT-qPCR unit per reaction to the highest dilution showing a positive threshold cycle (CT) value (27). A no-template control was included in all analyses to overcome any uncertainty of false-positive responses due to cross-contamination or reaction failure.
(iii) Sequencing and genotyping noroviruses.
The norovirus samples were genotyped on a bimonthly basis starting from July 2013 until May 2014 to identify the predominant strains present in the influent and effluent waters. The ORF1-ORF2 junction (region C) of the NoV viral genome was amplified by a heminested PCR on a C1000 thermal cycler (Bio-Rad, Hercules, CA) by utilizing a OneStep RT-PCR kit (Qiagen), 0.4 μM each oligonucleotide primer, and 5 U of SUPERase in RNase inhibitor (Ambion, Foster City, CA) in a final reaction volume of 25 μl. Primer sequences were obtained from the previous studies (25, 28). For the first PCR, 5 μl of extracted RNA was amplified by incorporating COG1F/G1SKR and COG2F/G2SKR primers for NoV GI and GII, respectively. RT-PCR conditions were the following: RT for 30 min at 42°C, heat activation of Taq DNA polymerase for 15 min at 95°C, PCR consisting of 40 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s, with a final extension for 7 min at 72°C. The PCR amplicons were purified by electrophoresis using 2% agarose gel (4.83 V/cm) containing 0.5 μg/ml ethidium bromide and were extracted by utilizing a QIAquick gel extraction kit per the manufacturer's instructions (Qiagen, Valencia, CA). Extracted DNA (1 μl) was subjected to a second PCR with G1SKF/G1SKR primers for GI and G2SKF/G2SKR primers for GII under the same conditions but excluding the RT step (28, 29). PCR amplicons (330 bp for GI and 344 bp for GII) were gel purified and cloned into a pCR2.1-TOPO TA vector using a TOPO TA cloning kit with TOP10 E. coli (Life Technologies).
Sequences were read on an ABI Prism BigDye Terminator cycle sequencing reaction kit (Applied Biosystems) on an ABI Prism 3130 automated sequencer genetic analyzer (Life Technologies) and processed on a CLC Sequence Viewer (version 7.5; CLC Bio, Aarhus, Denmark). Phylogenic trees of the partial capsid gene sequences (ORF2 region; 264 nucleotides for GI and 252 nucleotides for GII) were inferred by the maximum likelihood analysis based on the Tamura-Nei model (30) using MEGA 6 software (31). Reference strains were retrieved from the GenBank sequence database at NCBI (14, 32).
Data analysis.
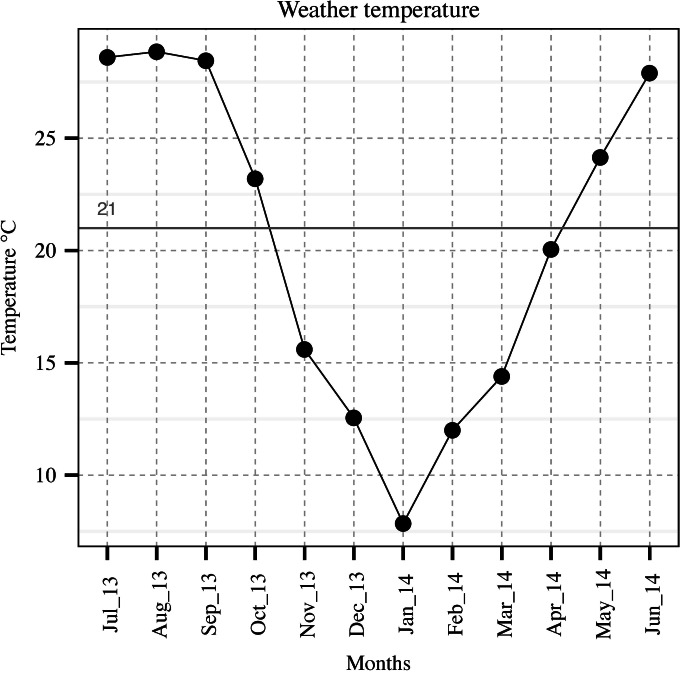
The logarithmic removal was calculated by subtracting logarithmic concentrations of variables in effluent from influent according to Francy et al. (9). Significant differences among mean ranks and multiple comparisons were evaluated using Kruskal-Wallis test at α = 0.05. A univariate logistic regression model was used to evaluate the contribution of each microbial group to the occurrence of others. The Pearson product-moment correlation coefficient (r) was used to assess the correlation among variables. Software RStudio (version 0.98.1091; RStudio Inc., Boston, MA) was used for the statistical analyses and data visualization. Local minimum and maximum weather temperature data were obtained from the National Oceanic and Atmospheric Administration's National Climatic Data Center (NOAA-NCDC), Carrollton station (latitude, 29.934; longitude, −90.136), and are reported as midrange values. A threshold of 21°C was considered to categorize the data into warm months (May to October) and cold months (November to April). Seasons were classified as spring (March to May), summer (June to August), fall (September to November), and winter (December to February).
Nucleotide sequence accession numbers.
The NoV sequences identified in our study can be found in GenBank under accession numbers KP868574 to KP868614.
RESULTS
PCR performance.
Analysis of serially diluted standard RNA showed RT-qPCR efficiencies of 87.22%, 96.25%, and 95.29% for NoV GI, NoV GII, and EV, respectively; the coefficients of determination (R2) ranged from 0.986 to 0.992 (data not shown). Promising results were obtained from the endpoint heminested PCR analysis of the NoV viral genome. Even though the RT-PCR did not yield bright or visible bands in some samples, the agarose gel purification of the amplicons yielded 100% positive reactions following the second PCR (data not shown).
Pathogenic enteric viruses and microbial indicators.
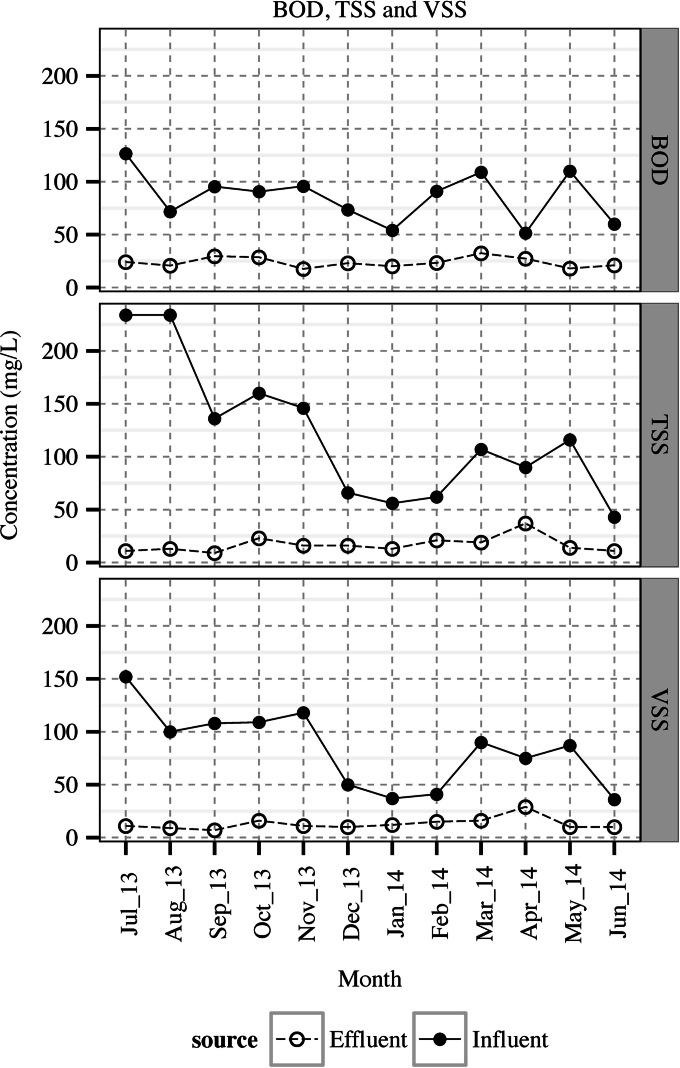
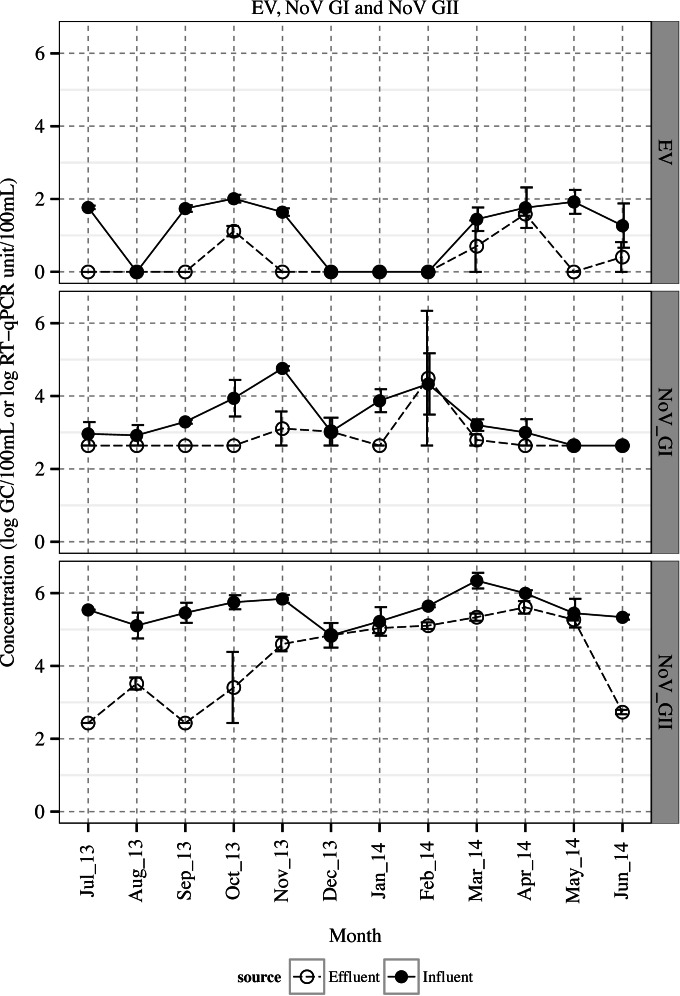
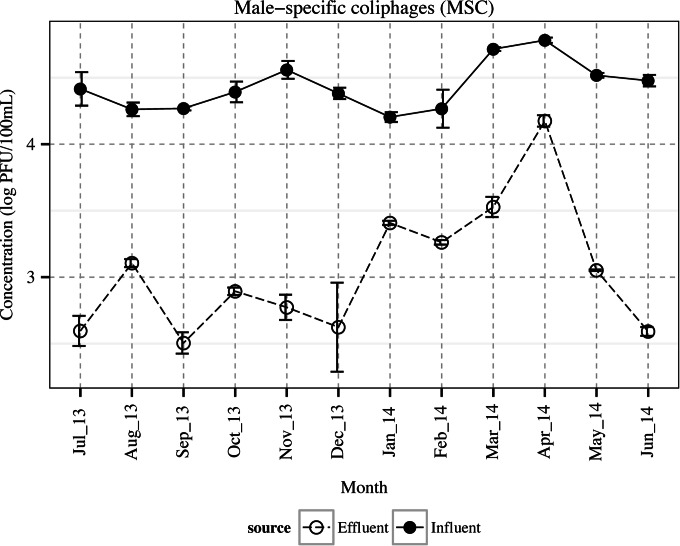
Monthly composite influent and effluent waters were obtained when no flooding or heavy rain had occurred within 24 h prior to each sample collection. Figures 1 and 2 represent the weather temperature and BOD, TSS, and VSS content of wastewater samples, respectively. Enteric viruses were detected in both influent and effluent nearly all year round (Fig. 3). Overall, 83.3% (10/12) of the influent samples and 33.3% (4/12) of the effluent samples were positive for NoV GI, whereas NoV GII and EV were more prevalent and were detected similarly in 100% (12/12) of the influent and 83.3% (10/12) of the effluent samples. The concentration of NoV GII was greater than those of NoV GI and EV in both influent and effluent waters (P < 0.05). Fecal coliforms, E. coli, and MSC were detected in all of the influent and effluent samples, while enterococci were found in 100% (12/12) of the influent and 66.7% (8/12) of the effluent waters, all with a slight fluctuation over time (Table 1 and Fig. 4). The overall mean levels of fecal coliforms, E. coli, and enterococci in the influent were 6.1 ± 0.1, 6.0 ± 0.1, and 5.1 ± 0.1 log10 CFU/100 ml, respectively. Following the treatment and chlorine disinfection, their corresponding densities decreased to 1.8 ± 0.2, 1.8 ± 0.2, and 0.4 ± 0.1 log10 CFU/100 ml, respectively. The mean concentration of MSC reached 4.4 ± 0.0 and 3.0 ± 0.1 log10 PFU/100 ml in the influent and effluent waters, respectively.
FIG 1.
Midrange weather temperature in New Orleans, LA, for each sample collection day.
FIG 2.
Concentrations of biochemical oxygen demand (BOD), total suspended solids (TSS), and volatile suspended solids (VSS) in the wastewater influent and effluent waters.
FIG 3.
Concentrations of NoV GI, GII, and EV in the wastewater influent and effluent (means ± standard errors [SE]).
TABLE 1.
Concentrations of indicator bacteria in the influent and effluent of the wastewater treatment plant
| Date | Bacterial concna (log10 CFU/100 ml) |
|||||
|---|---|---|---|---|---|---|
| Fecal coliforms |
E. coli |
Enterococci |
||||
| Influent | Effluent | Influent | Effluent | Influent | Effluent | |
| July 2013 | 5.58 ± 0.13E | 2.62 ± 0.38AB | 6.22 ± 0.08A | 2.50 ± 0.00B | 4.99 ± 0.18ABC | 0.17 ± 0.12A |
| August 2013 | 6.04 ± 0.07ABCD | 1.81 ± 0.34CDF | 6.17 ± 0.05AB | 1.67 ± 0.07D | 5.22 ± 0.19A | 0.60 ± 0.30B |
| September 2013 | 6.25 ± 0.22ABC | 1.38 ± 0.35DEFG | 6.22 ± 0.01A | 1.52 ± 0.02E | 5.08 ± 0.13ABC | 0.13 ± 0.00C |
| October 2013 | 6.14 ± 0.32ABCD | 1.90 ± 0.03BCD | 6.24 ± 0.06A | 2.17 ± 0.09C | 5.08 ± 0.01AB | 0.83 ± 0.02AB |
| November 2013 | 6.27 ± 0.27AB | 1.11 ± 0.08EFG | 6.22 ± 0.02A | 1.44 ± 0.13E | 5.09 ± 0.16ABC | 0.13 ± 0.00C |
| December 2013 | 6.06 ± 0.21ABCDE | 3.99 ± 0.08A | 6.07 ± 0.26AB | 3.47 ± 0.09E | 5.37 ± 0.12A | 0.92 ± 0.09AB |
| January 2014 | 5.86 ± 0.03BCDE | 0.54 ± 0.15G | 5.78 ± 0.02BC | 0.57 ± 0.03H | 4.59 ± 0.21C | 0.13 ± 0.00C |
| February 2014 | 5.70 ± 0.06DE | 0.81 ± 0.25FG | 5.43 ± 0.17C | 1.03 ± 0.03GH | 4.59 ± 0.24BC | 0.13 ± 0.00C |
| March 2014 | 6.20 ± 0.209ABC | 2.43 ± 0.14AB | 6.13 ± 0.09ABC | 2.79 ± 0.12AB | 5.50 ± 0.50A | 0.43 ± 0.03B |
| April 2014 | 5.82 ± 0.06CDE | 2.19 ± 0.19ABC | 5.49 ± 0.10C | 2.09 ± 0.01C | 4.77 ± 0.05BC | 0.17 ± 0.04C |
| May 2014 | 6.34 ± 0.15AB | 1.55 ± 0.02DEF | 5.94 ± 0.04ABC | 1.15 ± 0.01FG | 4.98 ± 0.03ABC | 0.13 ± 0.00C |
| June 2014 | 6.41 ± 0.17A | 1.83 ± 0.26BCD | 6.22 ± 0.03A | 1.23 ± 0.06F | 5.35 ± 0.04A | 0.13 ± 0.00C |
Within each microbial group and source (influent or effluent), means with the same letter are not significantly different. All values are reported as means ± SE.
FIG 4.
Concentrations of male-specific coliphages (MSC) in the wastewater influent and effluent (means ± SE).
Trends and seasonality.
Among the indicator bacteria, only E. coli densities in influent were affected by seasonality (P = 0.016), reaching the highest concentrations in summer and fall (average, 6.2 ± 0.0 log10 CFU/100 ml), with higher loads in warmer months (average, 6.2 ± 0.0 log10 CFU/100 ml) than cold months (average, 5.9 ± 0.1 log10 CFU/100 ml) (P = 0.03). Stronger fluctuations were observed in the densities of pathogenic enteric viruses (NoV GI, NoV GII, and EV), with distinct trends over time in NoV GII and EV (Fig. 3). The concentrations of NoV GII showed a seasonal trend, being substantially higher in winter and spring (cold months) and reaching the highest values in spring, at 5.9 ± 0.2 log10 GC/100 ml. Similarly, the concentration of MSC and EV in the influent increased during spring (March to May), resulting in average values of 4.7 ± 0.1 log10 PFU/100 ml and 1.7 ± 0.2 log10 RT-qPCR U/100 ml, respectively. In addition, EV showed another increase in fall, with an average value of 1.8 ± 0.1 log10 RT-qPCR U/100 ml.
Only in the cases of NoV GI and GII did the monthly concentration of viruses in the influent reflect their concentrations in the effluent (r = 0.41 and 0.45, respectively; P < 0.05). Among the pathogenic enteric viruses, NoV GI and EV densities in the influent and NoV GII in the effluent were correlated with the weather temperature (r = −0.56, 0.42, and −0.59, respectively; P < 0.05). The strongest correlations (r > 0.45) between enteric pathogens and microbial indicators were found between NoV GII and MSC in both the influent and effluent (r = 0.48 and 0.76, respectively; P < 0.05). In addition, univariate logistic regression analyses indicated the occurrence of NoV GII on the level of MSC in the effluent (odds ratio [OR], 1.45; 97.5% confidence interval [CI], 1.06 to 1.98; P = 0.030). EV was correlated with NoV GII in the influent (r = 0.45); however, the presence or absence of EV was unrelated to the NoV GII densities (P > 0.05). Among the indicator bacteria, enterococci showed the strongest correlation with the fecal coliforms (r > 0.63) and E. coli (r > 0.77), followed by fecal coliforms and E. coli (r > 0.55) in both influent and effluent waters. In this regard, the presence or absence of enterococci in the effluent was in accordance with the concentration of E. coli (OR, 1.48; CI, 1.22 to 1.79; P = 0.0007) and coliforms (OR, 1.41; CI, 1.19 to 1.67; P = 0.0006).
Microbial removal.
Secondary treatment of wastewater resulted in significant removal in all microbial groups. In general, log removal was more pronounced in indicator bacteria (4.36 log10; P > 0.05), followed by NoV GII, EV, and MSC (1.48 log10; P > 0.05) and then NoV GI (0.95 log10). Log removals in indicator bacteria and NoV GI were similar during warm versus cold months; however, for the rest of the microbial groups, higher removal was observed in warm months. The concentrations of NoV GII in the effluent waste were at the lowest levels in summer and fall, resulting in the highest virus removal among enteric viruses. Close similarity in monthly log removal was found between MSC and NoV GII (r = 0.72) and between MSC and EV (r = 0.73). Among all of the microbial groups, microbial removal in NoV GII and EV was correlated with weather temperature (r = 0.70 and 0.57, respectively). In general, log removal in the indicator bacteria did not have strong seasonality (P > 0.05), whereas it was higher in MSC during fall and summer (1.65 log10), in NoV GI during fall and winter (1.17 log10), in NoV GII in summer and fall (2.32 log10), and in EV in summer (1.74 log10).
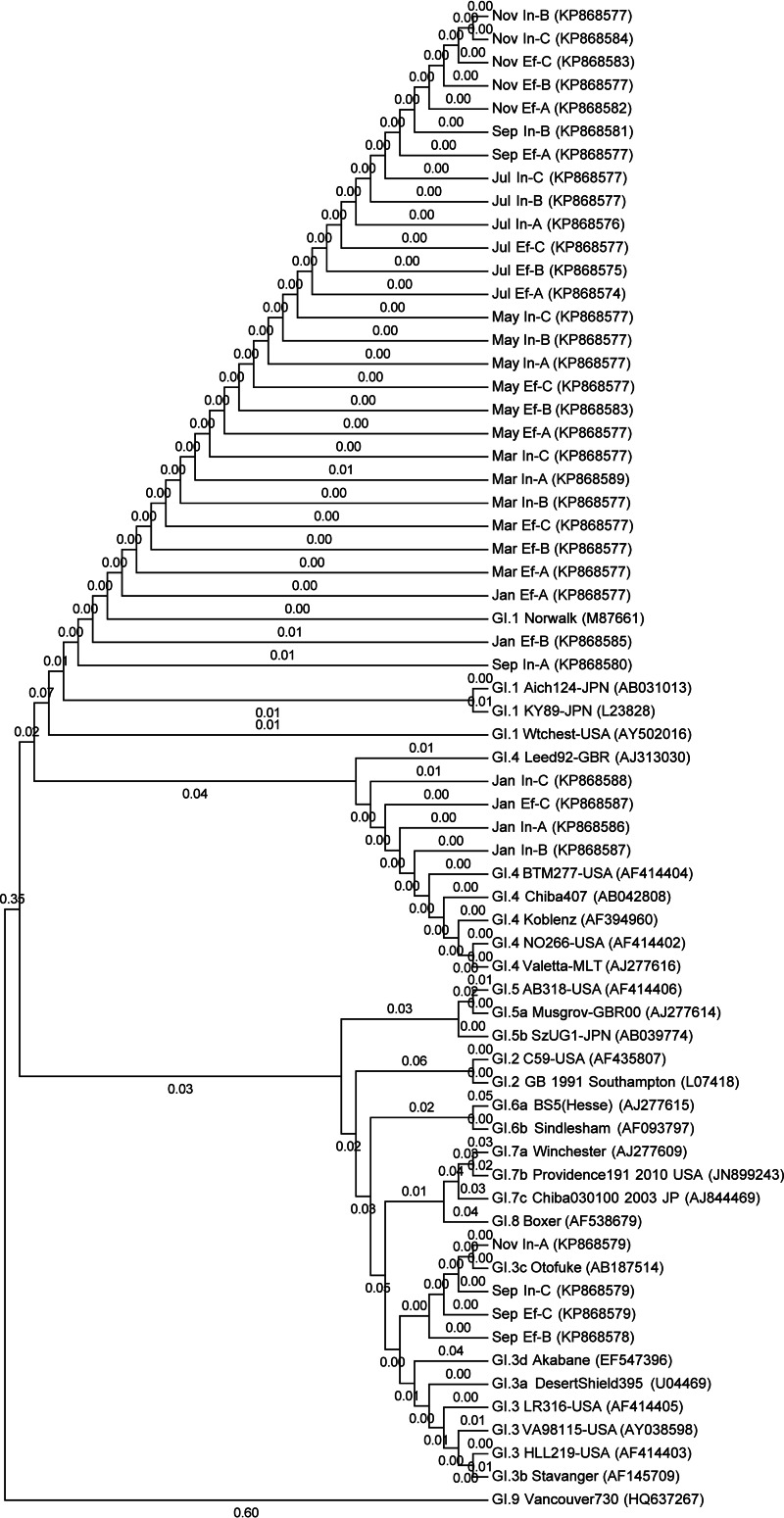
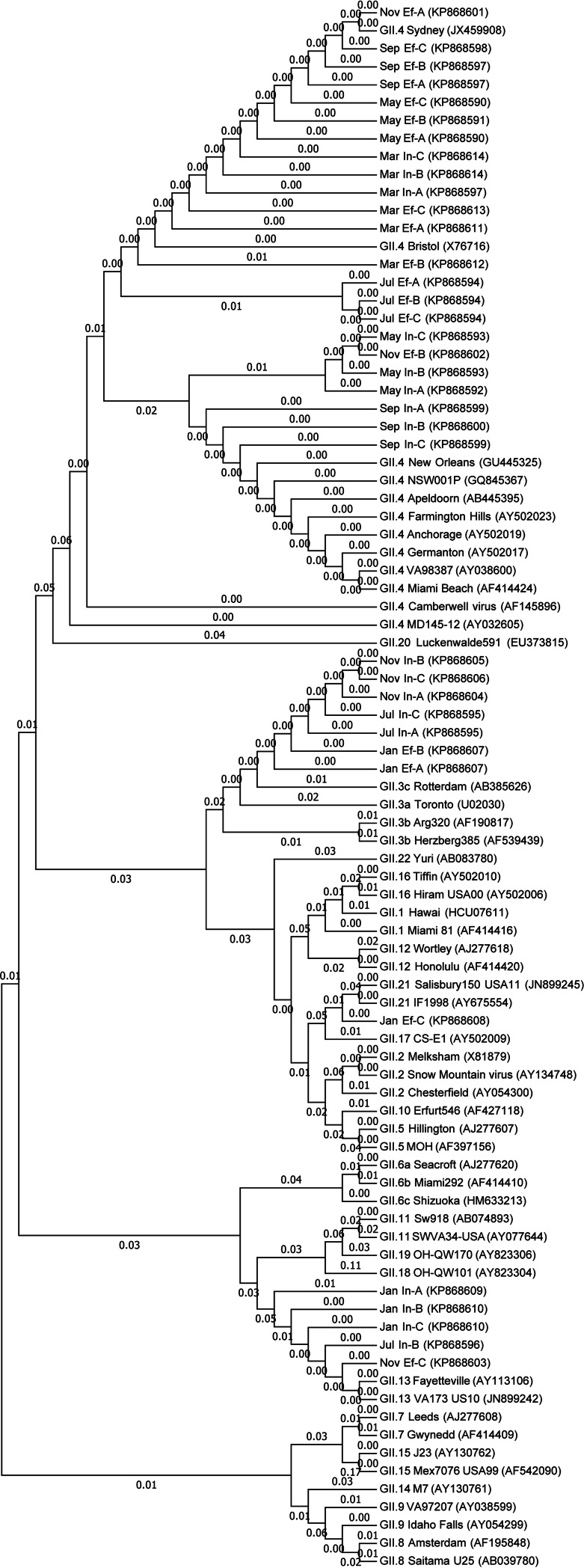
Genetic diversity of noroviruses.
Seven norovirus genotypes were identified from a total of 72 clones (Fig. 5 and 6 and Table 2). NoV GI.1 Norwalk was present in all samples except for the influent water in January, comprising 72.2% (13/18) and 83.3% (15/18) of the influent and effluent clones, respectively, followed by GI.3 and GI.4, which each comprised 11.1% (4/36) of the GI clones. NoV GII.4 Sydney dominated NoV GII genotypes and was identified in 22.2% (4/18) of the influent and 50.0% (9/18) of the effluent clones. The rest of the GII.4 genotypes (30.6%, 11/18) could not be distinguished at the strain level. Other genotypes were GII.3 (19.4%, 7/36), GII.13 (13.9%, 5/36), and GII.21 (2.78%, 1/36).
FIG 5.
Phylogenetic tree for norovirus GI strains using 264 nucleotides of the partial capsid gene sequences. Reference strains start with GI followed by the GenBank accession numbers (14, 32). Samples start with the months followed by the source (In, influent; Ef, effluent), replicates (A to C), and GenBank accession numbers.
FIG 6.
Phylogenetic tree for norovirus GII strains using 252 nucleotides of the partial capsid gene sequences. Reference strains start with GII followed by the GenBank accession numbers (14, 32). Samples start with the months, followed by the source (In, influent; Ef, effluent), replicates (A to C), and GenBank accession numbers.
TABLE 2.
Summary of the dominant norovirus GI and GII strains identified in the influent and effluent of wastewater
| Date and source | NoVa |
|
|---|---|---|
| GI | GII | |
| July 2013 | ||
| Influent | GI.1 Norwalk (3/3) | GII.3c Rotterdam (2/3), GII.13 (1/3) |
| Effluent | GI.1 Norwalk (3/3) | GII.4 (3/3) |
| September 2013 | ||
| Influent | GI.1 Norwalk (2/3), GI.3c Otofuke (1/3) | GII.4 (3/3) |
| Effluent | GI.1 Norwalk (1/3), GI.3c Otofuke (2/3) | GII.4 Sydney (3/3) |
| November 2013 | ||
| Influent | GI.1 Norwalk (2/3), GI.3c Otofuke (1/3) | GII.3c Rotterdam (3/3) |
| Effluent | GI.1 Norwalk (3/3) | GII.4 Sydney (1/3), GII.4 (1/3), GII.13 (1/3) |
| January 2014 | ||
| Influent | GI.4 (3/3) | GII.13 (3/3) |
| Effluent | GI.1 Norwalk (2/3), GI.4 (1/3) | GII.3c Rotterdam (2/3), GII.21 (1/3) |
| March 2014 | ||
| Influent | GI.1 Norwalk (3/3) | GII.4 Sydney (3/3) |
| Effluent | GI.1 Norwalk (3/3) | GII.4 Sydney (2/3), GII.4 (1/3) |
| May 2014 | ||
| Influent | GI.1 Norwalk (3/3) | GII.4 (3/3) |
| Effluent | GI.1 Norwalk (3/3) | GII.4 Sydney (3/3) |
The numbers in parentheses denote the number of clones that were identified.
DISCUSSION
Because of the inability to cultivate human NoV in vitro, RT-qPCR has been used extensively for the detection and quantification of NoV in environmental matrices and clinical specimens, particularly where the virus densities usually are low (1, 13, 14, 33). Wastewater contains humic compounds, divalent cations, and salts, which may occur in the PCR template, negatively impact the amplification efficiency, and underestimate quantification or cause false-negative results (8, 34). The internal amplification control used in the RT-qPCR was able to monitor any inhibitory effect. In this regard, the virus and RNA extraction protocols were efficient in removing the majority of the inhibitory compounds. The potential effects of the remaining inhibitory substances were mitigated by serial dilution of the RNA template prior to analysis.
The concentration of NoV GII in the influent was higher than that of GI, which is in accordance with previous reports and epidemiological trends in the United States and globally (3, 8, 35–38). Epidemiological data in the United States and worldwide have shown that NoV outbreaks peak in the colder winter months, with 55% of the cases in the United States occurring in December to February (35, 39), and is in agreement with our observation of high NoV GII densities (5.6 log10 GC/100 ml) in the influent in November to March and with previous reports (3, 37, 40, 41). These observations indicate that a significant portion of NoV GII in the wastewater is associated with NoV infections in the community. Similarly, Maunula et al. (36) reported a simultaneous occurrence of NoV in the river water and the upstream wastewater that were in coincidence with the NoV epidemiological peaks in the community. The prevalence of NoV in the freshwater rivers affected by the discharge of treated municipal wastewaters can vary between 30% and 75% (36, 42). A recent study estimated that the 50% human infectious dose (HID50) of NoV is similar to that of other RNA viruses and ranges between 3.01 and 3.45 log10 virions (43). Regarding high densities of NoV GI and GII observed in the effluent discharged into the river water, we can imply that the exposure of drinking water, irrigation water, shellfish harvesting, or recreational waters with sewage-contaminated surface waters can pose a potential health risk to humans (37, 38, 42, 44). The health risk can be higher during the aftermath of a heavy rain or system failure that causes overflow of raw or partially treated wastewater into the environment (45).
The dominant GI genotypes, in order of prevalence, were GI.1 Norwalk, GI.3c, and GI.4. We did not identify any GI.6 strains; however, it was the dominant GI genotype isolated from 49 WTTP in the Netherlands (44). GI.4 was the dominant genotype in wastewater effluent, and oysters were contaminated with wastewater in Ireland (38); however, we found this genotype only in January in 17% (3/18) of the influent and 5% (1/18) of the effluent clones. The GI.1 genotype was the dominant NoV strain in the influent; however, this genotype has been associated with less than 1% of the outbreaks in the United States during 2009 to 2013, as opposed to GI.6 (49.7%), GI.3 (24.6%), and GI.4 (9.9%) (46). It suggests that nonepidemic or subclinical gastroenteritis incidences in the community contributed to the high prevalence of GI.1 strains that were reflected in the influent samples (4). Other NoV genotypes that have been reported in WWTP are GI.2, GI.5 to GI.9, GI.11, GI.12, and GI.14 and GII.1, GII.2, GII.5 to GII.12, and GII.15 to GII.17 (5, 38).
NoV GII.4 has been largely associated with gastroenteritis outbreaks in the United States and worldwide and is the most frequently identified genotype in food-borne NoV outbreaks (13) and wastewater (3, 5, 38). According to epidemiological studies, about 94% of all outbreaks in the United States during 2009 to 2013 have been typed as NoV GII.4 New Orleans or GII.4 Sydney. These two strains have been known as emerging recombinants and are implicated in severe gastroenteritis outbreaks worldwide (13, 46). The GII.13 and GII.21 genotypes that were detected in our study have been rarely reported and have been responsible for less than 1% of the NoV outbreaks in the United States from 2009 to 2013 (46). The presence of multiple genotypes in wastewaters may contribute to the emergence of recombinant strains with more severe health risks (44, 46).
In general, the concentrations of EV in the wastewater samples were lower than what Hewitt et al. (8) reported for multiple WWTP, in which EV exceeded NoV GI and GII concentrations and reached 1.8 to 5.7 and 0.5 to 4.3 log10 GC/100 ml in the influent and effluent, respectively. Katayama et al. (40) found that adenovirus concentrations in the influent of a WWTP in Japan was higher than those of NoV and EV. The concentration of EV in the influent was within the range reported by others (40). Epidemiological studies of enterovirus infection cases in the United States have shown remarkable seasonality, increasing sharply during summer and fall, with a peak in August (15). We observed that the highest concentrations of EV occurred in fall and spring, and the lowest concentrations in spring and winter (Fig. 3).
High prevalence of enteric pathogens in wastewater throughout the year reflects the circulation of the pathogens in a population. Therefore, even when the rates of epidemiological and clinical cases are low, the potential health risks still exist (4, 8, 40, 41, 44, 47–49). Secondary treatment has been shown to be effective in reducing the microbial load but is inefficient at complete removal. In accordance with previous reports, log removal was lower in pathogenic enteric viruses than in indicator bacteria (41). Microbial removal in EV was 1.4 log10, which is similar to results of previous reports (8). The NoV removal through the treatment was 1.0 to 1.6 log10 and was lower than 2.7 log10, as observed by van den Berg et al. (44).
NoV removal was relatively higher in warmer months, as similarly observed for MSC. These findings indicate that virus removal was impacted negatively by the initial viral loads and positively by the environmental temperature. Virus removal fluctuated over time and was in accordance with previous studies (6, 36, 37, 49). Compared with NoV GII strains, the GI strains seemed to be approximately 4.8 times more resistant to the two-stage wastewater treatment. These results are similar to those of previous reports in which NoV GI showed lower removal than GII following wastewater treatment (47, 49). Furthermore, MSC removal seemed to have the potential to indicate the efficiency of secondary treatment on removing NoV GII in WWTP. Recently, MSC (along with E. coli) has been suggested as a good indicator of virus removal across conventional secondary disinfection processes (9, 37). Assuming a comparable abundance and survival of NoV and MSC, we could conclude that at least a portion of NoV GII strains detected in the effluent maintained their infectivity due to simultaneous detection of viable MSC plaques and NoV GII viral genomes, as suggested by Hewitt et al. (8), who reported the parallel occurrence of cultivable enteric viruses and PCR concentrations. However, similar to fecal coliforms and E. coli, not all of the coliphage types are human specific. Nonhuman fecal matter may pollute water but not necessarily pose health risks associated with enteric viruses. Among four genogroups of MSC, human sewage usually contains groups II and III, in which group II has correlated well with human fecal pollution in water (50, 51).
A recommended way to monitor viral contamination is the direct detection of pathogens without using indicators (1). Regarding the high prevalence of some enteric viruses in the influent, direct monitoring of enteric viruses, such as adenoviruses, enteroviruses, NoV, and, preferably, picobirnaviruses, has been suggested as a viral indicator of fecal contamination (6). Whether the detected levels of NoV or EV in the WWTP samples are considered a health hazard is rather complicated, because current RT-PCR assays may not reflect virus infectivity (29, 33). Additionally, a strong correlation between the removal of the viral genome (particularly RNA) and loss of infectivity has been reported that could be due to the rapid degradation of RNA in the environment (reviewed in reference 1). Assuming similar survival and abundance of NoV and enteroviruses, Hewitt et al. (8) suggested that at least a proportion of NoV detected in the effluents could be infectious due to the simultaneous occurrence of cultivable enteric viruses and PCR concentrations. Therefore, the secondary-treated wastewater can contribute to the release of infectious virus particles in the environment and increase the harmful health effects of human exposure to enteric pathogens. Implementing effective virus removal methods in the wastewater treatment processes is recommended to reduce the circulation of pathogenic enteric viruses between humans and the environment.
Conclusions.
According to our monthly surveillance of a municipal WWTP (July 2013 to June 2014, New Orleans, LA), we found that the occurrences of pathogenic enteric viruses in wastewater can reflect the circulation of the pathogens in a population but may not follow the epidemiological and clinical trends. Viruses were more resistant than bacteria to the secondary treatment of wastewater; however, the health risks associated with surviving infectious virions could not be confirmed. In general, NoV GII strains were more abundant than NoV GI and EV strains. Male-specific coliphages may be more reliable than bacterial indicators for monitoring NoV GII load in the wastewater and treatment removal process. This relationship indicates the likelihood of infectious NoV GII occurrence in the effluent water. Therefore, direct monitoring of infections pathogens are still preferred over bacterial indicators, particularly when the effluent water is to be further utilized for human activities.
ACKNOWLEDGMENTS
This project was supported by Agriculture and Food Research Initiative competitive grant no. 2011-6800-30395 from the USDA National Institute of Food and Agriculture.
We thank William Burkhardt and Jacquelina Woods (FDA Gulf Coast Seafood Laboratory), Jan Vinjé and team (CDC), Christine Moe and team (Emory University), Jennifer Cannon (University of Georgia), Lee-Ann Jaykus and team (North Carolina State University), operating personnel in the wastewater treatment plant, LSU Department of Experimental Statistics, LSU Agricultural Center Biotechnology Laboratory, and LSU School of Veterinary Medicine.
We have no conflicts of interest to report.
REFERENCES
- 1.Fong T-T, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lees D. 2000. Viruses and bivalve shellfish. Int J Food Microbiol 59:81–116. doi: 10.1016/S0168-1605(00)00248-8. [DOI] [PubMed] [Google Scholar]
- 3.Skraber S, Langlet J, Kremer JR, Mossong J, De Landtsheer S, Even J, Muller CP, Hoffmann L, Cauchie H-M. 2011. Concentration and diversity of noroviruses detected in Luxembourg wastewaters in 2008-2009. Appl Environ Microbiol 77:5566–5568. doi: 10.1128/AEM.00632-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myrmel M, Berg EM, Grinde B, Rimstad E. 2006. Enteric viruses in inlet and outlet samples from sewage treatment plants. J Water Health 4:197–209. [PubMed] [Google Scholar]
- 5.Kitajima M, Haramoto E, Phanuwan C, Katayama H, Furumai H. 2012. Molecular detection and genotyping of human noroviruses in influent and effluent water at a wastewater treatment plant in Japan. J Appl Microbiol 112:605–613. doi: 10.1111/j.1365-2672.2012.05231.x. [DOI] [PubMed] [Google Scholar]
- 6.Symonds EM, Griffin DW, Breitbart M. 2009. Eukaryotic viruses in wastewater samples from the United States. Appl Environ Microbiol 75:1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitton G. 2011. Wastewater microbiology, 4th ed John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 8.Hewitt J, Leonard M, Greening GE, Lewis GD. 2011. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res 45:6267–6276. doi: 10.1016/j.watres.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 9.Francy DS, Stelzer EA, Bushon RN, Brady AM, Williston AG, Riddell KR, Borchardt MA, Spencer SK, Gellner TM. 2012. Comparative effectiveness of membrane bioreactors, conventional secondary treatment, and chlorine and UV disinfection to remove microorganisms from municipal wastewaters. Water Res 46:4164–4178. doi: 10.1016/j.watres.2012.04.044. [DOI] [PubMed] [Google Scholar]
- 10.Keswick BH, Satterwhite TK, Johnson PC, DuPont HL, Secor SL, Bitsura JA, Gary GW, Hoff JC. 1985. Inactivation of Norwalk virus in drinking water by chlorine. Appl Environ Microbiol 50:261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez RA, Bounty S, Beck S, Chan C, McGuire C, Linden KG. 2014. Photoreactivation of bacteriophages after UV disinfection: role of genome structure and impacts of UV source. Water Res 55:143–149. doi: 10.1016/j.watres.2014.01.065. [DOI] [PubMed] [Google Scholar]
- 12.Green KY. 2007. Caliciviridae: the noroviruses, p 949–979. In Knipe DM, Howley PM (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Pringle K, Lopman B, Vega E, Vinje J, Parashar UD, Hall AJ. 2015. Noroviruses: epidemiology, immunity and prospects for prevention. Future Microbiol 10:53–67. doi: 10.2217/fmb.14.102. [DOI] [PubMed] [Google Scholar]
- 14.Vinjé J. 2015. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 53:373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA. 2006. Enterovirus surveillance—United States, 1970-2005. Morb Mortal Wkly Rep Surveill Summ 55:1–20. [PubMed] [Google Scholar]
- 16.US Census Bureau. 2013. State and county QuickFacts. http://quickfacts.census.gov/qfd/states/22/2255000.html Accessed 5 March 2015. [Google Scholar]
- 17.Rice EW, Baird RB, Eaton AD, Clesceri LS. 2012. Standard methods for the examination of water and wastewater, 22nd ed American Public Health Association, Washington, DC. [Google Scholar]
- 18.US Environmental Protection Agency. 2003. Standard method 9222D. Fecal coliform membrane filter procedure. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 19.US Environmental Protection Agency. 2009. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC), EPA-821-R-09-007 US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 20.US Environmental Protection Agency. 2002. Method 1600: enterococci in water by membrane filtration using membrane-enterococcus indoxyl-β-d-glucoside agar (mEI). US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 21.US Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 22.US Food and Drug Administration. 2010. Concentration and extraction of total enteric viruses from shellfish by ultracentrifugation—Gulf Coast Seafood Laboratory Protocol. Gulf Coast Seafood Laboratory, US Food and Drug Administration, Dauphin Island, AL. [Google Scholar]
- 23.Burkhardt W, Woods JW, Nordstrom J, Hartman G. 2006. A real-time RT-PCR protocol for the simultaneous detection of norovirus and enteroviruses (LIB #4369). US Food and Drug Administration, Gulf Coast Seafood Laboratory, Dauphin Island, AL. [Google Scholar]
- 24.Nordstrom JL, Vickery MC, Blackstone GM, Murray SL, DePaola A. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol 73:5840–5847. doi: 10.1128/AEM.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol 41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson KA, Griffin DW, Paul JH. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res 36:2505–2514. doi: 10.1016/S0043-1354(01)00479-1. [DOI] [PubMed] [Google Scholar]
- 27.Richards GP, Watson MA, Frankhauser RL, Monroe SS. 2004. Genogroup I and II noroviruses detected in stool samples by real-time reverse transcription-PCR using highly degenerate universal primers. Appl Environ Microbiol 70:7179–7184. doi: 10.1128/AEM.70.12.7179-7184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods 100:107–114. doi: 10.1016/S0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 29.Mattison K, Grudeski E, Auk B, Charest H, Drews SJ, Fritzinger A, Gregoricus N, Hayward S, Houde A, Lee BE, Pang XL, Wong J, Booth TF, Vinjé J. 2009. Multicenter comparison of two norovirus ORF2-based genotyping protocols. J Clin Microbiol 47:3927–3932. doi: 10.1128/JCM.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Knight A, Li D, Uyttendaele M, Jaykus LA. 2013. A critical review of methods for detecting human noroviruses and predicting their infectivity. Crit Rev Microbiol 39:295–309. doi: 10.3109/1040841X.2012.709820. [DOI] [PubMed] [Google Scholar]
- 34.Toze S. 1999. PCR and the detection of microbial pathogens in water and wastewater. Water Res 33:3545–3556. doi: 10.1016/S0043-1354(99)00071-8. [DOI] [Google Scholar]
- 35.Hall AJ, Wikswo ME, Pringle K, Gould LH, Parashar UD, Division of Viral Diseases–National Center for Immunization Respiratory Diseases–CDC. 2014. Vital signs: foodborne norovirus outbreaks—United States, 2009-2012. MMWR Morb Mortal Wkly Rep 63:491–495. [PMC free article] [PubMed] [Google Scholar]
- 36.Maunula L, Soderberg K, Vahtera H, Vuorilehto V-P, von Bonsdorff C-H, Valtari M, Laakso T, Lahti K. 2012. Presence of human noro- and adenoviruses in river and treated wastewater, a longitudinal study and method comparison. J Water Health 10:87–99. doi: 10.2166/wh.2011.095. [DOI] [PubMed] [Google Scholar]
- 37.Pouillot R, Van Doren JM, Woods J, Plante D, Smith M, Goblick G, Roberts C, Locas A, Hajen W, Stobo J. 2015. Reduction of norovirus and male-specific coliphage concentrations in wastewater treatment plants: a meta-analysis. Appl Environ Microbiol 81:4669–4681. doi: 10.1128/AEM.00509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajko-Nenow P, Waters A, Keaveney S, Flannery J, Tuite G, Coughlan S, O'Flaherty V, Dore W. 2013. Norovirus genotypes present in oysters and in effluent from a wastewater treatment plant during the seasonal peak of infections in Ireland in 2010. Appl Environ Microbiol 79:2578–2587. doi: 10.1128/AEM.03557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed SM, Lopman BA, Levy K. 2013. A systematic review and meta-analysis of the global seasonality of norovirus. PLoS One 8:e75922. doi: 10.1371/journal.pone.0075922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katayama H, Haramoto E, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res 42:1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 41.Victoria M, Guimaraes FR, Fumian TM, Ferreira FF, Vieira CB, Shubo T, Leite JP, Miagostovich MP. 2010. One year monitoring of norovirus in a sewage treatment plant in Rio de Janeiro, Brazil. J Water Health 8:158–165. doi: 10.2166/wh.2009.012. [DOI] [PubMed] [Google Scholar]
- 42.Ueki Y, Sano D, Watanabe T, Akiyama K, Omura T. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res 39:4271–4280. doi: 10.1016/j.watres.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 43.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Ramani S, Hill H, Ferreira J, Graham DY. 2014. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis 209:1016–1022. doi: 10.1093/infdis/jit620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Berg H, Lodder W, van der Poel W, Vennema H, de Roda Husman AM. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Res Microbiol 156:532–540. doi: 10.1016/j.resmic.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Astrom J, Pettersson TJ, Stenstrom TA, Bergstedt O. 2009. Variability analysis of pathogen and indicator loads from urban sewer systems along a river. Water Sci Technol 59:203–212. doi: 10.2166/wst.2009.860. [DOI] [PubMed] [Google Scholar]
- 46.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. 2014. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol 52:147–155. doi: 10.1128/JCM.02680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.da Silva AK, Le Saux JC, Parnaudeau S, Pommepuy M, Elimelech M, Le Guyader FS. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl Environ Microbiol 73:7891–7897. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haramoto E, Katayama H, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S. 2006. Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci Technol 54:301–308. doi: 10.2166/wst.2006.888. [DOI] [PubMed] [Google Scholar]
- 49.Cheng H-WA, Lucy FE, Broaders MA, Mastitsky SE, Chen C-H, Murray A. 2012. Municipal wastewater treatment plants as pathogen removal systems and as a contamination source of noroviruses and Enterococcus faecalis. J Water Health 10:380–389. doi: 10.2166/wh.2012.138. [DOI] [PubMed] [Google Scholar]
- 50.Brion GM, Meschke JS, Sobsey MD. 2002. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res 36:2419–2425. doi: 10.1016/S0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- 51.Havelaar AH, Furuse K, Hogeboom WM. 1986. Bacteriophages and indicator bacteria in human and animal faeces. J Appl Bacteriol 60:255–262. doi: 10.1111/j.1365-2672.1986.tb01081.x. [DOI] [PubMed] [Google Scholar]