Abstract
Cocoa pulp fermentation is a spontaneous process during which the natural microbiota present at cocoa farms is allowed to ferment the pulp surrounding cocoa beans. Because such spontaneous fermentations are inconsistent and contribute to product variability, there is growing interest in a microbial starter culture that could be used to inoculate cocoa pulp fermentations. Previous studies have revealed that many different fungi are recovered from different batches of spontaneous cocoa pulp fermentations, whereas the variation in the prokaryotic microbiome is much more limited. In this study, therefore, we aimed to develop a suitable yeast starter culture that is able to outcompete wild contaminants and consistently produce high-quality chocolate. Starting from specifically selected Saccharomyces cerevisiae strains, we developed robust hybrids with characteristics that allow them to efficiently ferment cocoa pulp, including improved temperature tolerance and fermentation capacity. We conducted several laboratory and field trials to show that these new hybrids often outperform their parental strains and are able to dominate spontaneous pilot scale fermentations, which results in much more consistent microbial profiles. Moreover, analysis of the resulting chocolate showed that some of the cocoa batches that were fermented with specific starter cultures yielded superior chocolate. Taken together, these results describe the development of robust yeast starter cultures for cocoa pulp fermentations that can contribute to improving the consistency and quality of commercial chocolate production.
INTRODUCTION
Microbial fermentation is a crucial step in the production process for many foods and beverages, including chocolate, beer, wine, bread, and cheese. The quality of these products strongly depends on the microbes present, with even slight deviations in the microbial population yielding marked differences in product characteristics. For thousands of years, these fermentation processes were conducted spontaneously, relying on the inoculation of a complex mixture of microbes present in the environment. However, since the development of techniques to isolate and maintain pure microbial cultures in the late 19th century, an increasing number of producers have adopted the idea of using a defined starter culture (1). This practice greatly increased the reproducibility and efficiency of the fermentation process and resulted in augmented product consistency. However, starter cultures were not adopted in all fermentation industries. One of the most striking examples is the cocoa industry, where the production (about 4 × 106 tons of beans per year) largely depends on the complex and highly variable microbial population present at cocoa farms, where the cocoa beans are exposed to the natural environment to start the fermentation process (2).
The species diversity of spontaneous cocoa pulp fermentations has received much scientific attention, with several studies describing the microbiome across the world (3–9). Interestingly, the variable environment encountered during cocoa pulp fermentations results in a complex and dynamic microbial population. However, despite the geographical distance between cocoa-producing countries and despite differences in climate between different harvest seasons, spontaneous cocoa pulp fermentations show a clear and consistent general trend (8, 10). Specifically, only a very limited number of different bacterial species are consistently isolated in high numbers, namely, lactic acid bacteria (LAB) (mainly Lactobacillus fermentum, Lactobacillus plantarum, and Leuconostoc pseudomesenteroides) and acetic acid bacteria (AAB) (mainly Acetobacter pasteurianus). In contrast, the fungal diversity is often much broader (8). Generally, a wide diversity of apiculate, fast-glucose-fermenting species (including Hanseniaspora spp.) is found at the start of the fermentation process. In a second stage, more robust, stress-resistant species (mainly Saccharomyces and/or Pichia spp.) are found, but the exact species composition varies significantly with the season, geographical location, agricultural practices, and other factors (4, 5, 8, 11).
Yeasts play several crucial roles in the cocoa pulp fermentation process, including the production of ethanol and organic acids, which are believed to arrest germination of the cocoa seeds and contribute to essential chemical conversions inside the cocoa beans (12). Moreover, fermenting yeasts also produce myriad volatile aroma compounds (13, 14). Lastly, yeasts producing pectinolytic enzymes are also believed to play a central role in the degradation of the viscous pectin-rich pulp (15, 16). Hence, yeasts are crucial for cocoa pulp fermentations and for development of cocoa flavors and fermentation efficiency (17, 18), and the observed diversity results in cocoa beans of inconsistent quality and causes huge economic losses (19–21).
Since the introduction of commercial fermentation starter cultures for the food industry, Saccharomyces cerevisiae (or a close relative) has often been the organism of choice. This can be explained by several interesting physiological features of Saccharomyces spp. that make them very suitable for industrial fermentations (22). First, S. cerevisiae is able to outcompete most other yeasts in industrial fermentations of wine (23), beer (24), cider (25), and also cocoa pulp (7). This fitness advantage can be attributed to several characteristics, including high stress tolerance (e.g., against ethanol and temperature), fast and efficient carbon metabolism (e.g., high glycolytic flux and glucose repression), and the ability to grow under aerobic and anaerobic conditions. Interestingly, these features are widespread among several yeast species but are uniquely combined in S. cerevisiae (22, 26). Second, S. cerevisiae yeasts produce many desirable flavor compounds, such as volatile esters and higher alcohols, and only a few off flavors (13). Lastly, S. cerevisiae cells do not produce specific toxins that would be harmful to humans, ensuring their safe use in food fermentations, as illustrated by the generally recognized as safe (GRAS) and qualified presumption of safety (QPS) status of S. cerevisiae (27, 28).
Given what is known about cocoa pulp fermentations, it does not come as a surprise that the use of starter cultures consisting of S. cerevisiae, LAB, and AAB has been suggested to increase the consistency and overall quality of chocolate. In 1998, a complex mixture of an indigenous S. cerevisiae strain; two LAB species, Lactobacillus lactis and L. plantarum; and two AAB species, Acetobacter aceti and Gluconobacter oxydans, was proposed (19). The added inoculum could mimic natural cocoa pulp fermentations and yielded cocoa of acceptable, but not superior, quality. In 2012, a starter culture with a similar composition, consisting of indigenous S. cerevisiae, L. fermentum, and A. pasteurianus strains, was applied (18). This study also highlighted the importance of yeast in a cocoa pulp starter culture, since inoculating with a starter culture consisting solely of LAB and AAB resulted in chocolate with a bitter, unfermented flavor. Despite the production of chocolate with a consistent flavor, qualitative advantages seemed limited, and the starter culture composition was too complex for efficient commercial application. Another paper reported on inoculation with L. fermentum, A. pasteurianus, and a non-Saccharomyces species, either Pichia kluyveri or Kluyveromyces marxianus (29). However, while pronounced differences from spontaneous fermentations were observed by gas chromatography-mass spectrometry (GC-MS) analysis, sensory analysis by a consumer panel revealed no significant differences, probably due to the inability of the inoculated yeast strains to dominate in the fermentation process (30). Finally, several papers describe the application of high-pectinolytic (often genetically modified) yeast starter cultures to increase liquid drainage (31–33). While these inoculated fermentations sometimes led to increased drainage, they did not result in clear benefits for cocoa quality. Moreover, legislation and consumer preferences currently do not allow genetically modified organisms to be used in an industrial setting.
In this study, new hybrid yeast strains able to perform an efficient and consistent inoculated cocoa pulp fermentation process were developed. First, two S. cerevisiae strains (one indigenous cocoa and one industrial strain), both performing well in a cocoa pulp environment, were selected as parental strains. Breeding of these strains ultimately yielded a large set of in- and outbred hybrids, some of which showed increased competitiveness in cocoa pulp. This resulted in the consistent production of high-quality cocoa liquor and chocolate, which makes them excellent candidates for use as a single-species starter culture.
MATERIALS AND METHODS
Yeast strains and storage conditions.
The most important strains used in this study are listed in Table 1. A full description of the industrial yeast strain collection was provided previously (34) and is summarized in Table S1 in the supplemental material. All strains were stored at −80°C using a glycerol-based storage medium (25% [wt/vol] glycerol, 2% [wt/vol] Bacto peptone, 1% [wt/vol] yeast extract, 2% [wt/vol] glucose).
TABLE 1.
Overview of the yeast strains used in this study
| Strain code | Strain genotype | Strain origin |
|---|---|---|
| Natural strains | ||
| Y115 | Wild type | This study |
| Y397 | Wild type | This study |
| Y927 | Wild type | 8 |
| Hybrids selected for pilot-scale fermentations | ||
| H19 | Hybrid (inbred Y927) | This study |
| H28 | Hybrid (outbred Y115 × Y927) | This study |
| H37 | Hybrid (inbred H28) | This study |
| Competition experiments | ||
| Y115-Hyg | Contains hygromycin B resistance marker integrated in Chr.XV:39657.0.39803 | This study |
| H28-Hyg | Contains hygromycin B resistance marker integrated in Chr.XV:39657.0.39803 | This study |
| Screening reference strains | ||
| EX436 (Klus) | Wild type | 63 |
| L2323 | Wild type | 40 |
| NCYC 441 (K28) | Wild type | NCYC 441 |
| NCYC 1001 (K2) | Wild type | 38 |
| NCYC 1407 (K1) | Wild type | NCYC 1407 |
| Y173 (killer sensitive) | Wild type | This study |
| Y481 (killer sensitive) | Wild type | This study |
| W303 | Wild type | 36 |
Temperature tolerance.
Screening for temperature tolerance was performed by plate assays. Strains were pregrown in 150 μl 2% YPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto peptone, 2% [wt/vol] glucose) for 16 h (30°C; 900 rpm). Next, the yeast strains were spotted at an initial optical density at 600 nm (OD600) of 0.1 on solid YPD agar (2% [wt/vol] agar) using a high-density-array robot (Singer Rotor HAD; Singer Instruments, United Kingdom). The plates were subsequently incubated at 30, 37, and 39°C for 48 h and 40, 41, 41.5, and 42°C for 96 h. The colony size was quantified using ScreenMill software (35) and ImageJ (NIH). To score the temperature tolerance of the strains (−, +, ++, and +++), the colony size at 40°C was normalized to W303, a widely used model organism known to be temperature tolerant (36); ++ indicates a colony size comparable to that of W303, whereas − indicates that no growth was observed.
Killer activity.
Killer activity (see below) was measured by the seeded-agar method, slightly adjusted from that of Woods and Bevan (37). In short, a killer-sensitive strain (lacking killer activity) (Table 1) was seeded at a concentration of 104 CFU ml−1 in YPD agar, supplemented with 0.003% methylene blue with the pH adjusted to 4.5. Next, 5 μl of the concentrated yeast cultures (approximately 109 CFU ml−1), including six reference strains (four with known killer activity and two that are known to be killer sensitive) (Table 1), was spotted on the agar plates. The plates were incubated at 23°C for 96 h. The killer activity was scored in a semiquantitative manner (−, +, ++, or +++) relative to the reference strain, NCYC 1001 (killer type K2 [38]), with ++ indicating a halo size comparable to that of NCYC 1001 and − indicating no killer activity.
Polygalacturonase activity.
Strains were screened for polygalacturonase activity (a trait that allows them to degrade pectin) using a semiquantitative plate assay described previously (39). Activity was quantified in a standardized fashion using ImageJ (NIH) and normalized to L2323, a polygalacturonase-positive reference strain (40) present on every plate to correct for plate variability.
Sporulation, tetrad dissection, and mating type characterization.
Yeast strains were sporulated, followed by tetrad dissection, to determine the hetero- or homothallic nature of the strains (34), as described previously (41). The mating type was determined by PCR executed in a C1000 thermal cycler (Bio-Rad), using MAT-A (5′-ACTCCACTTCAAGTAAGAGTT-3′), MAT-α (5′-GCACGGAATATGGGACTACTTCG-3′), and MAT-R (5′-AGTCACATCAAGATCGTTTATGG-3′) as primers and a temperature profile consisting of 30 cycles of 45 s at 94°C, 45 s at 50°C, 40 s at 72°C, and a final extension of 5 min at 72°C (42).
Genetic fingerprinting.
Genetic fingerprinting was applied to reveal genetic diversity within S. cerevisiae and to confirm the hybrid genotype of newly formed hybrids. First, genomic DNA was extracted in a 96-well format using the standard ether extraction protocol, as described previously (43), or using colony extraction with 0.02 M NaOH and a boiling step (10 min; 100°C). When the latter was used, e.g., for determining the intraspecific S. cerevisiae diversity in spontaneous fermentations, the ether extraction protocol was used for confirmation. Next, a transposon-based amplified fragment length polymorphism (AFLP) approach, called interdelta analysis, was used (44). PCRs were executed with primers delta12 (5′-TCAACAATGGAATCCCAAC-3′) and delta21 (5′-CATCTTAACACCGTATATGA-3′) and a temperature profile described previously (44). The PCR products were visualized using QIAxcel Advanced Systems (Qiagen, Netherlands).
Mass mating.
Strains selected for the mass mating procedure were sporulated on acetate medium (1% [wt/vol] potassium acetate, 2% [wt/vol] agar) and incubated at 25°C. After 5 to 10 days, all spores were harvested, and the remaining vegetative cells were lysed by overnight incubation at 35°C (80 rpm) in distilled water (dH2O) supplemented with zymolyase (100 mg liter−1; Seikagaku Corporation, Japan), chloramphenicol (50 mg liter−1), and 2-mercaptoethanol (2 ml liter−1). Next, the remaining vegetative cells were lysed by adding sterile glass beads (0.425 to 0.600 mm) and vigorous vortexing for 3 min. The remaining cells were resuspended in dH2O supplemented with 0.75% Triton X-100. To break the asci down into single spores, the cultures were cooled to 4°C and sonicated for 30 s (amplitude, 50%) in a digital sonifier (Branson, Connecticut, USA), repeated three times in total. Next, the spore suspension was washed twice with 1.5% Triton X-100, after which it was cooled and sonicated (20 s; amplitude, 50%) two more times. The spores were washed with dH2O and resuspended in 1 ml 1× phosphate buffer solution. To initiate mating, spore suspensions of both parental strains were mixed in 50 ml GNA (3% [wt/vol] Bacto peptone, 1% [wt/vol] yeast extract, and 5% [wt/vol] glucose) at a starting density of 106 spores ml−1 and incubated at 30°C overnight (80 rpm). In a final step, high-temperature-tolerant hybrids were identified by plating approximately 103 cells on YPD agar and subsequent incubation at 42°C for 5 days. The genotypes of the strains able to grow under these conditions were determined by mating type PCR and interdelta analysis (see above).
Backcrossing.
Sporulation was induced on acetate medium incubated at 25°C (see above). The ascus wall was digested with 2 mg ml−1 zymolyase (Seikagaku Corporation, Japan) suspension and incubated for 3 to 10 min at room temperature. Tetrads were dissected with a micromanipulator (Singer SMS Manual; Singer Instruments, United Kingdom) on YPD agar. Temperature-tolerant haploid segregants of both parental strains were first selected by screening individual segregants by spot assays at 42°C or, in cases where there was no growth at 42°C, at 41°C and 40°C (see “Temperature tolerance” above). Next, two segregants of opposite mating types were picked, mixed on YPD agar with 10 μl dH2O, and incubated at room temperature for 24 h. Then, a small fraction of the spot was streaked to single colonies and incubated for 48 h. Four single colonies from each spot were subsequently checked for mating type (see above) to identify backcrossed hybrids.
Transformation of yeast for competition experiments.
Antibiotic-resistant strains were developed for the competition experiments. A hygromycin B resistance cassette was amplified from a plasmid analogous to pUG6 (45) but containing Hph instead of KanMX, conferring resistance to hygromycin B. IntE_F (5′-ATATAATAACGAAATTGAGTTTTCTATAAGTAACATCAGCCCAGCTTGCCTTGTCCCCG-3′) and IntE_R (5′-AGGTAAGACACTGGAGCAAAAGAGTATAGTATTCTATAGAACTCGACACTGGATGGCGG-3′) were used to target Chr.XV:39657.0.39803, an intergenic region conserved between strains that was shown not to alter the yeast's phenotype when disrupted (J. F. Christiaens, T. Snoek, and K. J. Verstrepen, unpublished data). To amplify the cassettes, a temperature profile consisting of an initial denaturation of 3 min at 95°C; 9 cycles of 20 s at 95°C, 30 s at 55°C, and 2 min 30 s at 72°C; 25 cycles of 20 s at 95°C, 30 s at 58°C, and 2 min 30 s at 72°C; and a final elongation phase of 10 min at 72°C was used. A standard transformation protocol for S. cerevisiae was used (46). To assess the potential influence of the selection marker on the yeast's fitness at high temperatures, the growth rate of the transformed strain was compared with that of the nontransformed strain using Bioscreen C (Oy Growth Curves AB, Finland). Six biological replicates were used per strain, and cells were inoculated in 12% YPD at an OD600 of 0.002 and incubated at 37°C (24 h) and 41°C (48 h).
Competition experiments in a cocoa pulp environment.
Yeast propagation was performed in two propagation stages: first, an overnight pregrowth (30°C with shaking) in 5 ml 2% YPD, after which 500 μl of this culture was transferred to 50 ml 2% YPD and incubated overnight at 30°C (200 rpm). This culture was used to inoculate the fermentations. Competition experiments were performed in 250 ml Schott bottles using 100 ml cocoa pulp (shipped from Ivory Coast), which was pasteurized for 5 min at 105°C. Two strains (one transformed with a hygromycin B resistance cassette) were simultaneously inoculated into the fermentations at an OD600 of 0.25. After inoculation, the fermentation bottles were vigorously mixed to ensure homogeneous distribution in the fermentation medium. At this point, initial samples were taken and plated out on YPD agar and YPD-plus-hygromycin B (200 mg liter−1) agar to determine the relative population size of each strain. After 48 h, the competition experiment was ended, and final samples were taken and plated out on YPD agar and YPD-plus-hygromycin B (200 mg liter−1) agar. The change in the population ratio was calculated as follows: ln [(cell count test strain)(cell count transformed reference strain)−1]final − ln [(cell count test strain)(cell count transformed reference strain)−1]initial. Analysis of variance (ANOVA), combined with the Tukey or Dunnett test to correct for multiple testing, was used to determine significant differences in dominance between strains.
Pilot scale cocoa pulp fermentations.
Pilot scale cocoa pulp fermentations were performed during the main harvest season at the Barry Callebaut cocoa research facility, Pahang, Malaysia, in October and November 2013. Two spontaneous reference fermentations were also performed. The fermentations were started as described previously (8). Cocoa pods were harvested and opened after 3 days. Fermentations were performed in baskets (0.70 m by 0.50 m by 0.60 m) containing 50 kg of beans and covered with banana leaves. All fermentations were turned once after 48 h and stopped after 4 days. The temperature and pH in the middle of the fermenting mass were continuously measured using a digital pH meter (pH 3310 set 2; SenTix 41; WTW GmbH, Weilheim, Germany). Cocoa bean samples (200 g) from the fermentations were aseptically taken after 0 (fresh cocoa beans right after opening the pods; only for the spontaneous reference fermentations), 4, 24, 48, 72, and 96 h, each time at the same depth in the fermenting bean mass (20 cm below the surface). Next, the samples were cooled to 4°C and further analyzed within 1 h, as described previously (8).
Microbial isolation and identification during pilot scale field trials.
For microbiological analysis, six LAB, three AAB, and 16 random yeast isolates were picked at all time points in all fermentations. A total of 382 LAB, 144 AAB, and 757 yeasts were isolated. These isolates were further identified to the species level by sequencing and fingerprinting, as described previously (8). All yeast isolates were identified by 26S rDNA sequencing (47) and comparing all sequences to type strain sequences using nucleotide-nucleotide BLAST (blastn). Species were identified based on the highest identity (percent) of the BLAST output. In cases of <100% identity, percentages of identity between the sequenced isolates and the available type strains are reported. In the case of ambiguities concerning the identity based on 26S rDNA sequencing, ACT1 and internal transcribed spacer (ITS) regions were additionally amplified and compared to type strain sequences for the isolates. Available type strain sequences were complemented with the ACT1 sequence of Pichia manshurica H4S7K13 (FM199999) (11) and the ITS sequence of Hanseniaspora opuntiae CBS8820 (AJ512440) (48). For higher resolution and identification to strain level, all S. cerevisiae strains isolated from inoculated cocoa pulp fermentations were genetically fingerprinted using interdelta analysis. Bacterial isolates were first genetically fingerprinted [using (GTG)5 repetitive-element sequence-based PCR (rep-PCR)] and clustered, after which the 16S rDNA of several representatives of each observed cluster was sequenced. Similar to the yeast isolates, all bacterial sequences were compared to type strain sequences, and percentages of identity are reported when they were <100%.
Starter culture propagation for pilot scale field trials.
Yeast propagation was performed in two propagation stages; first, an overnight pregrowth (30°C with shaking) in 5 ml 2% YPD, after which the 5-ml culture was transferred to 300 ml 10% YPD and incubated overnight at 30°C (180 rpm). This culture was used to inoculate the fermentations at an initial density of 106 CFU g−1. The inoculum was prepared by vacuum filtration over a 0.45-μm filter (Millipore) and dilution of the cells in 0.85% NaCl. The inoculum size was verified by cell counting. Starter cultures were inoculated by pouring them onto the fermentation, and they were evenly distributed throughout the fermenting mass by manual mixing.
Chocolate production and sensory analysis.
Fermented cocoa beans were used for cocoa liquor and chocolate production by Barry Callebaut (Wieze, Belgium). Cocoa beans were roasted using medium roasting at 122°C, according to the general practices used for commercial production. Liquor from each fermentation batch was sampled and stored for GC-MS analysis (see below). Next, cocoa liquors from the fermentation duplicates were pooled (in order to obtain sufficient mass for chocolate production), refined, supplemented with cocoa butter, and conched for 4 h. Chocolate with 60% (wt/wt) cocoa solids was made for sensory analysis. Paired preference tests were performed by a consumer panel consisting of 16 people. Additionally, the panelists were asked to describe the samples (49, 50).
GC-MS analysis of cocoa liquor.
Volatiles of the cocoa liquor were extracted by liquefying 5.0 g of the samples at 50°C. The sample was spiked with 4 μl of a 4-heptanone internal-standard stock solution (0.32296 μg μl−1). After stirring, 100 μg was weighed in duplicate in a microvial and transferred to an empty thermal desorption unit (TDU) tube, which was closed. The volatiles were analyzed using GC-MS, consisting of a gas chromatograph (GC 7890A; Agilent Technologies, California, USA) in combination with a mass selective detector (MSD 5975C; Agilent Technologies) equipped with a MultiPurpose Sampler (MPS 2; Gerstel, Germany), a TDU (Gerstel), and a programmed temperature vaporizer (PTV) inlet (CIS4; Gerstel). Volatile compounds were thermally desorbed with the TDU temperature set at 70°C for 30 min. The desorbed volatile compounds were trapped at −10°C in the PTV inlet, after which the inlet was heated to 280°C (at a rate of 12°C min−1, held for 20 min). Volatile compounds were separated using a fused silica capillary column (HP-FFAP; 50 m by 0.32 mm by 0.50 μm; Agilent Technologies) preceded by a fused silica precolumn (2 m by 0.53 mm). The GC oven temperature was set at 35°C (held for 5 min) and increased to 240°C (at 5°C min−1, held for 4 min). Helium was used as a carrier gas with a flow rate of 2 ml min−1. Mass-to-charge ratios were scanned between 40 and 300 m/z, with the MSD operating at 70 eV in electron ionization mode. Volatile compounds were identified by comparing the mass spectra of the sample compounds with those in the Wiley 275L database. Compounds were semiquantified by comparing the peak area to the peak area of the 4-heptanone internal standard. Duplicate analyses of each sample were performed. Data were converted to Z-scores and visualized using principal-component analysis (PCA), performed with BioNumerics version 7.1 software (Applied Maths, Belgium).
Nucleotide sequence accession numbers.
Sequencing data for representative isolates have been deposited in GenBank (accession numbers KP190150, KP190151, KP190153 to KP190159, KP190161, KP190164 to KP190168, KP190170, KP190171, and KP190174).
RESULTS
This study aimed to develop a yeast starter culture that allows increasing the efficiency and/or consistency of cocoa pulp fermentations. Specifically, we focused on S. cerevisiae, because the species has been isolated from most spontaneous cocoa pulp fermentations and because it is directly applicable in food fermentations. Using sexual hybridization, we developed robust yeast hybrids that thrive in a cocoa pulp environment and are able to consistently produce superior high-quality chocolate.
Selection of a superior S. cerevisiae strain from spontaneous cocoa pulp fermentations.
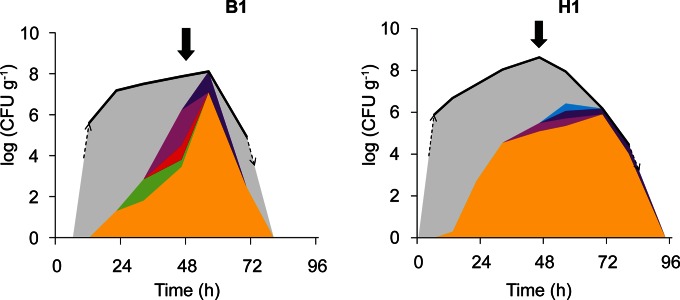
To evaluate the diversity of S. cerevisiae in spontaneous cocoa pulp fermentations and isolate dominant strains that could be candidates for application in starter cultures, we analyzed the indigenous S. cerevisiae diversity of two spontaneous fermentations (Fig. 1; see Fig. S1 in the supplemental material for details). Whereas S. cerevisiae has been isolated from various spontaneous cocoa pulp fermentations, the diversity within the species is only very rarely documented (7). Our analyses revealed the presence of six groups of closely related S. cerevisiae strains, with one group dominating both fermentations (Fig. 1). A single representative strain of the dominant group (Y927) was selected, and phenotyping revealed high temperature tolerance, killer activity (i.e., the ability to produce short peptides that inhibit the growth of other yeasts that do not show killer activity [51]), moderate polygalacturonase activity (i.e., the ability to degrade the pectin in the cocoa pulp), and a homothallic life cycle with efficient spore production (as determined by tetrad dissection and mating type PCR). Y927 was therefore selected as a potential starter culture and as a parental strain for further breeding experiments.
FIG 1.
Intraspecific diversity of the S. cerevisiae populations in two spontaneous Malaysian cocoa pulp fermentations as determined by interdelta analysis. Box fermentation 1 (B1) (October 2011) and heap fermentation 1 (H1) (October 2011) are described in reference 8. The total yeast count is represented by a thick black line. The dashed arrows indicate when the yeast cell counts rise to or drop below the detection limit. The solid arrows indicate turning. Clusters consisting of the same (or very closely related) strain(s) are represented using a color code (see Fig. S1 in the supplemental material). All non-S. cerevisiae strains are displayed in gray. Note that the scale for the total yeast count (y axes) is logarithmic, whereas individual S. cerevisiae strains and the non-S. cerevisiae moiety in the population at a given time point (indicated by the colors) are presented as the relative fraction of the total population on a linear scale.
Selection of a superior industrial strain from an in-house yeast collection.
Apart from indigenous yeast strains, strains currently applied to industrial fermentation processes (e.g., beer, wine, sake, or bioethanol fermentation) also provide an interesting source of potential cocoa starter cultures. After phenotypic evaluation of a large collection of industrial yeasts (318 strains), one strain (Y115, originating from the bioethanol industry) possessing very high thermotolerance and moderate polygalacturonase activity was selected as a potential starter culture. Indeed, a pairwise competition experiment between Y115 and the previously selected indigenous strain, Y927, revealed superiority of Y115 in a cocoa pulp environment (see below). Moreover, the strain showed efficient sporulation with stable haploid segregant production and could therefore be used as a parental strain in the breeding experiments.
Development of new hybrid strains with improved temperature tolerance by mass mating.
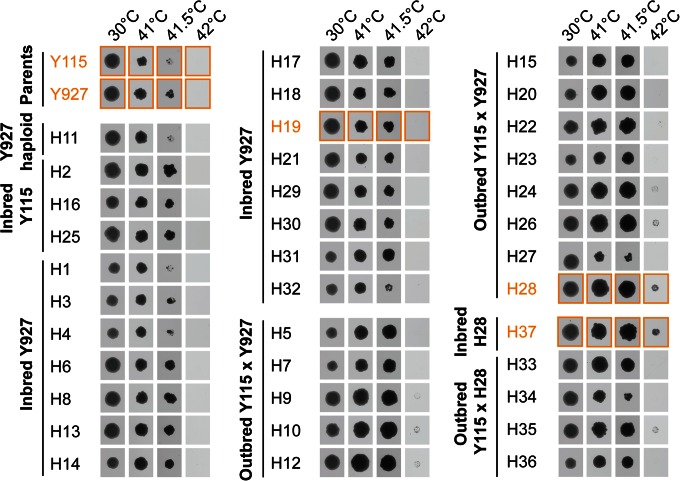
To generate superior strains that combine the characteristics of the dominant indigenous strain (Y927) and the industrial bioethanol strain (Y115), we set up a series of mass mating assays to generate a large set of hybrid yeasts. Following the mass mating procedure, the heterogeneous pool of newly formed hybrids was subjected to a 5-day incubation at high temperature (42°C) to select for hybrids with high temperature tolerance, a trait vital for dominance in a cocoa pulp environment. After this selection, 32 hybrids (H1 to H32) were selected and genetically fingerprinted. Diploids resulting from mating two cells of opposite mating types from the same parent, either germinating spores or haploid segregants, were classified as inbred hybrids. This resulted in the identification of 13 outbred hybrids, 3 Y115 inbred strains, 15 Y927 inbred hybrids, and 1 Y927 haploid (see Fig. S2 in the supplemental material). These strains were further tested for thermotolerance (Fig. 2) and killer activity (see Table S2 in the supplemental material) and compared to their parental strains. Based on the results of these tests, two hybrids were selected for further analysis, the outbred H28 and the Y927 inbred H19. H28 was selected because the strain showed the highest temperature tolerance (defined as the best growth at 42°C), and H19 was selected because it showed both high temperature tolerance and killer activity.
FIG 2.
Temperature tolerance tests of parental strains and hybrids. Strains were tested in duplicate, only one representative of which is shown. The parental strains Y115 and Y927 are displayed, together with H1 to H32 (first-generation hybrids resulting from mass mating of Y115 and Y927) and H33 to H37 (second-generation hybrids resulting from backcrossing H28 to its parent, Y115). Strains selected for laboratory and pilot scale fermentations are indicated in orange.
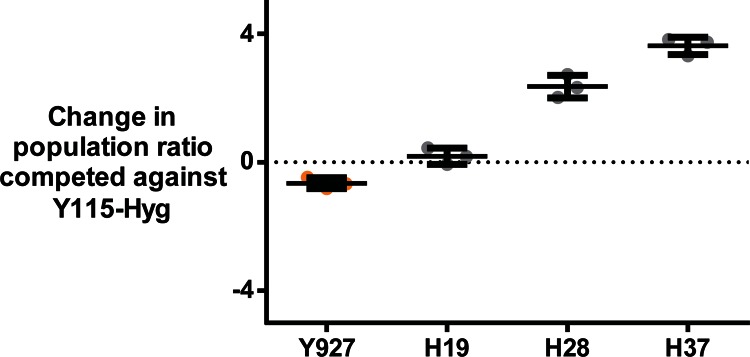
In order to obtain a better comparison of the competitive behavior of the newly formed hybrids and their respective parental strains in a cocoa pulp environment, the two selected hybrids (H19 and H28) and the parental strain Y927 were competed separately against Y115 in laboratory scale cocoa pulp fermentations. To quickly determine population ratios, Y115 was genetically transformed with a hygromycin B resistance marker cassette (no influence of the marker on the yeast's fitness was detected [see Fig. S3 in the supplemental material]). The competition experiments revealed that H28 showed the highest fitness, outcompeting its stronger parent, Y115, in cocoa pulp (P < 0.0001) (Fig. 3; see Table S3 in the supplemental material). Additionally, the comparison between the fitness scores of inbred H19 and the parental strain Y927 revealed significantly higher fitness of H19 (P = 0.0057; Dunnett's multiple-comparison test). Hybrids H19 and H28 were therefore selected for pilot scale cocoa pulp fermentation trials in Malaysia.
FIG 3.
Laboratory scale competition experiments in cocoa pulp. Selected strains and hybrids were competed in 100 ml cocoa pulp against a hygromycin (Hyg)-tagged transformant of the reference strain Y115. The error bars represent the 95% confidence intervals, calculated based on three biological replicates. The horizontal lines are means. See Materials and Methods for details.
Inbreeding of H28 further improves its fitness in laboratory scale cocoa pulp fermentations.
Since H28 showed the most pronounced phenotypic improvement of all hybrids compared to the parental strains, the strain was selected for further inbreeding and backcrossing to its stronger parent, Y115. As stable haploid segregants could be obtained for both parents, a total of 92 haploid segregants were screened for thermotolerance prior to the hybridization experiment, thereby increasing the chance of obtaining superior hybrids. The eight most temperature-tolerant haploids were subsequently crossed in eight different combinations, yielding four outbred hybrids between H28 and Y115 (H33 to H36) and one H28 inbred hybrid (H37). Screening for thermotolerance of these five second-generation hybrids revealed that the inbred H37 showed the highest temperature tolerance, outperforming both H28 and Y115 in spot assays. Additionally, competition experiments in cocoa pulp revealed that H37 showed the highest fitness score compared to Y115-Hyg (a hygromycin [Hyg]-tagged transformant of the reference strain Y115) of all tested strains, including H19 and H28 (P < 0.0001; Dunnett's multiple-comparison test) (Fig. 3). This was confirmed by competing a hygromycin B-transformed H28 directly with H37 (P = 0.0017; see Fig. S4 in the supplemental material). H37 was therefore also selected for further testing in inoculated pilot scale cocoa pulp fermentations.
Inoculated pilot scale fermentations reveal suitable yeast starter cultures.
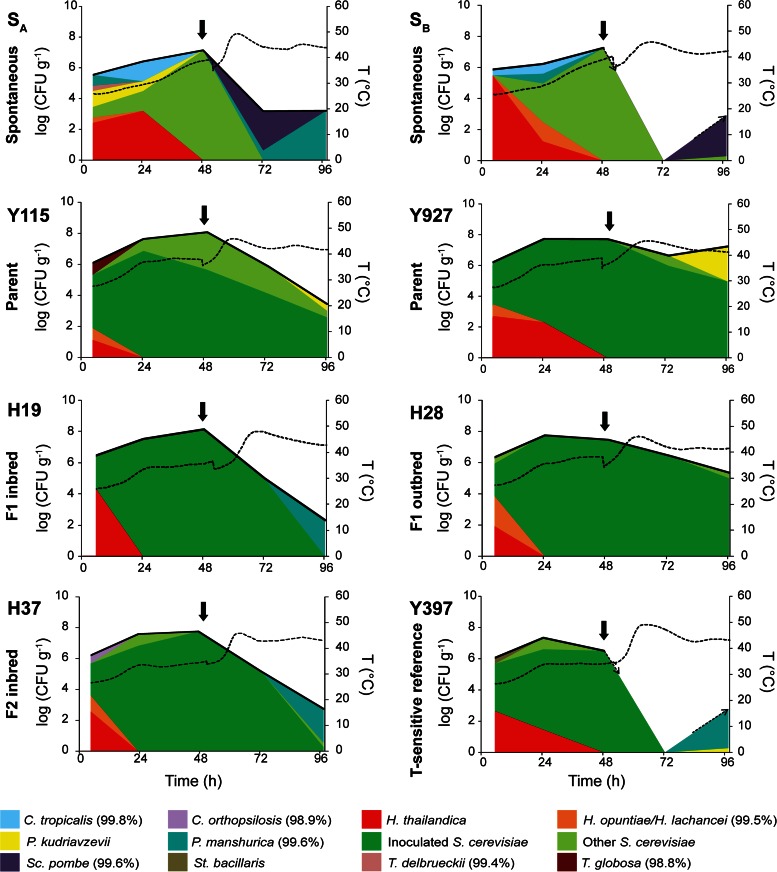
To evaluate the performance of the selected indigenous and industrial strains and the three selected hybrids under industrial conditions, pilot scale field experiments were performed at a Malaysian cocoa farm. Additionally, one temperature-sensitive reference strain (Y397) was included in the experiment (see Table S4 in the supplemental material). All experiments were performed in duplicate (labeled A and B), and temperature and pH were measured in real time (Fig. 4; see Table S5 in the supplemental material). Since the goal of these pilot scale experiments was to develop a simple and economically viable starter culture for commercial production, no bacterial culture was added. Importantly, the different yeasts were inoculated to a relatively low starting concentration (106 CFU g−1, i.e., about 10% of the normal inoculum size for beer fermentations), because higher inoculation numbers would be prohibitively expensive for large-scale commercial use.
FIG 4.
Yeast population dynamics during spontaneous and inoculated cocoa pulp pilot scale fermentations. Two spontaneous pilot scale fermentations (SA and SB) and 12 inoculated fermentations (six strains, performed in duplicate; A replicates are shown here, while B replicates are shown in Fig. S5 in the supplemental material) were monitored (see Materials and Methods for details). Temperature (right y axes) is indicated as a dashed black line. The dashed arrows indicate when the yeast cell count rises to or drops below the detection limit. Note that the scale for the total yeast count (left y axes) is logarithmic, whereas individual yeast species in the population at a given time point (indicated by the colors) are presented as the relative fraction of the total population on a linear scale. Absolute concentrations for individual species are given in Table S7 in the supplemental material. Percentages of identity are reported for <100% identity with type strain sequences (see Materials and Methods for details). Turning of the fermenting cocoa beans was carried out after 48 h of fermentation (solid vertical arrows). St., Starmerella; T., Torulaspora.
To assess the microbial diversity of the different pilot scale fermentations, samples were taken throughout the fermentation period. No significant differences in the LAB and AAB cell counts were observed between spontaneous and inoculated fermentations, except for AAB at 4 h and 24 h (P = 0.0448 and P = 0.0009, respectively). However, this was not linked to higher maximal or final temperatures or the time points at which the highest temperatures were reached in the inoculated fermentations. Moreover, even though no bacterial inoculum was added, only a few different LAB and AAB species were present in the fermentations. In fact, the bacterial microbiomes were very similar between different fermentations and also closely resembled the profiles found in spontaneous fermentations (see Table S6 in the supplemental material), suggesting that a bacterial starter culture is not vital to obtain a consistent microbiome.
In contrast, the yeast diversity in the inoculated fermentations was significantly reduced compared to the spontaneous fermentations (Fig. 4; see Fig. S5 in the supplemental material). In the spontaneous fermentations, six to eight different species were found per fermentation, with S. cerevisiae, Hanseniaspora thailandica, Hanseniaspora opuntiae/H. lachancei, Pichia manshurica, Candida tropicalis, and Schizosaccharomyces pombe occurring in both spontaneous fermentations. This number was significantly higher than that in the inoculated fermentations, where the numbers of different species per fermentation ranged from two to seven (P = 0.0385; Mann-Whitney-Wilcoxon test). The largest diversity in both spontaneous and inoculated fermentations was observed at the start. In all inoculated fermentations (except for Y397B), H. thailandica was present at the outset, with a relative population size between 13% and 87%. H. opuntiae/H. lachancei was also often reported at early stages (see Table S7 in the supplemental material). However, in contrast to the spontaneous fermentations, S. cerevisiae quickly became dominant in all inoculated fermentations, with relative population sizes ranging from 70% to 100% after 24 h compared to 20% to 40% in spontaneous fermentations.
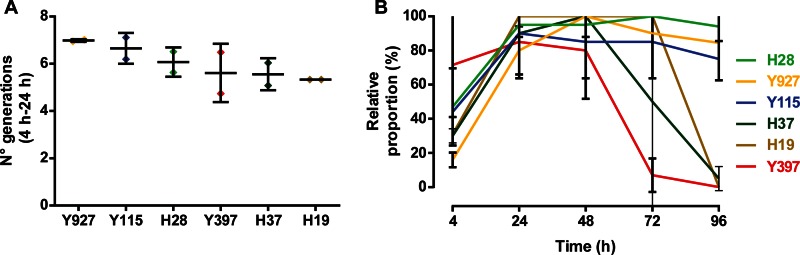
To determine which inoculated yeast strains effectively outcompeted wild contaminants, we analyzed the growth speeds of the inoculated strains during the first 24 h of the fermentation and their dominance (relative population size) during the course of the fermentation (Fig. 5). During the initial stages of the inoculated fermentations, some strains seemed to be better adapted to the initial fermentation environment than others, since they showed faster growth in the first 24 h. Interestingly, the best-performing strains in the initial, sugar-rich environment were the indigenous strain Y927 and the industrial bioethanol strain Y115, not the developed hybrids. However, all yeast strains reached an average relative population size between 80% (Y927) and 100% (H19) during the first 24 h. Over the next 24 h, the proportion of inoculated yeasts present in the pulp decreased slightly (Y115 and Y397), remained stable (H28 and H19), or increased to 100% (Y927 and H37). In the third stage of fermentation (between 48 h and 72 h), the average relative population sizes of all strains decreased, except for Y115 and H19 (stable at 85% and 100%, respectively) and H28 (increased to 100%). This third period was characterized by an increase in temperature due to turning of the fermenting beans. This rise in temperature may explain why the relative proportions of many yeast strains, and especially the temperature-sensitive Y397, decreased drastically. Interestingly, however, the temperature-tolerant strains Y115, Y927, and H28 remained present throughout the whole fermentation. H28 was shown to perform the best, dominating the yeast population for the complete course of the fermentation. Although no statistical differences were found between the dominance of H28, Y115, and Y927 at 96 h, they performed significantly better than all other tested strains (P ≤ 0.01). The performance of the yeast strains in early stages was not correlated with yeast growth in later stages of the fermentation process (Spearman's rank correlation coefficient [r] at 72 h = 0.00, P = 0.9833; at 96 h, r = 0.72, P = 0.1222) or with the temperature tolerance (r = 0.03, P > 0.9999). However, the temperature tolerance of the inoculated strains seemed to be an important predictor of growth at later stages, although not statistically significant (at 72 h, r = 0.52, P = 0.3000; at 96 h, r = 0.67, P = 0.1611).
FIG 5.
Analysis of the performance of inoculated yeast strains during pilot scale cocoa pulp fermentations. The relative population sizes were calculated based on a representative number of isolates taken at every time point (see Table S7 in the supplemental material for numerical values; see Materials and Methods for details). (A) Number of generations during the first 24 h of the fermentation, displayed as means ± standard deviations (SD). (B) Relative population sizes of the different inoculated strains during the fermentation. The values are represented as means ± SD.
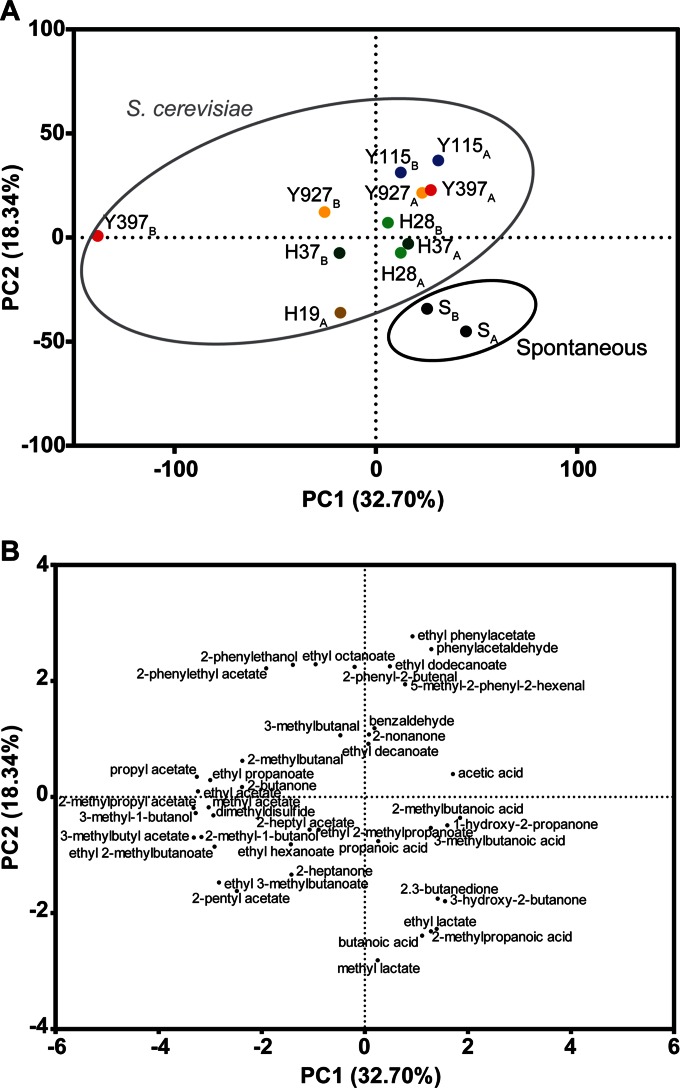
GC-MS analysis of cocoa liquor confirms the consistency of inoculated fermentations.
After fermentation, the beans were dried and further processed to cocoa liquor. GC-MS analysis of this liquor successfully identified 41 flavor-active compounds related to fermentation. To compare the flavor profiles of the fermentations and determine the reproducibility of duplicates, a two-dimensional PCA plot was constructed (Fig. 6). This analysis confirmed close grouping of liquor samples made from most of the duplicate fermentations, except for the temperature-sensitive strain Y397. Further, it revealed a clear separation between the aromatic profiles of the spontaneous and inoculated fermentations. The two spontaneous fermentations clustered together in the lower right part of the plot. This corresponded to the majority of volatile acids and lactic acid-related volatiles grouping together in the same quadrant (Fig. 6). Almost all ethyl and acetate esters were located on the negative axis of principal component 1 (PC1), indicating that these compounds were found to a lesser extent in the spontaneous than in the inoculated fermentations. Liquor produced from spontaneous fermentations contained significantly more 2.3-butanedione (diacetyl), 3-hydroxy-2-butanone (acetoin), ethyl lactate, 2-methylpropanoic acid, and butanoic acid than the inoculated fermentations (Wilk's lambda likelihood ratio test; P = 0.010). These compounds are mostly described as rancid (2-methylpropanoic acid and butanoic acid) and buttery (2.3-butanedione, butanoic acid, and 3-hydroxy-2-butanone) (52, 53).
FIG 6.
Principal-component analysis of volatiles in cocoa liquor produced from spontaneous and inoculated fermentations. (A) Score plot of two spontaneous fermentations (SA and SB) and fermentations inoculated with six S. cerevisiae strains (tested in duplicate, A and B). For hybrid H19, only one replicate was available. The values represent duplicate technical GC-MS analyses. (B) Corresponding loading plot of 41 compounds related to cocoa pulp fermentation identified using GC-MS.
Sensory analysis of chocolate confirms potential of yeast starter cultures.
Chocolates made from both spontaneous and inoculated fermentations were tasted by a consumer panel to determine the influence of adding the yeast starter culture (Table 2 shows all the comparisons). The tasting panel significantly preferred chocolates from fermentations inoculated with yeast hybrids over the identically executed spontaneous fermentations. The spontaneous reference strain was described as more bitter and sour, with a roasted flavor, while the inoculated fermentations were all described as more intense (Table 2). The consumer panel further mostly preferred the fermentations inoculated with the hybrids over the parental strains, with the strongest preference for H28.
TABLE 2.
Comparison by a consumer panel of chocolates made from inoculated pilot-scale cocoa pulp fermentationsa
| Strain | Consumer preferenceb |
||
|---|---|---|---|
| H19 (intense, strong, with sweet, fruity and sour hints) | H28 (intense, full, sweet, soft, smooth, nice aftertaste) | H37 (very aromatic, sweet, milky, on the edge of too sweet) | |
| Y115 | NA | 5/11 | 9/7 |
| Y927 | 8/8 | 3/13c | 6/10 |
| Spontaneous | 3/13c | 1/15c | 4/12c |
Sixteen panelists were asked to compare the spontaneous reference and parental strains (Y115 and Y927) with the hybrids (H19, H28, and H37). H19 is a Y927 inbred hybrid, H28 an outbred hybrid between Y115 and Y927, and H37 is an H28 inbred hybrid.
The number of panelists who preferred chocolate made with the parental strain is indicated on the left in each pair. NA, not applicable.
P ≤ 0.05. P values were calculated using the binomial test (probability = 0.50).
DISCUSSION
Our study demonstrates the use of selective breeding to generate robust yeast hybrids that are usable as starter cultures for cocoa pulp fermentations. The development of such starter cultures is particularly challenging, because cocoa pulp fermentations rely on a complex mixture of yeasts, LAB, and AAB and because cocoa pulp is not fermented in sterile, closed fermentors, but rather, in open heaps, boxes, or baskets. This implies that an appropriate starter culture must be able to outcompete microbes present in the environment that can contaminate the fermenting pulp.
The strategy we developed in this study relied on two basic pillars. First, we and others have observed that the bacterial microbiota appears to be very stable and similar between different fermentation batches, cocoa plantations, and seasons (8, 54, 55). This suggests that bacteria are not the main source of variability between cocoa pulp fermentations and that it may not be necessary to include bacteria in a starter culture. Second, we hypothesized that it would be possible to find or generate S. cerevisiae yeast strains that would be able to dominate the yeast population throughout the fermentation, thereby reducing the growth of other, unwanted fungi. Moreover, such a strain would not need to be added in high concentrations to become dominant, which makes the application of the starter culture economically feasible.
Over the past few years, several research groups have looked for optimal cocoa starter cultures. The combined inoculation of S. cerevisiae, LAB, and AAB has been proposed to increase the consistency of cocoa pulp fermentations and improve chocolate quality (18, 19). However, it is unclear whether the suggested starter cultures really yielded improved fermentations, and in addition, the commercial application of such complex mixed starter cultures is difficult and expensive. Starter cultures consisting of other, nonconventional yeast species, including K. marxianus and P. kluyveri, were also developed (29, 31). The downside of these nonconventional yeasts is that they often do not remain dominant throughout the fermentation process, and because they cannot suppress the growth of other yeasts, they may not yield reproducible fermentations. Therefore, a commercially viable starter culture for the consistent production of high-quality cocoa is still elusive.
To obtain an optimal yeast strain for cocoa pulp fermentations, we first isolated and identified large numbers of yeasts from spontaneous fermentations (8). This detailed analysis revealed that cocoa pulp fermentations typically contain many different yeast species, with S. cerevisiae being one of the most dominant species. Further DNA fingerprinting to the strain level revealed in this study that each fermentation typically harbors several different S. cerevisiae strains. While this phenomenon has been described intensively for wine fermentations (56, 57), the intraspecific S. cerevisiae diversity in cocoa pulp fermentations is barely documented (7). We showed that one group of closely related strains was dominant in both fermentations and therefore selected a representative strain (Y927) as a potential starter culture and as a parental strain for further breeding experiments. Additionally, we explored the currently available diversity of industrial and natural S. cerevisiae strains as a potential source for cocoa starter cultures. It is interesting that the selected strain (Y115, originating from the bioethanol industry) outcompeted the indigenous strain, Y927, in a cocoa pulp environment. This might be explained by the similar niches encountered in cocoa pulp and bioethanol fermentations, including low pH; the presence of weak organic acids, such as acetic acid (58); high temperatures (59); and the presence of LAB (60).
Our study also demonstrates the power of classic breeding to generate novel, superior hybrids by either in- or outbreeding of selected parental strains. Temperature tolerance seems to be a crucial feature for cocoa pulp fermentations, with temperature-sensitive strains rapidly disappearing from the pulp when the temperature increases after the beans are mixed (turning), although further investigations are required. Interestingly, our results show that temperature resistance is a transgressive phenotype, and we were able to identify several hybrids that performed better at high temperatures than their parental strains. This could be due to a combination of reasons, including elimination of deleterious mutations present in the parental strains, dominance complementation, overdominance, or epistasis between genetic factors of both parents (61).
Pilot scale testing of the parental strains and several newly generated hybrids demonstrated that both are suitable candidates to serve as a basis for a starter culture, with hybrid H28 being the most promising candidate for commercial exploitation. Fermentations inoculated with H28 or the parental strains Y927 and Y115 showed a significant reduction in microbial variability between replicate fermentations and in the number of different yeasts that reached relatively large population sizes. Species such as H. opuntiae/H. lachancei, H. thailandica, Pichia kudriavzevii, and P. manshurica were still occasionally observed in the inoculated fermentations, mainly at the start and end. These species were also observed in the spontaneous reference fermentations and have been reported before in Malaysia (8). Interestingly, the same dominant bacterial strains, L. fermentum and A. pasteurianus, were present in both spontaneous and yeast-inoculated fermentations. This suggests that bacteria are not essential for a cocoa pulp fermentation starter culture. The growth of AAB in the inoculated fermentations was slightly delayed compared to the spontaneous fermentations. To what extent this specific parameter impacted cocoa flavor was not further investigated and could therefore be interesting for future research.
Surprisingly, we did not find a correlation between polygalacturonase or killer activity and dominance in cocoa pulp fermentations, such as in the case of temperature tolerance and dominance in the fermentation. Polygalacturonase activity and the breakdown of pectin have been the main focus of previously developed starter cultures for cocoa pulp fermentations (see above), yet this study indicates that other phenotypes are more important. The limited influence of the killer activity might be explained by the harsh environmental conditions encountered in cocoa pulp fermentations, which may inhibit the killer-positive behavior. It has been shown that all S. cerevisiae killer-positive strains lose their ability to produce toxins at high temperatures, starting from 37°C (62). Moreover, the optimal pH for killer toxin production is generally higher than the pH of the cocoa pulp (37, 63, 64). However, it is important to note that temperature tolerance is not the sole factor determining dominance in a cocoa pulp environment. The second-generation inbred hybrid H37 displayed less dominance at later phases of the fermentation, despite showing superior temperature tolerance. This indicates that, in addition to temperature tolerance, there are other (although probably minor) factors determining dominance toward the end of the fermentation.
The inoculation of a yeast starter culture clearly affected the volatile fraction of the cocoa liquor. Liquors produced from spontaneous fermentations were shown to contain less acetate and ethyl esters. These pleasant aroma compounds are typically formed during S. cerevisiae metabolism and are responsible for the fruity flavor in fermented beverages (13, 34). Using spontaneously fermented beans also resulted in significantly more rancid and buttery aromas in the liquor, such as those of 2-methylpropanoic acid, butanoic acid, 2.3-butanedione (diacetyl), and 3-hydroxy-2-butanone (acetoin). The inoculation of S. cerevisiae might have excluded the growth of other species that can produce these undesired compounds. Therefore, we hypothesize that the S. cerevisiae starter culture not only altered the cocoa flavor directly by producing pleasant aroma-active compounds, such as esters, but also indirectly, by preventing the growth of other species that produce unwanted aromas. Testing the most promising yeasts on a larger scale is still required to validate the results. The differences in cocoa liquor volatiles between spontaneous and inoculated fermentations were reflected in the consumer panel's significant preference for the inoculated fermentations. The consumer panel significantly preferred chocolate produced with H28, identifying this newly developed hybrid as the most promising candidate for a commercial starter culture.
Together, our results demonstrate the potential of carefully selected and improved yeast strains as starter cultures for cocoa pulp fermentations. The use of such starter cultures results in more consistent fermentation profiles and ultimately yields high-quality chocolate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Niels Vanhoudt, Tom Wenseleers, and Verstrepen laboratory members for their help and suggestions. Special thanks are due to the Barry Callebaut innovation team and in particular Herwig Bernaert for their support in the execution and successful conclusion of this project.
Research in the laboratory of K.J.V. is supported by Barry Callebaut, ERC Starting Grant 241426, VIB, the EMBO YIP program, FWO, and IWT.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00133-15.
REFERENCES
- 1.Barnett JA, Lichtenthaler FW. 2001. A history of research on yeasts 3: Emil Fischer, Eduard Buchner and their contemporaries, 1880-1900. Yeast 18:363–388. doi:. [DOI] [PubMed] [Google Scholar]
- 2.International Cocoa Organization. 29 May 2015, posting date Quarterly bulletin of cocoa statistics (production of cocoa beans), vol XLI, no. 2, cocoa year 2014/15 International Cocoa Organization, London, United Kingdom: http://www.icco.org/about-us/icco-news/282-may-2015-quarterly-bulletin-of-cocoa-statistics.html. [Google Scholar]
- 3.Papalexandratou Z, Falony G, Romanens E, Jimenez JC, Amores F, Daniel HM, De Vuyst L. 2011. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional Ecuadorian spontaneous cocoa bean fermentations. Appl Environ Microbiol 77:7698–7714. doi: 10.1128/AEM.05523-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen DS, Honholt S, Tano-Debrah K, Jespersen L. 2005. Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE). Yeast 22:271–284. doi: 10.1002/yea.1207. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. 2007. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol 114:168–186. doi: 10.1016/j.ijfoodmicro.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Camu N, De Winter T, Verbrugghe K, Cleenwerck I, Vandamme P, Takrama JS, Vancanneyt M, De Vuyst L. 2007. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl Environ Microbiol 73:1809–1824. doi: 10.1128/AEM.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jespersen L, Nielsen DS, Honholt S, Jakobsen M. 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res 5:441–453. doi: 10.1016/j.femsyr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Meersman E, Steensels J, Mathawan M, Wittocx PJ, Saels V, Struyf N, Bernaert H, Vrancken G, Verstrepen KJ. 2013. Detailed analysis of the microbial population in Malaysian spontaneous cocoa pulp fermentations reveals a core and variable microbiota. PLoS One 8:e81559. doi: 10.1371/journal.pone.0081559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ardhana M, Fleet GH. 2003. The microbial ecology of cocoa bean fermentations in Indonesia. Int J Food Microbiol 86:87–99. doi: 10.1016/S0168-1605(03)00081-3. [DOI] [PubMed] [Google Scholar]
- 10.Lima LJ, Almeida MH, Nout MJ, Zwietering MH. 2011. Theobroma cacao L., “The food of the Gods”: quality determinants of commercial cocoa beans, with particular reference to the impact of fermentation. Crit Rev Food Sci Nutr 51:731–761. doi: 10.1080/10408391003799913. [DOI] [PubMed] [Google Scholar]
- 11.Daniel HM, Vrancken G, Takrama JF, Camu N, De Vos P, De Vuyst L. 2009. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res 9:774–783. doi: 10.1111/j.1567-1364.2009.00520.x. [DOI] [PubMed] [Google Scholar]
- 12.Hansen CE, del Olmo M, Burri C. 1998. Enzyme activities in cocoa beans during fermentation. J Sci Food Agric 77:273–281. doi:. [DOI] [Google Scholar]
- 13.Verstrepen KJ, Derdelinckx G, Dufour J, Winderickx J, Thevelein J, Pretorius I, Delvaux F. 2003. Flavor-active esters: adding fruitiness to beer. J Biosci Bioeng 96:110–118. doi: 10.1016/S1389-1723(03)90112-5. [DOI] [PubMed] [Google Scholar]
- 14.Verstrepen KJ, Derdelinckx G, Dufour JP, Winderickx J, Pretorius IS, Thevelein JM, Delvaux FR. 2003. The Saccharomyces cerevisiae alcohol acetyl transferase gene ATF1 is a target of the cAMP/PKA and FGM nutrient-signalling pathways. FEMS Yeast Res 4:285–296. doi: 10.1016/S1567-1356(03)00166-1. [DOI] [PubMed] [Google Scholar]
- 15.Schwan RF, Rose AH., Board RG. 1995. Microbial fermentation of cocoa beans, with emphasis on enzymatic degradation of the pulp. J Appl Bacteriol 79(Symp Suppl):96S–107S. [Google Scholar]
- 16.Lopez SA, Dimick PS. 1995. Cocoa fermentation, p 562–577. In Reed G, Nagodawithana TW (ed), Biotechnology: enzymes, biomass, food and feed, 2nd ed, vol 9 Wiley-VCH, New York, NY. [Google Scholar]
- 17.Ho VT, Zhao J, Fleet G. 2014. Yeasts are essential for cocoa bean fermentation. Int J Food Microbiol 174:72–87. doi: 10.1016/j.ijfoodmicro.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Lefeber T, Papalexandratou Z, Gobert W, Camu N, De Vuyst L. 2012. On-farm implementation of a starter culture for improved cocoa bean fermentation and its influence on the flavour of chocolates produced thereof. Food Microbiol 30:379–392. doi: 10.1016/j.fm.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Schwan RF. 1998. Cocoa fermentations conducted with a defined microbial cocktail inoculum. Appl Environ Microbiol 64:1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrian DW, Patterson GR. 1984. Cocoa fermentation, p 529–575. In Reed G. (ed), Biotechnology, vol 5 Verlag Chemie, Basel, Switzerland. [Google Scholar]
- 21.Lanaud C, Montamayor JC, Sounigo O. 1999. Le cacaoyer, p 56 In Maon P, Perner X, Glaszmann J (ed), Diversité génétique des plantes tropicales cultivées. CIRAD, Paris, France. [Google Scholar]
- 22.Steensels J, Verstrepen KJ. 2014. Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu Rev Microbiol 68:61–80. doi: 10.1146/annurev-micro-091213-113025. [DOI] [PubMed] [Google Scholar]
- 23.Pretorius IS. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729. doi:. [DOI] [PubMed] [Google Scholar]
- 24.Bokulich NA, Bamforth CW, Mills DA. 2012. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PLoS One 7:e35507. doi: 10.1371/journal.pone.0035507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pando Bedriñana R, Querol Simón A, Suárez Valles B. 2010. Genetic and phenotypic diversity of autochthonous cider yeasts in a cellar from Asturias. Food Microbiol 27:503–508. doi: 10.1016/j.fm.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. 2006. How did Saccharomyces evolve to become a good brewer? Trends Genet 22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 27.European Food Safety Authority. 2013. Scientific opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA J 11:3449. [Google Scholar]
- 28.U.S. Environmental Protection Agency. 27 September 2012, posting date. Saccharomyces cerevisiae final risk assessment. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 29.Crafack M, Mikkelsen MB, Saerens S, Knudsen M, Blennow A, Lowor S, Takrama J, Swiegers JH, Petersen GB, Heimdal H, Nielsen DS. 2013. Influencing cocoa flavour using Pichia kluyveri and Kluyveromyces marxianus in a defined mixed starter culture for cocoa fermentation. Int J Food Microbiol 167:103–116. doi: 10.1016/j.ijfoodmicro.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Crafack M, Keul H, Eskildsen CE, Petersen MA, Saerens S, Blennow A, Skovmand-Larsen M, Swiegers JH, Petersen GB, Heimdal H, Nielsen DS. 2014. Impact of starter cultures and fermentation techniques on the volatile aroma and sensory profile of chocolate. Food Res Int 63:306–316. doi: 10.1016/j.foodres.2014.04.032. [DOI] [Google Scholar]
- 31.Leal GAJ, Gomes LH, Efraim P, de Almeida Tavares FC, Figueira A. 2008. Fermentation of cacao (Theobroma cacao L.) seeds with a hybrid Kluyveromyces marxianus strain improved product quality attributes. FEMS Yeast Res 8:788–798. doi: 10.1111/j.1567-1364.2008.00405.x. [DOI] [PubMed] [Google Scholar]
- 32.Dzogbefia VP, Buamah R, Oldham JH. 1999. The controlled fermentation of cocoa (Theobroma cacao L) using yeasts: enzymatic process and associated physicochemical changes in cocoa sweatings. Food Biotechnol 13:1–12. doi: 10.1080/08905439609549958. [DOI] [Google Scholar]
- 33.Buamah R, Dzogbefia VP, Oldham JH. 1997. Pure yeast culture fermentation of cocoa (Theobroma cacao L): effect on yield of sweatings and cocoa bean quality. World J Microbiol Biotechnol 13:457–462. doi: 10.1023/A:1018536519325. [DOI] [Google Scholar]
- 34.Steensels J, Meersman E, Snoek T, Saels V, Verstrepen KJ. 2014. Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl Environ Microbiol 80:6965–6975. doi: 10.1128/AEM.02235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dittmar JC, Reid RJ, Rothstein R. 2010. ScreenMill: a freely available software suite for growth measurement, analysis and visualization of high-throughput screen data. BMC Bioinformatics 11:353. doi: 10.1186/1471-2105-11-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warringer J, Zorgo E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, Forsmark A, Durbin R, Omholt SW, Louis EJ, Liti G, Moses A, Blomberg A. 2011. Trait variation in yeast is defined by population history. PLoS Genet 7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woods DR, Bevan EA. 1968. Studies on the nature of the killer factor produced by Saccharomyces cerevisiae. J Gen Microbiol 51:115–126. doi: 10.1099/00221287-51-1-115. [DOI] [PubMed] [Google Scholar]
- 38.Young TW, Yagiu M. 1978. A comparison of the killer character in different yeasts and its classification. Antonie Van Leeuwenhoek 44:59–77. doi: 10.1007/BF00400077. [DOI] [PubMed] [Google Scholar]
- 39.Masoud W, Jespersen L. 2006. Pectin degrading enzymes in yeasts involved in fermentation of Coffea arabica in East Africa. Int J Food Microbiol 110:291–296. doi: 10.1016/j.ijfoodmicro.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 40.van Wyk H, Divol B. 2010. Recovery of endo-polygalacturonase activity in wine yeast and its effect on wine aroma. FEMS Yeast Res 10:58–71. doi: 10.1111/j.1567-1364.2009.00588.x. [DOI] [PubMed] [Google Scholar]
- 41.Snoek T, Picca Nicolino M, Van den Bremt S, Mertens S, Saels V, Verplaetse A, Steensels J, Verstrepen KJ. 2015. Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol Biofuels 8:32. doi: 10.1186/s13068-015-0216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huxley C, Green ED, Dunham I. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6:236. doi: 10.1016/0168-9525(90)90190-H. [DOI] [PubMed] [Google Scholar]
- 43.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1994. Current protocols in molecular biology. John Wiley and Sons, New York, NY. [Google Scholar]
- 44.Legras J-L, Karst F. 2003. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol Lett 221:249–255. doi: 10.1016/S0378-1097(03)00205-2. [DOI] [PubMed] [Google Scholar]
- 45.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gietz RD, Woods RA. 2001. Genetic transformation of yeast. Biotechniques 30:816–820, 822–826, 828. [DOI] [PubMed] [Google Scholar]
- 47.Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35:1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cadez N, Poot GA, Raspor P, Smith MT. 2003. Hanseniaspora meyeri sp. nov., Hanseniaspora clermontiae sp. nov, Hanseniaspora lachancei sp. nov. and Hanseniaspora opuntiae sp. nov., novel apiculate yeast species. Int J Syst Evol Microbiol 53:1671–1680. doi: 10.1099/ijs.0.02618-0. [DOI] [PubMed] [Google Scholar]
- 49.Meilgaard M, Civille GV, Carr BT. 1999. Sensory evaluation techniques, 3rd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 50.Lawless HT, Heymann H. 2010. Sensory evaluation of food. Principles and practices, 2nd ed Springer, New York, NY. [Google Scholar]
- 51.Wickner RB. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev 60:250–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frauendorfer F, Schieberle P. 2006. Identification of the key aroma compounds in cocoa powder based on molecular sensory correlations. J Agric Food Chem 54:5521–5529. doi: 10.1021/jf060728k. [DOI] [PubMed] [Google Scholar]
- 53.Afoakwa EO, Paterson A, Fowler M, Ryan A. 2008. Flavor formation and character in cocoa and chocolate: a critical review. Crit Rev Food Sci Nutr 48:840–857. doi: 10.1080/10408390701719272. [DOI] [PubMed] [Google Scholar]
- 54.Papalexandratou Z, Camu N, Falony G, De Vuyst L. 2011. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol 28:964–973. doi: 10.1016/j.fm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 55.Papalexandratou Z, Vrancken G, De Bruyne K, Vandamme P, De Vuyst L. 2011. Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol 28:1326–1338. doi: 10.1016/j.fm.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Schuller D, Cardoso F, Sousa S, Gomes P, Gomes AC, Santos MA, Casal M. 2012. Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS One 7:e32507. doi: 10.1371/journal.pone.0032507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Querol A, Barrio E, Ramon D. 1994. Population dynamics of natural Saccharomyces strains during wine fermentation. Int J Food Microbiol 21:315–323. doi: 10.1016/0168-1605(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 58.Almeida JRM, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. Int J Chem Technol Biotechnol 82:340–349. doi: 10.1002/jctb.1676. [DOI] [Google Scholar]
- 59.Sun Y, Cheng J. 2002. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83:1–11. doi: 10.1016/S0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 60.Skinner KA, Leathers TD. 2004. Bacterial contaminants of fuel ethanol production. J Ind Microbiol Biotechnol 31:401–408. doi: 10.1007/s10295-004-0159-0. [DOI] [PubMed] [Google Scholar]
- 61.Marullo P, Bely M, Masneuf-Pomarede I, Pons M, Aigle M, Dubourdieu D. 2006. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res 6:268–279. doi: 10.1111/j.1567-1364.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- 62.Wickner RB. 1974. “Killer character” of Saccharomyces cerevisiae—curing by growth at elevated temperature. J Bacteriol 117:1356–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Cousino N, Maqueda M, Ambrona J, Zamora E, Esteban R, Ramirez M. 2011. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl Environ Microbiol 77:1822–1832. doi: 10.1128/AEM.02501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitt MJ, Tipper DJ. 1990. K28, a unique double-stranded RNA killer virus of Saccharomyces cerevisiae. Mol Cell Biol 10:4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.